Abstract
The intestinal microbiome is a critical determinant of human health. Alterations in its composition have been correlated with chronic disorders, such as obesity and inflammatory bowel disease in adults, and may be associated with neonatal necrotizing enterocolitis in premature infants. Increasing evidence suggests that strain-level genomic variation may underpin distinct ecological trajectories within mixed populations, yet there have been few strain-resolved analyses of genotype–phenotype connections in the context of the human ecosystem. Here, we document strain-level genomic divergence during the first 3 wk of life within the fecal microbiota of an infant born at 28-wk gestation. We observed three compositional phases during colonization, and reconstructed and intensively curated population genomic datasets from the third phase. The relative abundance of two Citrobacter strains sharing ~99% nucleotide identity changed significantly over time within a community dominated by a nearly clonal Serratia population and harboring a lower abundance Enterococcus population and multiple plasmids and bacteriophage. Modeling of Citrobacter strain abundance suggests differences in growth rates and host colonization patterns. We identified genotypic variation potentially responsible for divergent strain ecologies, including hotspots of sequence variation in regulatory genes and intergenic regions, and in genes involved in transport, flagellar biosynthesis, substrate metabolism, and host colonization, as well as differences in the complements of these genes. Our results demonstrate that a community genomic approach can elucidate gut microbial colonization at the resolution required to discern medically relevant strain and species population dynamics, and hence improve our ability to diagnose and treat microbial community-mediated disorders.
Keywords: human microbiome, metagenomics, strain variation, succession, assembly
Intestinal microbes influence human health through harvesting of energy from dietary substrates, production of essential nutrients, and protection against colonization by pathogens (1, 2). Although the adult gut microbiota is highly variable between individuals, it displays limited diversity at the phylum level: only two bacterial phyla (Bacteroidetes and Firmicutes) contribute ∼90% of all microbes (3). In infants, early assembly of the gut microbiota has been linked to development of innate immune responses and terminal differentiation of intestinal structures (4). The dynamic process of colonization has been well studied at high taxonomic levels (5) and seems predictable based on competitive interactions between and within the dominant phyla (6). Yet at lower taxonomic levels, and at early stages of development, our knowledge of this process is incomplete.
Strain-level analyses of clinical isolates using multilocus sequence typing (MLST) and comparative genomics have been used to differentiate closely related organisms (7, 8). However, important contextual information may be lost when interpreting genomic variation between strains isolated from different communities. Microbial population dynamics can be strongly influenced by synergism and competition with coexisting microorganisms and through phage predation (9). The mobile element pool, which is generally excluded when analyzing isolates, can rapidly give rise to the genomic variation that underpins strain differentiation (10).
Cultivation-independent genomic analyses of time-series samples provide a way to link shifts in population abundance to genetic characteristics that underlie physiological traits, such as virulence. Here, we analyzed human intestinal colonization during the neonatal period. We conducted a 16S rRNA gene-based survey of fecal samples collected daily during the first 3 wk of life of a premature infant and reconstructed and manually curated population genomic datasets for the dominant gut microorganisms in the third of three colonization phases. We chose to focus on the premature infant microbiome because, in addition to its medical relevance, the limited number of dominant bacterial species in the community allows for deep sequence coverage of multiple subpopulations.
Results and Discussion
Study Subject.
We studied fecal samples from a female infant delivered by caesarean section at 28-wk gestation due to premature rupture of membranes. She was treated empirically with broad-spectrum antibiotics (ampicillin/gentamicin) for the first 7 d of life but did not receive antibiotics during the remainder of the study period. She received enteral feedings with maternal breast milk between the fourth and ninth days of life. Feedings were withheld between days 9 and 13 because of abdominal distension. On day 13, feedings were slowly resumed with artificial infant formula (Similac Special Care 20 cal/fl oz; Abbott Nutrition). She also received parenteral nutrition until caloric intake from enteral nutrition was adequate (day 28). She had no major illnesses during her hospitalization and was discharged to home at 64 d of life. Fecal samples were collected daily as available between days 5 and 21.
Day-to-Day Dynamics of Community Composition.
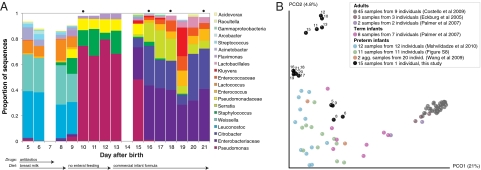
Sequencing of amplified bacterial 16S rRNA genes (SI Materials and Methods and Table S1 A and B) from 15 fecal samples collected on different days during the first 3 wk revealed three distinct community configurations demarcated by rapid transitions. This finding is consistent with previously reported colonization patterns in term infants: relative stability over days to months punctuated by rapid compositional change (5, 11). Marked shifts in abundant lineages around days 9 and 15 seemed to follow dietary adjustments. On days 5 through 9, communities were largely composed of Leuconostoc, Weissella, and Lactococcus (Fig. 1A). The genera Pseudomonas and Staphylococcus, which were relatively scarce on days 8 and 9, became abundant by day 10. On days 10 through 13, species richness and evenness were relatively low (Table S1) and Pseudomonadaceae predominated (Fig. 1A). After resuming feedings on day 13, taxa characteristic of the next phase appeared (Fig. 1A). On days 16 through 21, species richness and evenness recovered (Table S1) and the family Enterobacteriaceae and its constituent genera Citrobacter and Serratia came into the majority. Sample clustering based on community-wide similarity in membership and structure (Fig. 1B and Fig. S1 C–F) further delineated three microbiome configurations. Bacterial community membership and structure were significantly more similar within, than between these colonization phases (P < 0.001; PERMANOVA with Monte Carlo). A cross-study comparison suggests that the infant studied here harbored similar bacteria to those found in other premature infants surveyed using equivalent methods, especially during the first and third colonization phases (Fig. 1B) (5, 12–19).
Fig. 1.
Multiple stable compositional states in the developing gut microbiota of the premature infant. (A) Relative abundance of the 20 most dominant bacterial taxa in 15 fecal samples collected between days 5 and 21. Sequences were classified to the highest taxonomic level to which they could be confidently assigned. Dots indicate metagenomic survey dates. Relevant clinical features are shown along the x axis. (B) Principal coordinates analysis of unweighted UniFrac distances between fecal microbiotas shown in A and those from recently published surveys of adults (3, 5, 47), term infants (5), and preterm infants (17, 19), and from a survey of gut microbes from premature infants in a companion study (Fig. S8). Each circle corresponds to a collection of 16S rRNA gene sequences colored according to study. Samples from this work (black circles) are labeled by day. The percentage of variation explained by the plotted principal coordinates is indicated on the axes. Large-scale alterations in the infant's gut microbiota composition occurred around days 9 and 15.
Metagenomic Data Processing.
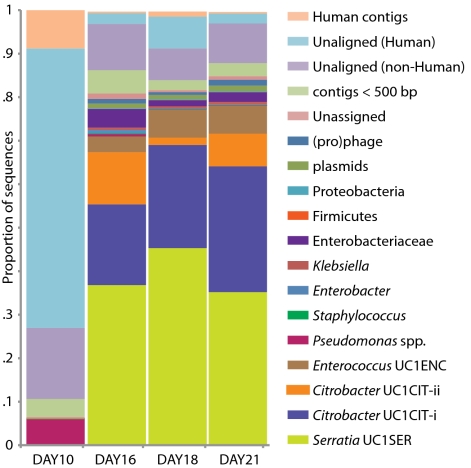
Genome-wide sequencing of DNA from fecal samples collected on days 10, 16, 18, and 21 yielded 245 Mbp of metagenomic sequence data. These data were coassembled using Newbler, keeping track of each read's sample of origin for quantification. Quantification of community composition based on read abundance can be confounded by DNA extraction and sequencing biases (20). However, we could analyze relative abundance shifts across the third colonization phase because the same biases were expected in all samples (Fig. 2). We identified three major sequence “bins” for Serratia, Citrobacter, and Enterococcus, which dominated the third phase of colonization (Figs. 1A and 2). Projecting the smaller contig data (500–1,500 bp) onto an emergent self-organizing map generated based on tetranucleotide frequencies of contigs >1,500 bp and reference genomes allowed us to assign additional fragments to Enterococcus and provide partial coverage for one or more Pseudomonas populations from the day 10 sample (SI Materials and Methods and Fig. S2). Most fragments from other minor populations were assigned to higher taxonomic levels (mostly Enterobacteriaceae) (Table S3 in Dataset S1). We also identified multiple plasmid and phage populations, some of which were completely sequenced (Table S4 in Dataset S1).
Fig. 2.
Population dynamics based on metagenomic profiling. Distribution of the reads over the curated sequence bins across each library (as percentage of all reads in the libraries from day 10, 16, 18, and 21, respectively).
Assembly of a Near-Clonal Serratia Genome and Comparative Genomics.
Manual curation resulted in a Serratia genome (strain UC1SER) with nine gaps, seven of which involve rRNA operons. Based on the sequence coverage of Serratia (∼17×) compared with other bacterial contigs (Table S2), UC1SER dominated the community genomic datasets from the formula-fed (third) phase. We detected remarkably low levels of nucleotide polymorphisms in the UC1SER sequences (close to the expected sequencing substitution error rate), and only very few regions in which gene content varied.
Serratia, a genus comprising motile, facultative anaerobes from the family Enterobacteriaceae, is found in many environments. The UC1SER genome assembled de novo from metagenomic data was compared with the publicly available genomes of Serratia proteamaculans (21) and Serratia marcescens (Sanger Institute, United Kingdom). S. marcescens is an important opportunistic pathogen and a known cause of nosocomial disease in neonatal intensive care units (22). S. proteamaculans is an endophytic bacterium rarely identified in human specimens. All curated UC1SER genome fragments (up to 2.36 Mb in length) share a syntenous backbone with the previously reported genomes, although numerous genomic differences were noted relative to the previously sequenced species (Table S5 in Dataset S1). For syntenous orthologs, UC1SER predicted proteins share 97.3% average amino acid identity (AAI) over 4,089 genes and 88.6% AAI over 3,672 genes with S. marcescens and S. proteamaculans, respectively. Given the overall synteny with S. marcescens and S. proteamaculans across reconstructed genome fragments, we ordered the nine UC1SER genome fragments according to the reference genomes (Table S5 in Dataset S1).
Within syntenous regions in UC1SER, there are small clusters of genes that occur elsewhere in S. marcescens and S. proteamaculans. These clusters encode proteins involved in protocatechuate utilization, fimbrial biosynthesis and export, nitrate reduction, general secretion, siderophore (enterobactin) synthesis and transport, tetrathionate reduction and regulation, osmoprotectant transport, and general metabolism, including amino acid biosynthesis. These rearranged or “indel” regions show elevated sequence divergence relative to syntenous orthologs (AAI of 77 and 58% relative to S. marcescens and S. proteamaculans, respectively). Thus, these regions may contribute to metabolic variation that differentiates these species.
Regions of the UC1SER genome that are absent in one or both of the other Serratia species encode factors involved in transport (most notably iron uptake) and regulation, outer membrane and exopolysaccharide biosynthesis, adhesion, antibiotic biosynthesis, virulence, quorum sensing, biosynthesis of the redox cofactor pyrroloquinoline quinone, arsenate resistance, and propanoate metabolism (Table S5 in Dataset S1). Only UC1SER contains pga operon genes involved in polysaccharide synthesis for biofilm adhesion and a regulon for allantoin utilization, which may be associated with virulence (23). It is also the only genome with yjf-sga operon genes (phosphotransferase system components sgaH, U, E), which enable some strains of gut bacteria to use vitamin C as an energy source (24). UC1SER also has a large nonribosomal peptide biosynthesis protein not found in the other genomes. In contrast to the other reconstructed genomes in this study, UC1SER contains few mobile element-derived sequences.
Analyses of Two Ecologically Distinct Citrobacter Subpopulations.
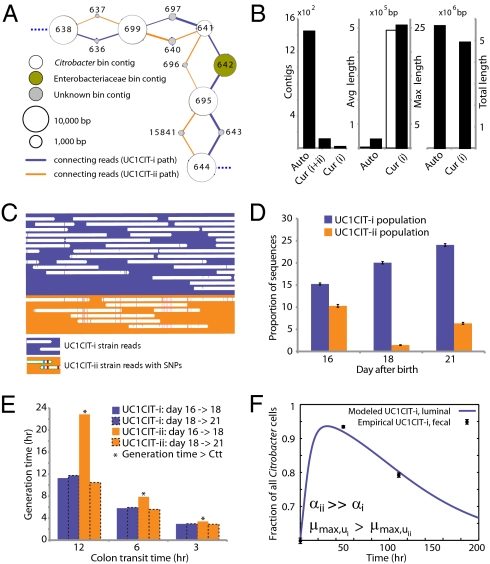
Based on 16S rRNA gene sequences on assembled contigs, Citrobacter in the third colonization phase is closely related to Citrobacter freundii. Despite average coverage of ∼13× on larger Citrobacter fragments, automated assembly resulted in a highly fragmented genome. Citrobacter contigs displayed many diallelic sites among their reads that were almost always linked (i.e., no evidence for homologous recombination), indicating the presence of two coassembled strain populations. Examination of most contig ends revealed path bifurcation (Fig. 3A) because of local strain sequence divergence, differences in gene content, and intergenic region length (see below).
Fig. 3.
Analyses of two ecologically divergent Citrobacter UC1CIT subpopulations. (A) Schematic representation of part of the fragmented UC1CIT assembly. At the ends of many Citrobacter contigs (e.g., contig number 699), reads that partially coassembled led to two sequence paths because they became too divergent (e.g., reads placed in contigs 697 vs. 640) or contained completely novel sequence (reads placed in 642 vs. 696). (B) Condensing of these paths by manual curation reduced the number of contigs, increased average (white bar, left axis) and maximum contig length (black bars, right axis), and reduced cumulative length of contigs (auto, automatic assembly, cur (i+ii), curated UC1CIT-i and -ii strain contigs; cur (i), curated UC1CIT-i contigs). (C) After curation, reads from the strains were grouped based on single nucleotide polymorphism patterns using Strainer. (D) Shifts in proportion (of the respective library total number of reads) of UC1CIT-i and UC1CIT-ii reads over time. Error bar indicates SD between UC1CIT contigs. (E) A chemostat model of the colon, only allowing for differences in growth rate, was used to predict generation time differences needed to explain the observed dynamics in D. (F) Simulation based on a model that incorporated intestinal wall attachment fitted to the observed strain abundances when strain UC1CIT-ii had a higher affinity α to the intestinal wall and the UC1CIT-i luminal maximum growth rate μmax,u was higher than the growth rate of UC1CIT-ii.
Manual curation resolved these bifurcations and reduced the number of Citrobacter contigs from ∼1,400 to 10 (the largest curated contig is 2.55 Mb) (Fig. 3B). The final contigs are generally syntenous with the Citrobacter 30_2 strain draft genome (Broad Institute, Cambridge, MA) and the complete Citrobacter koseri ATCC BAA-895 genome (Washington University, St. Louis, MO). Consequently, the fragments were oriented and ordered by reference to the C. koseri genome to generate a final genome representation for the dominant strain, UC1CIT-i (Table S6 in Dataset S2). Of the ten genome gaps, eight are the rRNA-encoding regions that could not be resolved, one is within a prophage, and one is in the intergenic region between genes on contig ends that are adjacent in both isolate genomes.
Citrobacter species are facultative anaerobes from the family Enterobacteriaceae and are commonly found as commensals within the mammalian intestinal tract. Like Serratia, they have been frequently documented as pathogens in premature newborns (25) (e.g., in cases of neonatal meningitis). Citrobacter 30_2 was isolated from a patient with Crohn disease, whereas C. koseri was isolated from an infant with meningitis. UC1CIT strains lack a “supercontig” of 402 genes reported as part of Citrobacter 30_2; based on our assembly and the functional annotation, we suspect this supercontig derives from a megaplasmid.
As expected based upon the known physiology of human-associated Citrobacter strains (25), the UC1CIT strains have numerous genes for uptake and utilization of a wide variety of substrates. Similar to C. koseri and Citrobacter 30_2, the UC1CIT strains are predicted to express curli and fimbriae that mediate biofilm formation and binding to host epithelial cells (26) (Table S6 in Dataset S2). Interestingly, the UC1CIT strains and C. koseri have dual flagellar systems but Citrobacter sp. 30_2 lacks a lateral flagellar apparatus (Table S7 in Dataset S2). Lateral flagella confer swarming motility in viscous fluids (e.g., mucus) and have been associated with virulence, adhesion, and biofilm formation (27, 28).
UC1CIT sequence variation occurs genome-wide, but one sequence type dominates at most loci (Table S6 in Dataset S2). Given evidence for clonal rather than recombined strains, we defined the minor strain type (UC1CIT-ii) by separating reads primarily using polymorphism patterns in Strainer (29) (Fig. 3C), which allowed for direct comparison of the two aligned strains. UC1CIT-ii sequence blocks (up to a few kilobases in length) share 98.5% average nucleotide identity with UC1CIT-i. In regions of shared gene content, ∼90% of the UC1CIT-ii genome was reconstructed. When the UC1CIT-ii strain blocks were linked and intervening gaps filled by UC1CIT-i sequence, the strains shared 99.1 ± 0.3% average nucleotide identity across their genomes (Table S8 in Dataset S2). The true level of similarity for orthologous sequences likely lies between these values.
Based on the relative frequency of strain-associated reads in the combined dataset for days 10, 16, 18, and 21, UC1CIT-i comprised 77% of the Citrobacter population (SI Materials and Methods and Table S8 in Dataset S2). However, the relative abundance of the strains changed dramatically during the third colonization phase (Fig. 3D and Table S8 in Dataset S2). Possible explanations for the strain abundance shifts include: (i) a bloom of a strain-specific phage that decimated the UC1CIT-ii population around day 18; (ii) a reduced growth rate of UC1CIT-ii when it was outcompeted for resources by UC1CIT-i, Serratia or Enterococcus populations; and (iii) a higher potential of UC1CIT-ii for intestinal wall colonization, leading to an observed decrease in the luminal (fecal) population.
A phage bloom is unlikely because we did not observe an increase in the abundance of Citrobacter phage sequences across the time series. To evaluate the other hypotheses, we constructed two models of bacterial growth in the colon (SI Materials and Methods and Fig. S3). First, using a simplified colon chemostat model, we calculated the differences in growth rates needed to fit the strain population abundance shifts from days 16 to 18 and days 18 to 21 (Fig. 3E). Assuming approximately equal numbers of cells per milliliter luminal content, the model predicts nearly constant generation times for UC1CIT-i. The UC1CIT-ii generation time estimates equaled those for UC1CIT-i between days 18 and 21, but increased above the colon transit time (CTT) between days 16 and 18, resulting in washout between days 16 and 18. Based on CTT in children (12–84 h) (30) and estimates for Escherichia coli generation times in animal models (∼2 h) (31), results from this model guided us to select parameters for a second model (SI Materials and Methods). The second model incorporated intestinal wall-associated growth and enabled fitting of the empirical data by assuming three orders of magnitude higher intestinal-wall affinity for UC1CIT-ii compared with UC1CIT-i (Fig. 3F and Fig. S3). In addition, to avoid rapid washout of UC1CIT-i, its maximum growth rate had to be doubled relative to UC1CIT-ii and the maximum growth rate of wall-adherent cells had to be lowered by an order of magnitude relative to luminal cells. Because these models were built upon a small amount of data, they are inherently limited in their ability to explain the Citrobacter strain behavior. However, they do strongly suggest that the strain shifts are not the result of random fluctuations. Regardless of whether the growth rates and intestinal niches differ, these Citrobacter strains are distinct in their ability to persist in, and interact with, the human host. The availability of genomic data for both strains provides the opportunity to identify possible metabolic characteristics upon which their physiological and ecological divergence is founded.
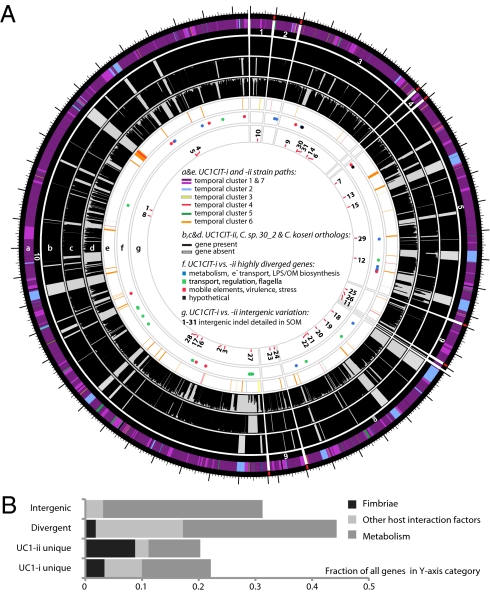
A prominent form of variation that differentiated the two UC1CIT strains involved insertions and deletions in intergenic regions (Fig. 4 and Table S9 in Dataset S2). In most of the 31 observed cases, intergenic regions differed in length between the strains by >10% and in most cases differed by ≥ 30%. Most variable intergenic segments were flanked by gene sequences that were nearly identical in both strains. Transcriptional regulators [25% of cases; e.g., the LexA repressor, and the NanR regulator of fimbrial adhesins previously shown to be affected by sequence variation (32)] and transporters (30% of cases) were common among the flanking genes. We identified strong predicted secondary structure for many divergent intergenic regions and shared sequence similarity with known E. coli sRNAs (Fig. S4).
Fig. 4.
(A) Citrobacter UC1CIT genomic overview. A larger version of this figure is included as Fig. S9. (a) Outside circle represents the ten contigs of the UC1CIT-i genome. Coloring indicates read temporal distribution clusters of the contigs condensed during curation. Genes unique to UC1CIT-i are generally located in areas colored in blue (Fig. S2 cluster 2, Table S11 in Dataset S2). (b) Orthologs to UC1CIT-i in UC1CIT-ii. (c and d) Orthologs to UC1CIT-i in Citrobacter sp. 30_2 and C. koseri. (e) UC1CIT-ii paths with gene content not shared by UC1CIT-i, colored based on read temporal distribution clusters (Table S12 in Dataset S2). (f) Highly divergent genes between the UC1CIT strains, colored by functional class. (Tables S6 and S10 in Dataset S2). (g) Intergenic regions marked by indels that differentiate the UC1CIT strains (Table S9 in Dataset S2). (B) Summary of genomic differences between the UC1CIT strains.
Hotspots of sequence variation that differentiated the UC1CIT strains (mostly substitutions rather than sequence insertions/deletions) also occurred within genes involved in transport, regulation, motility, cell-surface composition, carbohydrate metabolism, virulence, and stress response (Tables S6 and S10 in Dataset S2). Sequence polymorphisms that could potentially affect pathogenicity included the misL-like gene (autotransporter), fimbrial proteins, and a polysaccharide antigen-chain regulator. Interestingly, a large gene encoding RatA, believed to promote intestinal colonization, was a hotspot for microdiversity and was found to be absent in both Citrobacter sp. 30_2 and C. koseri. In Salmonella Typhimurium, RatB (and ShdA, see below) are associated with cecal colonization and fecal shedding, and the gene encoding this protein exhibits strain-associated sequence variation in the form of variable-number direct repeats (33). If RatA were associated with similar phenotypes, then sequence variation between the two strains could explain differences in niche-partitioning and fecal abundance. We also observed unusually high amino acid sequence divergence in lateral flagellar genes between the UC1CIT strains, which could impact interactions with host cell surfaces (Table S7 in Dataset S2) (34). High divergence between the UC1CIT strains in both copies of the gene encoding carbonic anhydrase is also notable because this gene is involved in pH homeostasis and has been identified as a colonization factor in some pathogens (35).
Finally, gene content differentiated the UC1CIT strains (Tables S6, S11, and S12 in Dataset S2). Although many strain-specific genes were clearly associated with phage, several may confer specific metabolic traits. Potentially important genes that were found in both UC1CIT-i and Citrobacter sp. 30_2 but not in UC1CIT-ii encoded (i) ShdA, a large virulence protein that is part of a pathogenicity island in Salmonella Typhimurium and essential for successful intestinal colonization (33); (ii) the inner membrane protein YjfL; (iii) a permease specific for transport of products of pectinolysis (KdgT); (iv) a cluster of four proteins involved in cyclic nucleotide metabolism; (v) fimbrial proteins; (vi) a cluster of 13 proteins involved in phenylacetate degradation; and (vii) genes involved in lipopolysaccharide and polysaccharide/O antigen biosynthesis (abequose). Genes unique to UC1CIT-ii included many fimbrial genes, and genes enabling fructose and other sugar import, streptomycin 3 biosynthesis, and acetoacetate metabolism.
In summary, comparative genomic analyses of the UC1CIT strains highlight metabolic and host interaction traits with the potential to influence strain ecology (Fig. 4B). The observation that both regulatory genes and large intergenic regions are hotspots for sequence divergence indicates that one basis for physiological differentiation involves gene regulation, consistent with prior studies implicating regulation as an evolutionary mechanism underlying early ecological differentiation (36, 37).
Enterococcus.
The Enterococcus population increased in abundance during the third phase of colonization (Figs. 1 and 2). The 16S rRNA gene sequence of strain UC1ENC (from our data) is identical to those of several E. faecalis isolates. UC1ENC shares 98.7% AAI with E. faecalis V583 (38). We mapped the UC1ENC contigs and reads to the V583 genome and recovered ∼81% of the latter (Fig. S5 and Table S14 in Dataset S1). The genome size is similar to that of E. faecalis T3 and T11 [available in high-quality draft (8)]. Absence of multiple UC1ENC contigs covering the same genomic region and low SNP frequency indicated that only one strain was present (Fig. S5).
We compared the sequences of seven UC1ENC genes to sequences of genes used in MLST analyses of clinical isolates (http://efaecalis.mlst.net/), and found that UC1ENC was identical at all seven MLST loci to a sequence type 179, the profile of an isolate recovered from a hospitalized patient's blood sample in The Netherlands. Furthermore, six out of seven loci were identical to sequence type 16 from an isolate found in a Norwegian infant's fecal sample (39). Consistent with physiological characterization of the latter isolate, we found genes linked to antibiotic transport or modification and genes encoding virulence factors including collagen-binding adhesin, aggregation substance, enterococcal surface protein, gelatinase (gelE), and cytolysin (39). Additional predicted virulence factors included an exfoliative toxin A and a serine protease known to be transcribed with gelE (40). Comparison with the V583 genome revealed the absence in UC1ENC of the mobile element containing the vancomycin resistance genes (except for vanZ), as well as small sections of the pathogenicity island and most of the plasmid regions and prophages (Fig. S5).
Mobile Elements and Minor Bacterial Populations.
Manual curation allowed for genomic reconstruction of a Citrobacter plasmid distinct from the above-mentioned megaplasmid of Citrobacter sp. 30_2, except for two shared regions encoding arsenate and Cu/Ag resistance (∼85% AAI). Unlike the UC1CIT plasmid, the putative Citrobacter sp. 30_2 megaplasmid encodes tellurite resistance genes, which have been speculated to confer protection against mammalian host defenses (e.g., by counteracting toxic substances produced by macrophages) (41). The UC1CIT plasmid (∼1.4 plasmid copies per cell) has two variants that differ slightly in gene content and have read distributions across the libraries matching the UC1CIT-i and UC1CIT-ii strains, suggesting that they are strain-specific (Table S4 in Dataset S1). Several phage-like contigs were also recovered, and some displayed boom-and-bust dynamics, indicative of a lytic phage. We also reconstructed two plasmids and two phage of Enterococcus with fluctuating copy numbers (Fig. S6 and Table S4 in Dataset S1). No plasmids or phages were linked to the Serratia population.
Low-abundance bacterial populations were genomically sampled as well. As predicted by the daily 16S rRNA screening (Fig. 1), genomic sequence-abundance data suggest that Pseudomonas peaked around day 10, whereas Enterobacter peaked on day 16, and the Klebsiella population fluctuated over time (Fig. 2 and Fig. S6). Several mobile elements have dynamics corresponding to the minor Klebsiella and Enterobacter populations and may derive from them (Fig. S6).
We performed a community-level analysis of functional potential using genomic information from all populations (Fig. S7). This analysis involved comparison of the microbiome of the preterm infant studied here to the core human microbiome (42). Most of the core adult orthologous groups missing from the UC1 infant communities have poorly characterized and unknown functions. There is also a depletion of functions related to carbohydrate metabolism in the infant studied, perhaps because of differences in diet and species composition, with a notable absence of lineages typical of adults from the phyla Firmicutes, Bacteroidetes, and Actinobacteria.
Conclusions
Attempts to correlate gut microbial community structure with onset of disease in premature infants have yielded conflicting results. For example, in some studies, infants with and without necrotizing enterocolitis (NEC) harbored similar species, whereas in other studies samples from infants with this disease were enriched for a particular species (e.g., Clostridium perfringens) or a particular phylum (e.g., Proteobacteria) (12, 19). In a recent study, Citrobacter was detected in fecal samples from three of four infants with NEC, but in none of the 17 control samples (17). Although it remains possible that Citrobacter is a causative agent for NEC, its presence in samples from the unaffected infant in this study highlights the difficulty in connecting a specific bacterium to disease.
We infer from the results of this study that substantial shifts in Citrobacter strain abundances arise as a result of strain-specific physiology, despite a level of sequence similarity that would typically result in classification of these species as functionally comparable. Given the differences in genetic, especially pathogenic, potential among the otherwise closely related Citrobacter strains reported here, it is perhaps not surprising that medical comparisons at the species or higher level are often inconclusive. The intriguing differences between the UC1CIT strains in size and sequence of a subset of intergenic regions with similarity to small regulatory RNAs, as well as sequence divergence in regulatory genes, emphasize the understudied importance of the evolution of gene expression in strain ecology (36).
Application of our approach to more complex microbial communities is feasible if organisms of interest within those communities can be deeply sampled, an objective that can be achieved with current platforms for high throughput sequencing. In fact, a recent study of adult gut communities that used ∼10 times more sequencing than did our study succeeded in deeply sampling several populations (43). Thus, ultimately, strain-resolved community genomic approaches can provide the resolution needed for appropriate diagnosis and treatment of a range of microbial community associated conditions.
Materials and Methods
Sample Collection.
The protocol for sample collection and processing was approved by the Institutional Review Board of The University of Chicago (IRB #15895A). The sampling method involved manual perineal stimulation with a lubricated cotton swab, which induced prompt defecation. Samples were placed at −80 °C within 10 min.
Sequence Analysis of 16S rRNA Genes.
Bacterial 16S rRNA genes were amplified using the broad-range bacterial primers 8–27F and 788–806R. Sequences were processed using the QIIME software package (44) (SI Materials and Methods, Fig. S1, and Table S1). Fecal 16S rRNA gene sequences from previous studies were obtained directly from GenBank or provided by the authors. Pairwise UniFrac distances were calculated and subjected to principal coordinates analysis (SI Materials and Methods).
Metagenomic Data Analyses.
Sequencing reads from the four libraries were coassembled using Newbler (GSassembler v. 2.0.01; Roche) after removal of replicated reads (SI Materials and Methods). We annotated contigs larger than 1,500 bp with an in-house annotation pipeline. Sequence bin assignments were based on a combination of manual assembly curation, blastn, blastp, GC%, sequencing depth, SNP density, and emergent self-organizing maps (eSOM) based on tetranucleotide frequency in combination with a K-means clustering of the temporal profiles of the reads of each contig (SI Materials and Methods). In cases of ambiguity, contigs were assigned to a higher phylogenetic category. Contigs of virus and plasmid origin were primarily identified based on boom-and-bust dynamics deduced from read temporal profiles, colocalization with plasmid/phage reference genome fragments on the eSOM map, and functional annotation information. Contigs between 500 and 1,500 bp were assigned to genomic bins based on an approach similar to that used for the large contigs, except for the use of eSOM projection. Contigs smaller than 500 nt that were not incorporated during manual assembly curation were not further analyzed.
Assemblies for the dominant bacterial, viral, and plasmid populations were manually curated in Consed (45). Sequences that matched the human genome (blastn e-value cutoff of 1e−35) were removed from the dataset. For each Citrobacter contig, sequence types were identified based on SNP patterns and separated for downstream analyses in Strainer (29). Details on the straining process and identification of variation hotspots is described in SI Materials and Methods. Modeling of Citrobacter strain dynamics relied on a simplified model of interstrain competition within the colon, assuming chemostat dynamics (46) (SI Materials and Methods and Fig. S6). The ORFs predicted on all contigs >500 bp were contrasted to the 4,055 core adult microbiome orthologous groups by blastp analysis using the same parameters and database used by Qin et al. (42).
Supplementary Material
Acknowledgments
The authors thank the Sanger Institute for S. marcescens genome data access; Dr. J. Raes for details on the adult core metagenome dataset; Dr. V. Mai for sharing 16S rRNA sequence data; Dr. C. Fischer for help with MatLab simulations; and C. Sun, N. Justice and Dr. C. Miller for comments on the manuscript. This work was supported in part by the Surgical Infection Society and the March of Dimes Foundation research Grant 5-FY10-103 (to M.J.M.), Department of Energy Genomic Science program Grant DE-FG02-05ER64134 (to J.F.B.), a Walter V. and Idun Berry Postdoctoral Fellowhip (to E.K.C.), National Institutes of Health Pioneer Award DP1OD000964 (to D.A.R.), and by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and Department of Health and Human Services (Contracts HHSN27220090018C and HHSN266200400001C; Broad Institute Citrobacter sp. 30_2 sequencing). D.A.R. is supported by the Thomas C. and Joan M. Merigan Endowment at Stanford University.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive (accession no. SRA026959) and GenBank database.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010992108/-/DCSupplemental.
References
- 1.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 3.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 5.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trosvik P, Stenseth NC, Rudi K. Convergent temporal dynamics of the human infant gut microbiota. ISME J. 2010;4:151–158. doi: 10.1038/ismej.2009.96. [DOI] [PubMed] [Google Scholar]
- 7.Hanage WP, Fraser C, Tang J, Connor TR, Corander J. Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science. 2009;324:1454–1457. doi: 10.1126/science.1171908. [DOI] [PubMed] [Google Scholar]
- 8.Palmer KL, et al. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J Bacteriol. 2010;192:2469–2470. doi: 10.1128/JB.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandaa R-A, et al. Viral control of bacterial biodiversity—Evidence from a nutrient-enriched marine mesocosm experiment. Environ Microbiol. 2009;11:2585–2597. doi: 10.1111/j.1462-2920.2009.01983.x. [DOI] [PubMed] [Google Scholar]
- 10.Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1000081107. 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Cochetiere MF, et al. Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: The putative role of Clostridium. Pediatr Res. 2004;56:366–370. doi: 10.1203/01.PDR.0000134251.45878.D5. [DOI] [PubMed] [Google Scholar]
- 13.Gewolb IH, Schwalbe RS, Taciak VL, Harrison TS, Panigrahi P. Stool microflora in extremely low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 1999;80:F167–F173. doi: 10.1136/fn.80.3.f167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 15.Magne F, et al. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol. 2006;57:128–138. doi: 10.1111/j.1574-6941.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- 16.Millar MR, et al. Application of 16S rRNA gene PCR to study bowel flora of preterm infants with and without necrotizing enterocolitis. J Clin Microbiol. 1996;34:2506–2510. doi: 10.1128/jcm.34.10.2506-2510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mshvildadze M, et al. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr. 2010;156:20–25. doi: 10.1016/j.jpeds.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwiertz A, et al. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr Res. 2003;54:393–399. doi: 10.1203/01.PDR.0000078274.74607.7A. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan JL, Darling AE, Eisen JA. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS ONE. 2010;5:e10209. doi: 10.1371/journal.pone.0010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taghavi S, et al. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar. Appl Environ Microbiol. 2008;75:748–757. doi: 10.1128/AEM.02239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voelz A, et al. Outbreaks of Serratia marcescens in neonatal and pediatric intensive care units: Clinical aspects, risk factors and management. Int J Hyg Environ Health. 2010;213:79–87. doi: 10.1016/j.ijheh.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Chou HC, et al. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect Immun. 2004;72:3783–3792. doi: 10.1128/IAI.72.7.3783-3792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campos E, et al. The yiaKLX1X2PQRS and ulaABCDEFG gene systems are required for the aerobic utilization of L-ascorbate in Klebsiella pneumoniae strain 13882 with L-ascorbate-6-phosphate as the inducer. J Bacteriol. 2008;190:6615–6624. doi: 10.1128/JB.00815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doran TI. The role of Citrobacter in clinical disease of children: Review. Clin Infect Dis. 1999;28:384–394. doi: 10.1086/515106. [DOI] [PubMed] [Google Scholar]
- 26.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavín R, et al. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol Microbiol. 2002;43:383–397. doi: 10.1046/j.1365-2958.2002.02750.x. [DOI] [PubMed] [Google Scholar]
- 28.Merino S, Shaw JG, Tomás JM. Bacterial lateral flagella: An inducible flagella system. FEMS Microbiol Lett. 2006;263:127–135. doi: 10.1111/j.1574-6968.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 29.Eppley JM, Tyson GW, Getz WM, Banfield JF. Strainer: Software for analysis of population variation in community genomic datasets. BMC Bioinformatics. 2007;8:398. doi: 10.1186/1471-2105-8-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagener S, Shankar KR, Turnock RR, Lamont GL, Baillie CT. Colonic transit time—What is normal? J Pediatr Surg. 2004;39:166–169. doi: 10.1016/j.jpedsurg.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Rang CU, et al. Estimation of growth rates of Escherichia coli BJ4 in streptomycin-treated and previously germfree mice by in situ rRNA hybridization. Clin Diagn Lab Immunol. 1999;6:434–436. doi: 10.1128/cdli.6.3.434-436.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohanpal BK, El-Labany S, Lahooti M, Plumbridge JA, Blomfield IC. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc Natl Acad Sci USA. 2004;101:16322–16327. doi: 10.1073/pnas.0405821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kingsley RA, et al. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype typhimurium: Identification of intestinal colonization and persistence determinants. Infect Immun. 2003;71:629–640. doi: 10.1128/IAI.71.2.629-640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lüneberg E, Glenn-Calvo E, Hartmann M, Bär W, Frosch M. The central, surface-exposed region of the flagellar hook protein FlgE of Campylobacter jejuni shows hypervariability among strains. J Bacteriol. 1998;180:3711–3714. doi: 10.1128/jb.180.14.3711-3714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bury-Moné S, et al. Roles of alpha and beta carbonic anhydrases of Helicobacter pylori in the urease-dependent response to acidity and in colonization of the murine gastric mucosa. Infect Immun. 2008;76:497–509. doi: 10.1128/IAI.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denef VJ, et al. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc Natl Acad Sci USA. 2010;107:2383–2390. doi: 10.1073/pnas.0907041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konstantinidis KT, et al. Comparative systems biology across an evolutionary gradient within the Shewanella genus. Proc Natl Acad Sci USA. 2009;106:15909–15914. doi: 10.1073/pnas.0902000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsen IT, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 39.Solheim M, Aakra A, Snipen LG, Brede DA, Nes IF. Comparative genomics of Enterococcus faecalis from healthy Norwegian infants. BMC Genomics. 2009;10:194. doi: 10.1186/1471-2164-10-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 41.Taylor DE. Bacterial tellurite resistance. Trends Microbiol. 1999;7:111–115. doi: 10.1016/s0966-842x(99)01454-7. [DOI] [PubMed] [Google Scholar]
- 42.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turnbaugh PJ, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 46.Ballyk MM, Jones DA, Smith HL. Microbial competition in reactors with wall attachment. Microb Ecol. 2001;41:210–221. doi: 10.1007/s002480000005. [DOI] [PubMed] [Google Scholar]
- 47.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.