Abstract
The hindbrain of larval zebrafish contains a relatively simple ground plan in which the neurons throughout it are arranged into stripes that represent broad neuronal classes that differ in transmitter identity, morphology, and transcription factor expression. Within the stripes, neurons are stacked continuously according to age as well as structural and functional properties, such as axonal extent, input resistance, and the speed at which they are recruited during movements. Here we address the question of how particular networks among the many different sensory-motor networks in hindbrain arise from such an orderly plan. We use a combination of transgenic lines and pairwise patch recording to identify excitatory and inhibitory interneurons in the hindbrain network for escape behaviors initiated by the Mauthner cell. We map this network onto the ground plan to show that an individual hindbrain network is built by drawing components in predictable ways from the underlying broad patterning of cell types stacked within stripes according to their age and structural and functional properties. Many different specialized hindbrain networks may arise similarly from a simple early patterning.
Keywords: development, locomotion, neuronal circuit, Mauthner neuron
The vertebrate hindbrain contains many different sensory-motor networks that control movements of structures in the body and the head. Our recent work shows that despite the diverse networks it contains, there is a remarkably orderly patterning of neurons that extends throughout the hindbrain in young zebrafish, across the well-studied segments, or rhombomeres (1–4). This pattern consists of a series of neurotransmitter stripes that represent broad neuronal classes that differ in transmitter identity, morphology, and transcription factor expression. Within these stripes, neurons are stacked continuously according to age as well as structural and functional characteristics, such as axonal extent, input resistance, and the speed at which they are recruited during movements.
The presence of such an orderly array of neurons suggests that networks are built by drawing components from particular stripes that contain cell types with the required structure and transmitter phenotype, and from the particular position within a stripe occupied by neurons with the appropriate functional properties, much like selecting parts from a catalog. Here we test this possibility through a study of the hindbrain network for the escape behavior initiated by the Mauthner cell (M-cell) (5–8). We conclusively identify neurons in the hindbrain escape network of the M-cell in zebrafish and map each cell type onto the arrangement into the stripes that we describe in the previous work. Our work reveals that this network is built from neurons in predictable locations that are consistent with the idea that the stripe patterning in hindbrain represents an orderly array of neurons from which network components are drawn. Many different hindbrain networks probably arise in a similar way by drawing neurons from a shared structurally and functionally ordered set of parts.
Results
Retrograde Labeling of Interneurons.
We set out to identify the locations of neuronal candidates for a role in the Mauthner escape network in zebrafish by backfilling them by injections of fluorescent dyes (Alexa 647 or Texas Red) targeted to regions of the M-cell on the basis of the innervation patterns of neurons in the network known from goldfish, which is diagrammed in Fig. 1 (5). These backfills were performed in a transgenic line, Tg(glyt2:GFP), in which the neurons in glycinergic stripes can be visualized by GFP expression (Fig. 1 A1–A3). Here we organize these backfilling data from sensory processing to motor output components of the escape circuit.
Fig. 1.
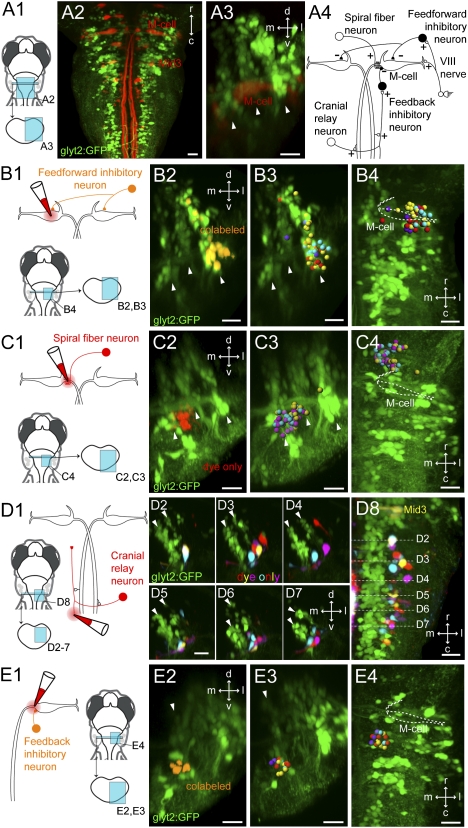
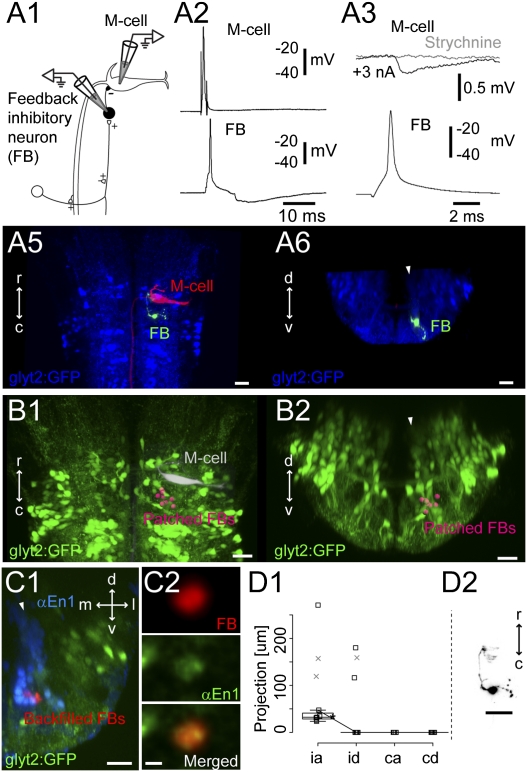
Retrograde labeling of prospective interneurons in the Mauthner circuit. (A) Location of the M-cell relative to glycinergic neurons in the hindbrain. (A1) Location of images on right. (A2) Dorsal view of a Tg(glyt2:GFP) line at 5 dpf with reticulospinal neurons backfilled from the spinal cord with Texas Red. (A3) Cross-section view of the M-cell and the three glycinergic stripes at rhombomere 4. White arrowheads mark the glycinergic stripes. (A4) Circuit diagram of the Mauthner circuit from goldfish. Excitatory neurons, white; inhibitory neurons, black. (B) Retrograde labeling of potential feedforward inhibitory neurons. (B1) Experimental diagram. (B2) Cross-section view of the backfilled neurons at rhombomere 4 in the Tg(glyt2:GFP) line at 5 dpf. Colabeled voxels are orange. Arrowheads mark glycinergic stripes. (B3) Cross-section from registrations showing the glycinergic neurons from four example fish, with different colored spheres for each. (B4) Top-down view of neurons in B3. White dashed line marks the M-cell. (C) Retrograde labeling of prospective spiral fiber neurons. (C1) Experimental diagram. (C2) Cross-section at rhombomere 3 in the Tg(glyt2:GFP) line at 5 dpf. Voxels that only contain the far red dye used to backfill are shown in red. (C3) Cross-section of the registered reconstructions for three different fish, as in B3. (C4) Top-down view of neurons in C3. M-cell marked by white dashed line. (D) Retrograde labeling of potential cranial relay neurons. (D1) Experimental diagram. (D2–D7) Backfilled neurons from four different fish shown in registered cross sections in rhombomere 7/8. Arrowheads mark medial and middle glycinergic stripes. (D8) Top-down view of the neurons in D2–D7. White lines mark corresponding cross sections. (E) Retrograde labeling of prospective feedback inhibitory neurons. (E1) Experimental diagram. (E2) Cross-section of glycinergic neurons at rhombomere 5 in the Tg(glyt2:GFP) line at 5 dpf. Orange indicates colabeling with GFP and the far red dye. White arrowhead marks medial glycinergic neurons. (E3) Cross-section of the registered reconstructions from four fish. (E4) Top-down view of the same neurons. r/c, rostral and caudal; d/v, dorsal and ventral; m/l, medial and lateral in this and later figures. (Scale bars, 20 μm.)
A balance between direct sensory excitation of the M-cell and a feed-forward inhibition regulates Mauthner firing and the initiation of the escape response (Fig. 1A4) (5, 9, 10). To localize potential glycinergic feedforward inhibitory neurons, we electroporated a red dye onto the cell body of one of the two M-cells in the line with GFP labeled glycinergic neurons [Tg(glyt2:GFP)] and looked for contralateral neurons that were both red (backfilled) and green (glycinergic/GFP) (Fig. 1B1; n = 8 fish). Nearly all of the colabeled neurons were located in the most lateral of the three glycine stripes (Fig. 1 B2 and B3; 64 of 66 cells) in hindbrain segment (rhombomere) 4 (Fig. 1 B2–B4), the segment that contains the M-cell body. The colabeled neurons were largely located in the ventral portion of the stripe (Fig. 1B3; 57 of 64 cells).
Another less-well-studied interneuron thought to play a role in sensory processing in the network is the spiral fiber neuron (Fig. 1 A4 and C) (11, 12). To identify potential spiral fiber neurons in zebrafish, we electroporated Alexa 647 or Texas Red into the axon cap of the M-cell in Tg(GlyT2:GFP) fish and screened for contralateral GFP-negative neurons that were filled with the red dye (Fig. 1C1; n = 4 fish). Almost all of these were located in rhombomere 3 between the medial and middle of the three glycinergic stripes, at their ventral ends (Fig. 1 C2 and C3; 67 of 68 cells). The middle of the three glutamatergic stripes is located in this region between the medial and the middle glycinergic stripes, suggesting that the spiral fiber neurons are located in the ventral part of the middle glutamatergic stripe.
The outputs of the M-cell in hindbrain are relayed to other neurons via an interneuron called a cranial relay neuron that is monosynaptically activated by the M-cell (13–15). To locate cranial relay (also called T-reticular) neurons in zebrafish, we backfilled from the medial longitudinal fasciculus in rhombomere 8, where the axons of these neurons run, and looked for red, GFP-negative neurons caudal to rhombomere 6 on the contralateral side in the Tg(glyt2:GFP) line (Fig. 1D1). Most of the backfilled neurons were located ventrally near the middle and the lateral glycinergic stripes (Fig. 1 D2–D8; 48 of 51 cells).
The M-cell fires single action potentials in goldfish because multiple firing is blocked by feedback inhibition (Fig. 1A4) (15).We located potential feedback inhibitory neurons in zebrafish larvae by electroporating red dye into the axon cap (initial segment) of the M-cell in the transgenic line containing GFP-labeled glycinergic neurons and then searching for the ipsilateral glycinergic neurons that were both red and green (Fig. 1E1; n = 4 fish). All of the colabeled glycinergic neurons ipsilateral to the backfill were located ventrally among the most medial glycinergic neurons (Fig. 1 E2 and E3; 15 neurons) and were in rhombomere 5, caudal to the M-cell (Fig. 1E4).
This backfilling served to identify candidate neurons in the M-cell network for patch recording, so we could both fill them with dye to examine their structure relative to neurons in the associated stripes and explore their connectivity and pharmacology in pairwise patch recording experiments to verify their roles in the network and map firmly identified neurons onto the hindbrain patterning.
Feedforward Glycinergic Neurons.
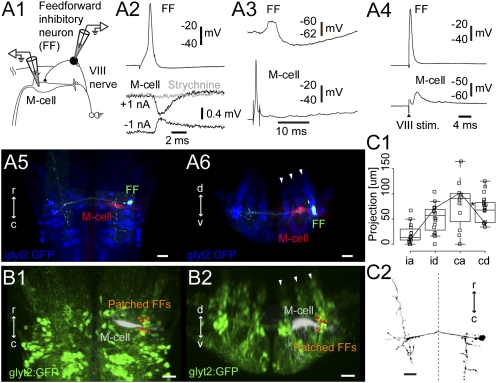
To study the structure and connectivity of putative feedforward inhibitory neurons, we conducted a series of pairwise patch recordings from the M-cell and from GFP-positive interneurons in the Tg(glyt2:GFP) line that were located in the region of the candidate feedforward neurons identified by backfilling (Fig. 2A1). We found 17 glycinergic neurons that when activated produced inhibitory postsynaptic potentials (IPSPs) in the ipsilateral M-cell but were not activated in response to firing of the M-cell by current injection (Fig. 2 A2 and A3). Activation of the M-cell did lead to weak subthreshold PSPs in these neurons (Fig. 2A3; n = 17 pairs). The pathway mediating these PSPs and the role of these connections onto the putative feedforward neurons are unknown. Firing the glycinergic neurons led to short-latency IPSPs (0.32 ± 0.10 ms, n = 17 pairs) in the M-cell, which were most visible when the membrane potential of M-cell was depolarized or hyperpolarized (Fig. 2A2), suggesting that the reversal potential was close to the resting membrane potential of the M-cell (−79.5 ± 1.8 mV, n = 17, junction potential corrected). The IPSPs were blocked by a glycinergic blocker (Fig. 2A2; strychnine, 1 μM, n = 10 pairs). The soma of the glycinergic neuron whose physiology is shown in Fig. 2 A2–A4 was located in the ventral part of the lateral glycinergic stripe, close to the lateral dendrite of the M-cell (Fig. 2 A5 and A6). Consistent with the short latency of IPSPs observed in the M-cell, the axon coming from this neuron had puncta directly apposed to the cell body of the M-cell. This neuron also had both an ipsilateral descending projection as well as a projection to the contralateral hindbrain that approached the other M-cell and then extended further rostrocaudally from there.
Fig. 2.
Feedforward inhibitory neurons. (A1) Pairwise whole-cell recording of a GFP-positive feedforward inhibitory neuron and the ipsilateral M-cell from a Tg(glyt2:GFP) × relaxed line at 4 dpf. (A2) Firing the feedforward neuron led to PSPs (average of five traces) in the M-cell that inverted below threshold (black) and were blocked by strychnine (1 μM, gray). (A3) Response of the feedforward inhibitory neuron to single spikes in the M-cell. (A4) Eighth nerve stimulation led to firing of the interneuron and an EPSP in the M-cell. (A5) Top-down view of the feedforward inhibitory neuron (green) and the M-cell (red) from A2 relative to the glycinergic neurons (blue). (A6) Cross-section of the same neurons, in rhombomere 4. Arrowheads mark glycinergic stripes. (B1 and B2) Location in top-down (B1) and cross-section views (B2) of 12 registered feedforward inhibitory neurons (orange spheres) identified by pairwise recordings. Arrowheads mark the glycinergic stripes. (C) Morphology of feedforward inhibitory neurons (16 cells). One cell is excluded owing to poor filling. (C1) Box-and-whisker plots showing the extent of axonal projections in four directions: ipsilateral ascending (ia), ipsilateral descending (id), contralateral ascending (ca), and contralateral descending (cd). The whiskers extend to the most extreme data point, which is no more than 1.5 times the interquartile range from the box. Asterisk marks the line for the neuron in C2. (C2) Example reconstruction; dotted line marks the midline. (Scale bars, 20 μm.)
To represent the location of all of the feedforward glycinergic neurons identified physiologically, we registered the confocal image stacks from different fish onto a template image stack of the Tg(glyt2:GFP) line. As shown in Fig. 2 B1 and B2, all of the functionally identified feedforward inhibitory neurons were located in the ventral part of the lateral glycinergic stripe at rhombomere 4.
To confirm that the glycinergic neurons in this region actually receive sensory inputs, we used dye electroporations into auditory and facial nuclei and found that they innervate the region containing cell bodies of the identified glycinergic neurons, although their afferent terminations were somewhat separate within this region. We then tested whether the glycinergic neurons receive excitatory input from the auditory nerve by stimulating the auditory nerve while doing pairwise patch recordings (n = 6 pairs; Fig. 2A4) from the M-cell and the interneuron. The stimulus evoked an action potential in these glycinergic neurons at strengths at which the M-cell only showed subthreshold PSPs (4/6), indicating an input pattern consistent with a feedforward identity.
We reconstructed all of the recorded neurons to examine their ipsilateral and contralateral projections and then quantified the extent of these projections in rostral and caudal directions (Fig. 2C). All of the neurons had ipsilateral descending and/or ascending projections and axons that crossed to the contralateral hindbrain, where they formed relatively long ascending and/or descending branches (n = 17). All of these neurons also had an axon close to the contralateral M-cell, suggesting bilateral M-cell connections as in goldfish. The inhibitory inputs to the contralateral M-cell were confirmed by pairwise patch recordings from an interneuron and a contralateral M-cell in separate experiments (n = 3 pairs). We conclude that these are indeed feedforward glycinergic neurons; they are located ventrally among the oldest cells in the lateral glycinergic stripe, and they have the ipsilateral and contralateral projections characteristic of neurons stochastically labeled from many different locations in this stripe in our previous work (2).
Spiral Fiber Neurons.
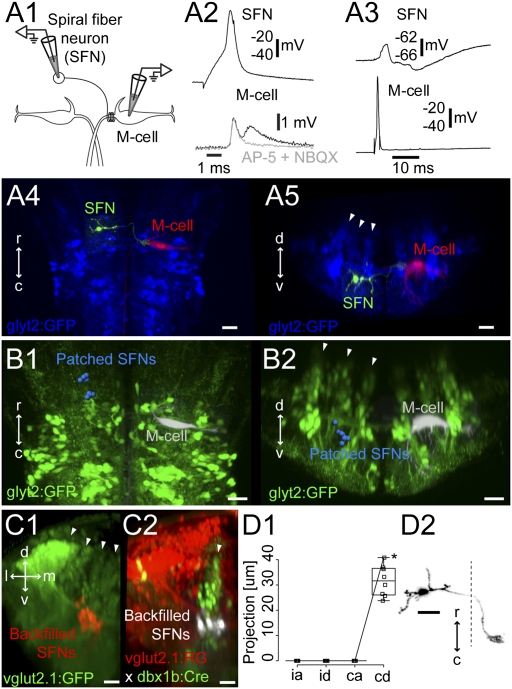
Paired patch recordings from the M-cell and contralateral GFP-negative cells in the Tg(glyt2:GFP) in the region containing putative spiral fiber neurons led to the identification of 10 GFP-negative neurons that had excitatory connections with the contralateral M-cell, characteristic of spiral fiber neurons (Fig. 3A1) (11). The majority of these neurons produced biphasic excitatory postsynaptic potentials (EPSPs) in the contralateral M-cell (Fig. 3A2; n = 9 of 10 pairs), with a short latency (peak to peak, 0.24 ± 0.03 ms, n = 10 pairs). The later component in the EPSP was blocked by glutamatergic blockers [Fig. 3A2; 10 μM 2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo[f]quinoxaline-2,3-dione (NBQX) and 50 μM d-(−)-2-Amino-5-phosphonopentanoic acid (d-AP5), n = 4 pairs), suggesting this slower component was glutamatergic. Injection of negative current pulses into one cell led to a hyperpolarizing response in the other neuron (n = 8 pairs), and the fastest early component of the PSP was resistant to high-frequency firing over 100 Hz (n = 6 pairs). Both of these observations indicate that the fast component was probably an electrical synapse mediated by gap junctions. An action potential in the M-cell only led to subthreshold PSPs in SFNs (Fig. 3A3; n = 10 pairs); the pathway mediating these PSPs and its functional role are unknown.
Fig. 3.
Spiral fiber neurons. (A1) Pairwise recording of a GFP-negative spiral fiber neuron at rhombomere 3 and the contralateral M-cell in the Tg(glyt2:GFP) × relaxed line at 4 dpf (Left). (A2) Firing the spiral fiber led to EPSPs (black) in the M-cell. The slow component of the EPSPs was blocked by the glutamatergic blockers 10 μM NBQX and 100 μM d-AP5 (gray). (A3) PSP in the spiral fiber neuron in response to firing the M-cell. (A4 and A5) Top-down and cross-section views of the spiral fiber neuron (green) and the M-cell (red) from A2 and A3 relative to the glycinergic neurons (blue). Arrowheads mark the glycinergic stripes. (B1 and B2) Summary of the locations of eight spiral fiber neurons (blue) in the glycinergic line in rhombomeres 4 and 5, laid out as in Fig. 2B. (C1) Cross-section view of the backfilled spiral fiber neurons (red) in the transgenic glutamatergic line [Tg(vglut2.1:GFP)] at rhombomere 3. Arrowheads mark the glutamatergic stripes. (C2) Cross-section of the backfilled spiral fiber neurons (white) in the Tg(vglut2.1:RG × dbx1b:Cre) line. Arrowhead indicates the dbx1b-positive middle glutamatergic stripe (green). (D1 and D2) Projections of the spiral fiber neurons shown as in Fig. 2C. Asterisk on D1 marks a point on the line for the neuron in D2. (Scale bars, 20 μm.)
The neuron shown in Fig. 3 A2 and A3, like all physiologically identified spiral fiber neurons, was located in rhombomere 3 between the medial and the middle glycinergic stripes, where the middle glutamatergic stripe is located (Fig. 3 A4, A5, B1, and B2; n = 8). To confirm their location in the middle glutamatergic stripe, we backfilled the neurons in transgenic fish [Tg(vglut2a:GFP)] in which the glutamatergic neurons are labeled with GFP, and found that the neurons were indeed in the middle glutamatergic stripe (Fig. 3C1). We also tested whether the neurons were in the dbx1b transcription factor stripe (dbx1b marks the middle glutamatergic and glycinergic stripes) by backfilling the spiral fiber neurons in fish from a cross of a dbx1b:cre line with a line in which the vesicular glutamate transporter drives expression of floxed red fluorescent protein, with GFP downstream. This cross produces fish in which the glutamatergic neurons that are also dbx1b positive are green and the other glutamatergic neurons are red. The spiral fiber neurons were in the ventral part of the dbx1b-positive middle glutamatergic stripe (Fig. 3C2; n = 4 fish, 75 of 77 cells). For reasons we do not understand, although they were in the glutamatergic stripe, they were not stained red in the glutamatergic transgenic line. The pharmacology of their synaptic connections, however, supported the conclusion that they are glutamatergic.
Reconstructions of all of the patched neurons (n = 8) showed that they had only contralateral and descending axonal projections running directly into the axon cap of the contralateral M-cell, with a characteristic axonal elaboration inside the cap (Fig. 3 D1 and D2). In summary, the evidence shows that the spiral fiber neurons are located ventrally in the middle glutamatergic stripes and have contralateral projections like those of neurons in the adjacent middle glycinergic stripe.
Cranial Relay Neurons.
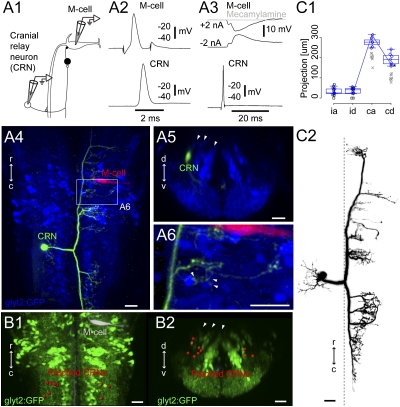
We conducted pairwise patch recordings between the M-cell and GFP-negative neurons in the Tg(glyt2:GFP) line in the region of putative cranial relay neurons and found 10 GFP-negative neurons that fired in response to the M-cell's activation and whose activation led to (polysynaptic) IPSPs in the M-cell (Fig. 4 A1–A3).These neurons fired an action potential in response to the activation of the M-cell (Fig. 4A2; n = 10 pairs) with a short latency (onset of EPSP: 0.15 ± 0.09 ms; peak of action potential: 0.70 ± 0.31 ms, n = 10 pairs). A single action potential in a contralateral neuron in this class led to a longer latency, presumably polysynaptic, IPSP in the M-cell (1.60 ± 0.28 ms, n = 7 pairs; Fig. 4A3). This IPSP was blocked by a cholinergic blocker (Fig. 4A3, Right, mecamylamine, 100 μM; n = 5 pairs), as well as a glycinergic blocker (strychnine, 1 μM; n = 3 pairs), suggesting that this class may excite glycinergic neurons via cholinergic transmission and the glycinergic neurons in turn feedback to inhibit the M-cell. Consistent with this possibility, the recorded putative cranial relay neurons innervated the region where the feedback glycinergic neurons were located (Fig. 4 A4–A6; n = 10).
Fig. 4.
Cranial relay neurons. (A1) Pairwise recordings of a GFP-negative cranial relay neuron at rhombomere 7/8 and the contralateral M-cell in the Tg(glyt2:GFP) × relaxed line at 4 dpf. (A2) Firing the M-cell led to an action potential in the cranial relay neuron. (A3) Firing the cranial relay neuron with current injection led to a long-latency IPSP in the M-cell (black) that was blocked by application of the cholinergic blocker mecamylamine (100 μM, gray). (A4) Top-down view of the recorded cranial relay neuron (green) and the M-cell (red) relative to glycinergic neurons (blue). (A5) Cross-section of the cranial relay neuron at rhombomere 8. Arrowheads mark glycinergic stripes. (A6) Close up of axonal terminations from the cranial relay neuron in region of the white rectangle in A4 showing apposition (arrowheads) to glycinergic neurons in the region. (B1 and B2) Locations of 10 cranial relay neurons at rhombomeres 7/8 (red) identified with pairwise recordings, in top-down (B1) and cross-section (B2) views. (C1 and C2) Summary of the morphology of seven cranial relay neurons, laid out as in Fig. 2C. Xs mark neurons whose whole axonal extent was not captured after recording. Box-and-whisker plots are from seven electroporated cells marked with blue diamonds whose whole axonal extent was captured. Asterisk marks a point on the line shown for the neuron in C2. (Scale bars, 20 μm.)
These cranial relay neurons also exhibited extensive innervation of the contralateral hindbrain (Fig. 4A4), including the regions where cranial motor nuclei (fifth, seventh, and 10th motor nuclei) and pectoral fin motoneurons are known to be located. We did several experiments to examine these connections. Paired recordings of an M-cell and a cranial relay neuron in the Tg(islet1:GFP) line in which motoneurons are GFP labeled (n = 3) showed that cranial relay neurons contributing to the feedback inhibition of M-cell projected to the cranial motor nuclei. Projections to the region of the pectoral fin motoneurons were confirmed morphologically by single-cell electroporation of cranial relay neurons in fish with fin motoneurons backfilled (n = 3). Finally, in the most direct approach, an excitatory connection to the fifth motor nuclei was observed electrophysiologically by paired recordings of cranial relay neurons and motoneurons in the fifth motor nuclei in the Tg(islet1:GFP) line (n = 7). The somata of the recorded neurons in this class were located below the middle or lateral glycinergic stripes, which contain neurons with commissural projections like the cranial relay neurons (Fig. 4 B1 and B2). Compared with their extensive contralateral innervation of motor pools, these relay neurons had only short or no ipsilateral axonal/dendritic processes (Fig. 4 A4, C1, and C2).
Feedback Glycinergic Neurons.
We used pairwise recording to identify nine glycinergic, feedback-inhibitory neurons that were activated by firing the ipsilateral M-cell and had inhibitory projections back to that M-cell (Fig. 5 A1–A6). These neurons fired an action potential in response to the M-cell's action potential (Fig. 5A2), with a relatively long latency (3.87 ± 2.60 ms, n = 9 pairs) and produced IPSPs in the same M-cell (Fig. 5A3) with a short latency (0.38 ± 0.15 ms, n = 9 pairs). The IPSPs were blocked by bath application of strychnine (1 μM, n = 4 pairs), indicating they were glycinergic (Fig. 5A3). The strychnine application completely blocked the feedback inhibition in the M-cell, suggesting that the feedback inhibition in the 4-dpf zebrafish was solely mediated by glycinergic neurons (n = 23). The IPSP amplitude in the M-cell produced by a single feedback-inhibitory neuron was smaller than the IPSP produced by firing a cranial relay neuron (Figs. 5A3 vs. 4A3), consistent with the possibility that a single cranial relay neuron activates multiple feedback-inhibitory cells.
Fig. 5.
Feedback-inhibitory neurons. (A1) Pairwise recordings from a feedback-inhibitory neuron at rhombomere 5 and the ipsilateral M-cell in the Tg(glyt2:GFP) × relaxed line at 4 dpf. (A2) Firing the M-cell led to an action potential in the feedback neuron. (A3) Firing the feedback inhibitory neuron led to an IPSP in the M-cell that was blocked with 1 μM strychnine (gray line). (A5 and A6) Top-down (A5) and cross-section (A6, at rhombomere 5) views of the recorded feedback-inhibitory neuron (green) and the M-cell (red) relative to the glycinergic neurons (blue). Arrowhead marks the most medial glycinergic neurons. (B1 and B2) Summary of the position of nine recorded feedback-inhibitory neurons (pink) in top-down (B1) and cross-section (B2) views at rhombomere 5. Arrowhead marks the medial glycinergic stripe. (C1) Hemicross-section of backfilled feedback-inhibitory neurons (red) in the Tg(glyt2:GFP) at 5 dpf, immunostained forengrailed-1 (En1, blue). (C2) Single electroporated feedback neuron (FB) stained for engrailed (αEn1), and the merged image. (D1 and D2) Morphology of feedback-inhibitory neurons (n = 7 cells) formatted as in Fig. 2 C1 and C2. Black Xs mark two cells whose whole axonal extent was not captured. Asterisk marks the line for the neuron in D2. (Scale bars, 2 μm in C2, 20 um in other panels.)
All of the electrophysiologically identified feedback inhibitory neurons were located among the most ventral and medial glycinergic populations in rhombomere 5 (Fig. 5 B1 and B2; n = 9). Because the stripe-like structure at rhombomere 5 in the glycinergic line was not always very crisp, we also examined the stripe location of the neurons by immunostaining for engrailed-1, which clearly marks the medial glycinergic stripe in other rhombomeres (2). The feedback-inhibitory neurons identified by backfilling were in the ventral part of the engrailed-1–positive stripe (Fig. 5C1; n = 14). The engrailed labeling of these cells was dim (probably owing to down-regulation with age), but in single-neuron filling experiments we found engrailed-positive staining of this cell type (Fig. 5C2).
Six of nine of these neurons had only ipsilateral ascending projections directly to the M-cell's cell body, ending at its initial segment. The other three cells had only ipsilateral and primarily ascending projections contacting the proximal part of the ventral dendrite of the M-cell (Fig. 5 D1 and D2).We also identified four additional glycinergic neurons in the same region that had inhibitory input to the ipsilateral M-cell but did not reach firing threshold in response to M cell's activation and so were excluded from the more detailed analysis. These also had only ipsilateral ascending projections directly to the axon cap. These several lines of morphological and electrophysiological evidence show that feedback-inhibitory neurons are located ventrally among the oldest cells in the medial glycinergic stripe and have the ipsilateral and primarily ascending projections characteristic of this stripe. Their features, like those of the other neurons in the Mauthner network we examined here, thus mapped exactly as we would have predicted on the basis of the overall ground plan in hindbrain.
Discussion
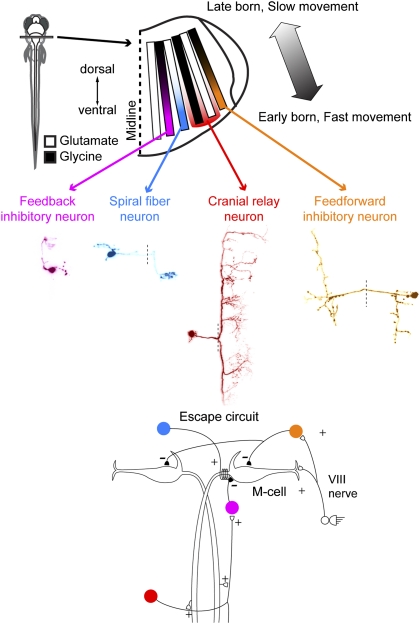
Our previous work showed that there is a very orderly structural and functional patterning extending rostrocaudally throughout the hindbrain segments (rhombomeres) in zebrafish at a young age, when their motor behaviors are functional and essential for survival (2). One important prediction from this observation is that sensory-motor networks in the hindbrain are built by drawing neurons from a simple plan in which different cell types are arranged into columns that are ordered by age as well as structural and functional properties. The organization of the Mauthner network, which includes neurons located in different rhombomeres with outputs across rhombomeres, confirms our predictions about the relationships between particular hindbrain networks and the broad neuronal patterning in hindbrain. Each of the neuronal types in the M-cell network occupies a location consistent with what one would have predicted on the basis of the overall stripe patterning in the brain, as summarized in Fig. 6. Not only are the neurons located in stripes that correspond to their morphological and neurotransmitter phenotypes, they all lie at the ventral end of the stripes, exactly where we would predict the fastest motor behaviors to map on the basis of the continuous map of swimming speed from slow to fast as one moves from dorsal to ventral along a stripe, as we described in our previous article (2). Here the behavior is not a rhythmic one but is the fastest motor behavior produced by the fish, and the neurons participating in it occupy the low input resistance, high threshold, fast end of the stripes, which also contain the oldest neurons. The Mauthner network thus draws neurons by stripe and position just as expected given their particular functional roles.
Fig. 6.
Summary of the construction of the Mauthner network from the ground plan in hindbrain. Neuronal members of the Mauthner circuit map onto the stripes as predicted by our previous study of overall hindbrain patterning. The colors of the neurons correspond to those on the stripe diagram and on the circuit diagram below.
The hindbrain of vertebrates contains a variety of motor networks controlling sensory-motor aspects of movements of the eyes, jaws, gills, and diaphragm (5, 16, 17). If the other networks follow the same general patterning as the Mauthner network, we predict that those neurons contributing to faster (more coarse) sensory-motor behaviors will be drawn from early differentiating cells located ventrally in those stripes that give rise to neurons with the appropriate morphological and transmitter phenotype. For example, a behavior that involves a range of movement speeds, such as eye movements, which include very fast, large saccadic movements as well as slow pursuit movements, would be expected to draw from a range of dorsoventral locations (and ages) in the stripes (16). Those neurons driving the fast saccadic eye movements would be expected to have ventral (older) locations in stripes, with slower eye movements driven by more dorsal (younger) cells.
We do not yet know how the several other hindbrain networks in zebrafish or in any other animals map onto the overall order we describe because physiologically defining even one network and its connectivity is a major challenge. Nonetheless, morphological and transcription factor expression studies suggest that the ground plan in zebrafish is likely to be shared across the vertebrate subphylum (17–24). The extent to which this plan presages age-related networks in other vertebrates that are akin to those in zebrafish remains to be determined. In humans, faster behaviors such as saccadic eye movements and startle responses develop before slower ones, consistent with a patterning like that in zebrafish (25, 26). Large Mauthner-like hindbrain neurons in the mammalian startle response differentiate early (27, 28), suggesting there might be similarities in patterning in mammals at the neuronal as well as behavioral level. We expect that other vertebrate networks, like that of the M-cell, will draw components in predictable ways from the orderly array of parts that underlies the construction of networks in hindbrain.
Experimental Procedures
Fish Care.
All experiments were performed on 4- to 5-dpf zebrafish (Daniorerio) obtained from a laboratory stock of wild-type, transgenic, and mutant adults. All procedures conform to the US National Institutes of Health guidelines regarding animal experimentation and were approved by Cornell University's Institutional Animal Care and Use Committee.
Transgenic Lines and Mutants.
The transgenic lines Tg(glyt2:GFP (29) and Tg(vglut2a:GFP) (30) were described previously. The transgenic lines Tg(vglut2a:loxPDsRed-GFP) and Tg(dbx1b:Cre) were constructed using bacterial artificial chromosomes (DKEY-145P24 and zK17G17, respectively) (31). Details will be described elsewhere. The mutant fish relaxed (red ts25a) was obtained from the Max Planck Institute in Tubingen, Germany (32).
Electrophysiology.
Patch-clamp recordings were performed as described previously, with minor modifications. We initially used cholinergic blockers α-bungarotoxin or d-tubocurarine to paralyze fish by blocking transmission at the neuromuscular junction (33), but this treatment also blocked the cholinergic transmission from the M-cell. Therefore, we primarily used the relaxed mutants, in which a mutation in the skeletal muscle dihydropyridine receptor b1 gene, cacnb1, eliminates muscle contraction. We also used formamide treatment to paralyze fish in some experiments (34, 35). These two preparations produced similar results, so we pooled the data.
Neuronal Labeling.
Four-day-old larvae were anesthetized and embedded in 1.7% agarose. Then, 20% Alexa Fluor Dextran, 10,000 MW, anionic, lysine fixable (Molecular Probes) was applied in extracellular solution through a patch micropipette by electroporating the dye into regions of interest with 20-s trains of 20-V, 1-ms pulses at 50 Hz. Confocal images were acquired at 5 dpf.
Immunohistochemistry.
Standard whole-mount antibody staining procedures were used (1, 36). A rabbit anti-Enhb1 antibody (a gift from A. Joyner, Sloan-Kettering Institute, New York, NY; 1:250–1:500) was detected with goat anti-rabbit antibody conjugated with HRP (Invitrogen; 1:200) and the Alexa Fluor 647-Tyramide system (Invitrogen).
Image Analysis.
To summarize the location of the recorded or backfilled neurons from multiple experiments, we registered volumes to a template image of the Tg(glyt2:GFP) line by using custom MATLAB (MathWorks) software to integrate the functionality of Imaris, including ImarisXT (Bitplane), with the coregistration/normalization functionality of SPM8 (http://www.fil.ion.ucl.ac.uk/spm). The quality of the registration was carefully examined by checking the alignment of stripes, and unsatisfactory registrations were discarded from the final visualization. Volumetric colocalization was done in Imaris.
Details of the above methods are provided in SI Experimental Procedures.
Supplementary Material
Acknowledgments
This work was supported by Japan Society for Promotion of Science Postdoctoral Fellowships for Research Abroad (to M.K.), Uehara Memorial Foundation Postdoctoral Fellowships (to M.K.), National Science Foundation Integrative Graduate Education and Research Traineeship 0333366 (to A.K.), National Institutes of Health (NIH) Training Grant T32 GM007469 (to A.K.), the Ministry of Education, Science, Technology, Sports and Culture of Japan (S.-i.H.), and NIH Grant NS26539 (to J.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012189108/-/DCSupplemental.
References
- 1.Higashijima S, Mandel G, Fetcho JR. Distribution of prospective glutamatergic, glycinergic, and GABAergic neurons in embryonic and larval zebrafish. J Comp Neurol. 2004;480:1–18. doi: 10.1002/cne.20278. [DOI] [PubMed] [Google Scholar]
- 2.Kinkhabwala A, et al. A structural and functional ground plan for neurons in the hindbrain of zebrafish. Proc Natl Acad Sci USA. 2011;108:1164–1169. doi: 10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hale ME, Kheirbek MA, Schriefer JE, Prince VE. Hox gene misexpression and cell-specific lesions reveal functionality of homeotically transformed neurons. J Neurosci. 2004;24:3070–3076. doi: 10.1523/JNEUROSCI.5624-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moens CB, Cordes SP, Giorgianni MW, Barsh GS, Kimmel CB. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development. 1998;125:381–391. doi: 10.1242/dev.125.3.381. [DOI] [PubMed] [Google Scholar]
- 5.Faber DS, Fetcho JR, Korn H. Neuronal networks underlying the escape response in goldfish. General implications for motor control. Ann N Y Acad Sci. 1989;563:11–33. doi: 10.1111/j.1749-6632.1989.tb42187.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu KS, Fetcho JR. Laser ablations reveal functional relationships of segmental hindbrain neurons in zebrafish. Neuron. 1999;23:325–335. doi: 10.1016/s0896-6273(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 7.Eaton RC, Bombardieri RA, Meyer DL. The Mauthner-initiated startle response in teleost fish. J Exp Biol. 1977;66:65–81. doi: 10.1242/jeb.66.1.65. [DOI] [PubMed] [Google Scholar]
- 8.Kimmel CB, Sessions SK, Kimmel RJ. Morphogenesis and synaptogenesis of the zebrafish Mauthner neuron. J Comp Neurol. 1981;198:101–120. doi: 10.1002/cne.901980110. [DOI] [PubMed] [Google Scholar]
- 9.Zottoli SJ, Faber DS. An identifiable class of statoacoustic interneurons with bilateral projections in the goldfish medulla. Neuroscience. 1980;5:1287–1302. doi: 10.1016/0306-4522(80)90201-8. [DOI] [PubMed] [Google Scholar]
- 10.Nakayama H, Oda Y. Common sensory inputs and differential excitability of segmentally homologous reticulospinal neurons in the hindbrain. J Neurosci. 2004;24:3199–3209. doi: 10.1523/JNEUROSCI.4419-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott JW, Zottoli SJ, Beatty NP, Korn H. Origin and function of spiral fibers projecting to the goldfish Mauthner cell. J Comp Neurol. 1994;339:76–90. doi: 10.1002/cne.903390108. [DOI] [PubMed] [Google Scholar]
- 12.Lorent K, Liu KS, Fetcho JR, Granato M. The zebrafish space cadet gene controls axonal pathfinding of neurons that modulate fast turning movements. Development. 2001;128:2131–2142. doi: 10.1242/dev.128.11.2131. [DOI] [PubMed] [Google Scholar]
- 13.Kimmel CB, Metcalfe WK, Schabtach E. T reticular interneurons: A class of serially repeating cells in the zebrafish hindbrain. J Comp Neurol. 1985;233:365–376. doi: 10.1002/cne.902330306. [DOI] [PubMed] [Google Scholar]
- 14.Hackett JT, Faber DS. Mauthner axon networks mediating supraspinal components of the startle response in the goldfish. Neuroscience. 1983;8:317–331. doi: 10.1016/0306-4522(83)90069-6. [DOI] [PubMed] [Google Scholar]
- 15.Hackett JT, Faber DS. Relay neurons mediate collateral inhibition of the goldfish Mauthner cell. Brain Res. 1983;264:302–306. doi: 10.1016/0006-8993(83)90829-6. [DOI] [PubMed] [Google Scholar]
- 16.Sparks DL. The brainstem control of saccadic eye movements. Nat Rev Neurosci. 2002;3:952–964. doi: 10.1038/nrn986. [DOI] [PubMed] [Google Scholar]
- 17.Gray PA. Transcription factors and the genetic organization of brain stem respiratory neurons. J Appl Physiol. 2008;104:1513–1521. doi: 10.1152/japplphysiol.01383.2007. [DOI] [PubMed] [Google Scholar]
- 18.Clarke JD, Lumsden A. Segmental repetition of neuronal phenotype sets in the chick embryo hindbrain. Development. 1993;118:151–162. doi: 10.1242/dev.118.1.151. [DOI] [PubMed] [Google Scholar]
- 19.Cepeda-Nieto AC, Pfaff SL, Varela-Echavarría A. Homeodomain transcription factors in the development of subsets of hindbrain reticulospinal neurons. Mol Cell Neurosci. 2005;28:30–41. doi: 10.1016/j.mcn.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Moreno N, Bachy I, Rétaux S, González A. LIM-homeodomain genes as territory markers in the brainstem of adult and developing Xenopus laevis. J Comp Neurol. 2005;485:240–254. doi: 10.1002/cne.20498. [DOI] [PubMed] [Google Scholar]
- 21.Sieber MA, et al. Lbx1 acts as a selector gene in the fate determination of somatosensory and viscerosensory relay neurons in the hindbrain. J Neurosci. 2007;27:4902–4909. doi: 10.1523/JNEUROSCI.0717-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ericson J, et al. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- 23.Placzek M, Yamada T, Tessier-Lavigne M, Jessell T, Dodd J. Control of dorsoventral pattern in vertebrate neural development: induction and polarizing properties of the floor plate. Development. 1991;2(Suppl 2):105–122. [PubMed] [Google Scholar]
- 24.Takahashi M, Osumi N. Pax6 regulates specification of ventral neurone subtypes in the hindbrain by establishing progenitor domains. Development. 2002;129:1327–1338. doi: 10.1242/dev.129.6.1327. [DOI] [PubMed] [Google Scholar]
- 25.Luna B, Velanova K, Geier CF. Development of eye-movement control. Brain Cogn. 2008;68:293–308. doi: 10.1016/j.bandc.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Vries JI, Visser GH, Prechtl HF. The emergence of fetal behaviour. I. Qualitative aspects. Early Hum Dev. 1982;7:301–322. doi: 10.1016/0378-3782(82)90033-0. [DOI] [PubMed] [Google Scholar]
- 27.Lingenhöhl K, Friauf E. Giant neurons in the rat reticular formation: A sensorimotor interface in the elementary acoustic startle circuit? J Neurosci. 1994;14:1176–1194. doi: 10.1523/JNEUROSCI.14-03-01176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altman J, Bayer SA. Development of the brain stem in the rat. IV. Thymidine-radiographic study of the time of origin of neurons in the pontine region. J Comp Neurol. 1980;194:905–929. doi: 10.1002/cne.901940411. [DOI] [PubMed] [Google Scholar]
- 29.McLean DL, Fan J, Higashijima S, Hale ME, Fetcho JR. A topographic map of recruitment in spinal cord. Nature. 2007;446:71–75. doi: 10.1038/nature05588. [DOI] [PubMed] [Google Scholar]
- 30.Bae YK, et al. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev Biol. 2009;330:406–426. doi: 10.1016/j.ydbio.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Kimura Y, Okamura Y, Higashijima S. alx, a zebrafish homolog of Chx10, marks ipsilateral descending excitatory interneurons that participate in the regulation of spinal locomotor circuits. J Neurosci. 2006;26:5684–5697. doi: 10.1523/JNEUROSCI.4993-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granato M, et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- 33.Drapeau P, Ali DW, Buss RR, Saint-Amant L. In vivo recording from identifiable neurons of the locomotor network in the developing zebrafish. J Neurosci Methods. 1999;88:1–13. doi: 10.1016/s0165-0270(99)00008-4. [DOI] [PubMed] [Google Scholar]
- 34.Wang M, Wen H, Brehm P. Function of neuromuscular synapses in the zebrafish choline-acetyltransferase mutant bajan. J Neurophysiol. 2008;100:1995–2004. doi: 10.1152/jn.90517.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono F, Higashijima S, Shcherbatko A, Fetcho JR, Brehm P. Paralytic zebrafish lacking acetylcholine receptors fail to localize rapsyn clusters to the synapse. J Neurosci. 2001;21:5439–5448. doi: 10.1523/JNEUROSCI.21-15-05439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLean DL, Fetcho JR. Ontogeny and innervation patterns of dopaminergic, noradrenergic, and serotonergic neurons in larval zebrafish. J Comp Neurol. 2004;480:38–56. doi: 10.1002/cne.20280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.