Abstract
Budding yeast noncoding RNAs (ncRNAs) are pervasively transcribed during mitosis, and some regulate mitotic protein-coding genes. However, little is known about ncRNA expression during meiotic development. Using high-resolution profiling we identified an extensive meiotic ncRNA expression program interlaced with the protein-coding transcriptome via sense/antisense transcript pairs, bidirectional promoters, and ncRNAs that overlap the regulatory regions of genes. Meiotic unannotated transcripts (MUTs) are mitotic targets of the conserved exosome component Rrp6, which itself is degraded after the onset of meiosis when MUTs and other ncRNAs accumulate in successive waves. Diploid cells lacking Rrp6 fail to initiate premeiotic DNA replication normally and cannot undergo efficient meiotic development. The present study demonstrates a unique function for budding yeast Rrp6 in degrading distinct classes of meiotically induced ncRNAs during vegetative growth and the onset of meiosis and thus points to a critical role of differential ncRNA expression in the execution of a conserved developmental program.
Keywords: Saccharomyces cerevisiae, sporulation, tiling arrays
Meiosis is a conserved developmental pathway during which cells replicate and recombine their DNA before they progress through two successive divisions to produce haploid gametes. In simple eukaryotes such as yeasts, the process is partially controlled by a complex expression program coordinating the activity of several hundred protein-coding genes (1–3).
Although most work on meiotic development has focused on protein-coding genes, some evidence for the expression of noncoding RNAs (ncRNAs) in sporulating budding yeast and fission yeast has been reported (4–7). The presence of noncoding transcripts of various sizes during the reproductive stages of the eukaryotic life cycle has been observed in many multicellular organisms. Moreover, the exosome, which includes the RNase D-type exoribonuclease Rrp6 involved in mitotic ncRNA turnover, is conserved from yeasts to mammals (8–11). Importantly, the fission yeast ortholog of Rrp6 was shown to be involved not only in the mitotic degradation of meiotic mRNAs but also of at least one meiotic noncoding transcript encoded by sme2 (12, 13).
There is a rapidly growing body of evidence that ncRNAs regulate many biological processes via mechanisms that involve either their synthesis or their posttranscriptional activity (14, 15). However, very little is known about the abundance, transcript boundaries, genomic localization, and possible roles of meiotically induced ncRNAs in the key experimental model organism Saccharomyces cerevisiae. Furthermore, the role of the budding yeast exosome component Rrp6 in coordinating the transition from mitosis to meiosis has not been investigated (16).
Here, we used high-resolution oligonucleotide tiling arrays to study the protein-coding and noncoding expression program underlying vegetative and reproductive phases of the haploid and diploid budding yeast life cycle, and we integrated the results with genomic, genetic, and biochemical analyses of Rrp6. Our data reveal extensively interleaved meiotic mRNA and ncRNA transcriptomes, and they identify meiosis-specific noncoding transcripts. Moreover, the present study provides clues for the importance of staggered ncRNA accumulation during the exit from mitotic growth and the transition through meiotic development via Rrp6’s role and the protein's posttranslational down-regulation after the onset of meiosis.
Results
High-Resolution Expression Profiling of the Budding Yeast Life Cycle.
We analyzed vegetatively growing and sporulating diploid MATa/α cells, a sporulation-deficient MATα/α control strain, and synchronized mitotic haploid MATa cells (SI Materials and Methods and Fig. S1 A and B) (17) using high-resolution oligonucleotide tiling arrays (18) as well as modified normalization and segmentation methods (Figs. S1C and S2A) (19). The goal of these experiments was to identify and characterize ncRNAs that are preferentially or specifically expressed in diploid cells undergoing meiotic development.
Previous work identified stable unannotated transcripts (SUTs) as ncRNAs being detectable during mitosis (20) and cryptic unstable transcripts (CUTs) as ncRNAs accumulating during vegetative growth only in the absence of Rrp6 (9). Following this nomenclature, we designated ncRNAs found in meiotic but not in fermenting or respiring cells as meiotic unannotated transcripts (MUTs), and we refer to transcripts typically showing peak expression in respiring or sporulating MATa/α cells as rsSUTs.
Detailed information about molecular biological and computational methods is available in SI Materials and Methods. Genome-wide normalized and log2-transformed expression data at the single oligonucleotide probe level are accessible via GermOnline's Saccharomyces Genomics Viewer (www.germonline.org) (21).
Genome-Wide Identification of Meiotically Induced ncRNAs and Their Sense/Antisense Configuration.
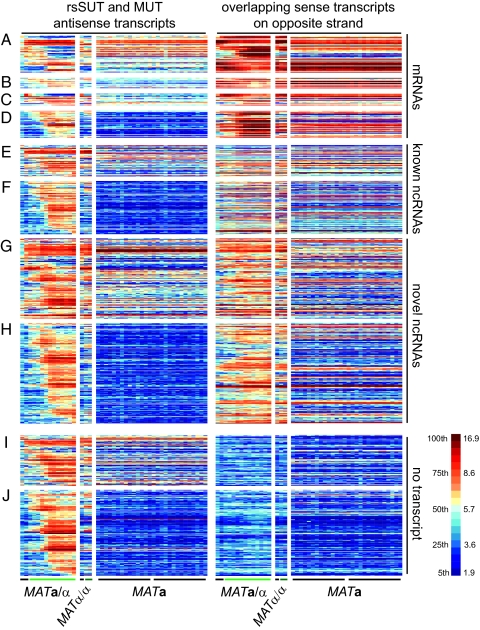
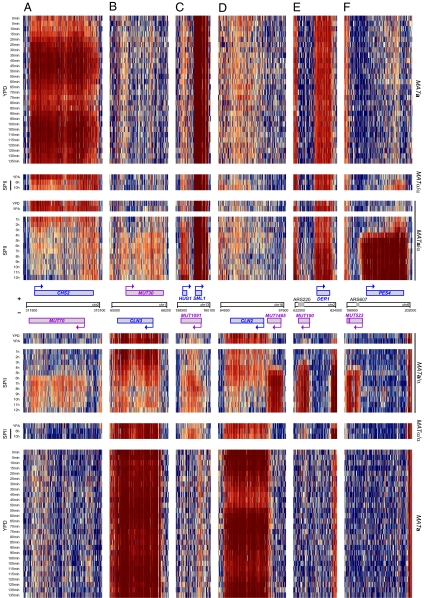
In this report we focus on 1,452 differentially expressed ncRNAs, which we organized into three categories: those that overlap with sense mRNAs on the opposite DNA strand, a second group that overlaps with other known or newly identified sense ncRNAs, and those that are not associated with any transcripts on the opposite strand (Fig. S2 B and C). A total of 878 ncRNAs (363 MUTs) were found to overlap with mRNAs. To explore the functional relevance of sense/antisense pairs, we selected rsSUTs and MUTs that overlapped with differentially expressed mRNAs and determined how their expression profiles correlated with each other (ncRNAs are “antisense” and mRNAs are “sense” transcripts; Dataset S1). Thirty-three rsSUTs (Fig. 1A) and six MUTs (Fig. 1B) showed significantly opposed patterns, whereas 11 rsSUTs (Fig. 1C) and 32 MUTs (Fig. 1D) displayed significantly similar profiles compared with the differentially expressed mRNAs. The group of protein-coding genes that showed opposed patterns to antisense rsSUTs was significantly enriched for Gene Ontology terms (22) such as sexual reproduction (GO: 0019953; five observed/one expected by chance p = 9.1 × 10−3) and cellular cell wall organization (0007047; seven observed/one expected by chance; p = 3.4 × 10−4). It is noteworthy that genes involved in cell division (CHS2, CLN3) or cell cycle progression (HUG1), which are down- or up-regulated during meiosis, respectively, overlap with meiotically induced antisense MUTs (Fig. 2 A–C).
Fig. 1.
Global expression profile of differentially expressed ncRNAs. Each line corresponds to an annotated segment, and each column represents the averaged values (computed for each segment) observed in duplicate samples. (A) Antisense rsSUTs or (B) MUTs and differentially expressed sense mRNAs showing opposed expression patterns. (C) rsSUTs or (D) MUTs and differentially expressed antisense mRNAs showing similar patterns. (E and F) Sense/antisense pairs of rsSUTs and MUTs and known ncRNAs. (G and H) Sense/antisense pairs of rsSUTs and MUTs. (I and J) rsSUTs or MUTs not overlapping with any transcript. MATa/α cells were cultured in YPD, YPA (black bar), and sporulation medium (SPII, hourly samples from 1 to 12 h; light green bar). The MATα/α strain was incubated in YPA (black bar) and SPII (6 and 10 h; dark green bar). MATa cells were grown in rich medium (YPD, samples were taken every 5 min from 0 to 135 min covering two cycles; black bars). Right: Scale for log2-transformed values, containing 20 colors corresponding to increments of 5 percentiles each.
Fig. 2.
Genomic ncRNA heatmaps. (A and B) Opposed sense/antisense profiles are shown for CSH2/MUT70 and CLN3/MUT30. (C) Correlated induction profiles for HUG1/MUT1091. (D) CLN2 mRNA and MUT1465 covering CLN2’s regulatory region. (E) Bidirectional promoter mediating expression of DER1 and MUT100 covering ARS220. (F) Bidirectional promoter of PES4 and MUT523 covering ARS607. Blue, purple, and gray boxes symbolize genes, MUTs, and ARS elements, respectively. Arrows indicate the direction of transcription. Averaged, normalized, and log2-transformed data at the oligonucleotide probe level covering the top (+) and bottom (−) strands are shown. Media, samples, and scale are as in Fig. 1.
Subsequently, we searched for 421 noncoding transcripts not associated with mRNAs to explore functions not directly related to the expression of protein-coding genes. Indeed, 120 transcripts (43 rsSUTs, Fig. 1E; 77 MUTs, Fig. 1F) overlapped with known antisense ncRNAs such as CUTs, SUTs, and small nucleolar RNAs (snoRNAs), whereas 301 (132 rsSUTs, Fig. 1G; 169 MUTs, Fig. 1H) overlapped with other antisense ncRNAs on the opposite strand. The remaining 202 ncRNAs (78 rsSUTs, Fig. 1I; 124 MUTs, Fig. 1J) were not associated with transcripts but in some cases with genome features such as promoter regions and DNA replication elements. For example, the highly induced MUT1465 covers the promoter region of CLN2 (Fig. 2D). Other MUTs—including some that are expressed from bidirectional promoters that control expression of ncRNAs and mRNAs in all possible combinations (Fig. S3 A and B)—cover autonomously replicating sequences (ARSs) such as ARS220 and ARS607 (Fig. 2 E and F). Finally, we identified numerous ncRNAs not associated with any of the genome features we investigated, including many that seem to be controlled by bidirectional promoters (Fig. S3C).
These results reveal a complex meiotic ncRNA expression program interlaced with the mRNA transcriptome via partially or totally overlapping sense/antisense transcript pairs, bidirectional promoters, and ncRNAs that cover developmentally regulated promoters of protein-coding genes.
Rrp6 Controls MUTs and rsSUTs During Vegetative Growth.
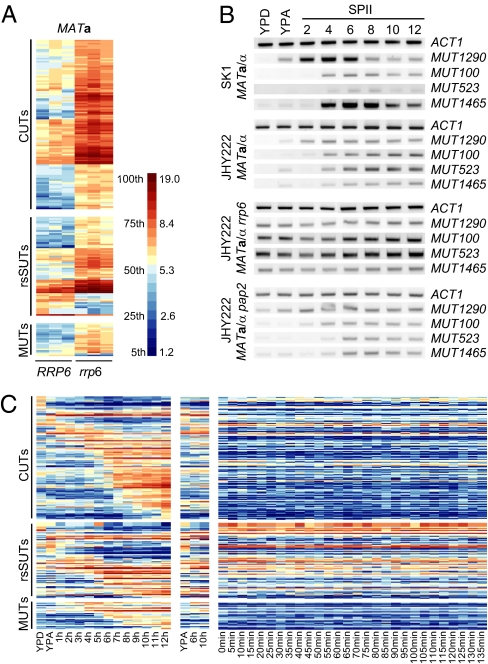
To determine whether Rrp6 controls the expression of meiotic ncRNAs during mitosis we compared tiling array expression data from MATa RRP6 vs. rrp6 strains and identified 22 MUTs and 59 rsSUTs that strongly accumulated in a pattern similar to 106 CUTs in the mutant (greater than threefold change), as shown in Fig. 3A (20). We conclude that Rrp6 is required for mitotic degradation of a subset of rsSUTs and MUTs detectable in haploid cells.
Fig. 3.
Rrp6 control of meiotic and mitotic ncRNAs. (A) Differentially expressed CUTs, rsSUTs, and MUTs in RRP6 vs. rrp6 strains. A scale for percentiles and log2-transformed data is given. (B) MUT concentrations determined by RT-PCR in fermenting (YPD), respiring (YPA), and sporulating (SPII, 2–12 h) cells. ACT1 was used as a standard. (C) Expression data for Rrp6-dependent MUTs, rsSUTs, and CUTs in sporulating (MATa/α) and starving (MATα/α) diploid cells and in a growing haploid strain (MATa). The scale is as in Fig. 1.
We next confirmed and extended these findings in two different diploid strains by RT-PCR: whereas MUT1290, -100, -523, and -1465 showed the expected pattern in fermenting (YPD), respiring (YPA), and sporulating (SPII 2–12 h) wild-type cells, respectively (Fig. 3B), we found that MATa/α rrp6 cells accumulated MUTs during vegetative growth in the presence of different carbon sources (fermentation, respiration) and sporulation (Fig. 3B). Whereas diploid JHY222 rrp6 cells displayed the temperature-sensitive phenotype previously reported (16), the diploid SK1 rrp6 mutant already failed to grow normally at the permissive temperatures (25 °C/30 °C; Fig. S4 A and B) and was therefore not further investigated.
We next examined the poly-A polymerase Pap2, a component of the TRAMP complex, which enhances Rrp6’s activity (23, 24). Although Pap2 was suggested to act in the same pathway as Rrp6 (9), an RT-PCR analysis of MUTs in MATa/α pap2 cells revealed no mitotic accumulation and showed their meiotic expression to be indistinguishable from the one observed in wild-type strains (Fig. 3B, Bottom). The ACT1 control revealed no fluctuations in RNA concentration throughout the sample sets.
These findings show that Rrp6 prevents MUTs from accumulating during mitosis. Pap2 is not essential for this process in the cases we examined, most probably because it is partially redundant with Trf5, a TRAMP subunit homologous to and synthetically lethal with Pap2 (25, 26).
Rrp6 Is Developmentally Regulated at the Posttranslational Level.
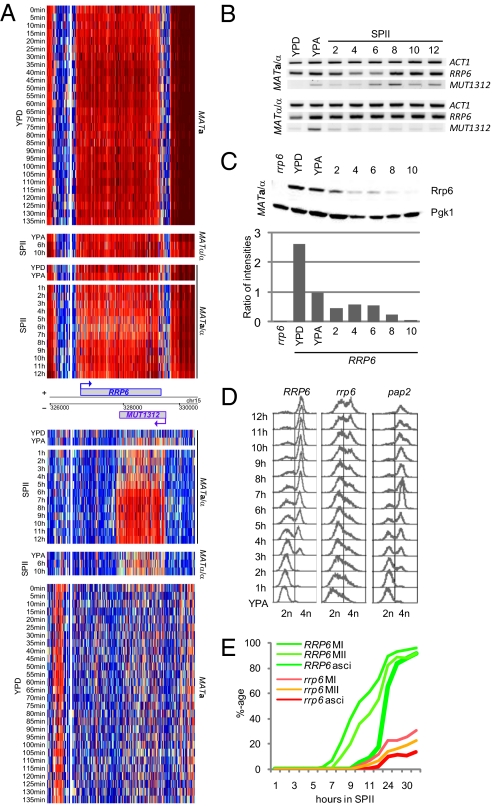
Further analysis of CUTs during meiosis revealed their staggered accumulation similar to SUTs and MUTs in sporulating MATa/α but typically not in starving MATα/α cells (Fig. 3C). This result prompted us to hypothesize that Rrp6 might be inactivated during meiosis and spore formation. Tiling array data and RT-PCR results shown in Fig. 4 A and B, respectively, reveal fairly constant RRP6 mRNA levels and transcript boundaries during fermentation, respiration, sporulation, and starvation (MATα/α in SPII); a transient drop in transcript concentration is detectable around 4–6 h into sporulation, but overall the data argue against a major transcriptional control mechanism acting on RRP6 expression.
Fig. 4.
Rrp6 regulation and function. (A) Genomic heatmap of expression data for RRP6/MUT1312 from strains and samples as given. The layout is as in Fig. 2, and the scale is as in Fig. 1. (B) RT-PCR expression data for RRP6/MUT1312 using samples as in Fig. 3B. (C) Western blot data for Rrp6 and Pgk1 detected in extracts prepared from a diploid rrp6 mutant (rrp6) and diploid growing (YPD, YPA) and sporulating wild-type cells (time points taken every 2 h from 2 to 10 h). Histogram below the bands shows relative Rrp6 protein amounts on the y-axis detected in the samples given on the x-axis. (D) DNA replication dynamics analyzed by fluorescence-activated cell sorting in diploid strains containing wild-type (RRP6) and mutant (rrp6, pap2) alleles. Respiring cells (YPA) were compared with hourly samples from sporulating cells (SPII, 1–12 h). DNA content is given at the bottom. (E) Wild-type (RRP6) and mutant (rrp6) strains undergoing MI, MI+II, and ascus formation. The x-axis shows the time of incubation in SPII sporulation medium in hours, and the y-axis displays the percentage of cells within the population that have completed landmark events. The average of three independent sporulation assays is shown.
We next investigated Rrp6’s stability and observed a ≈2.5-fold decrease in Rrp6 concentration as cells switched from fermentation to respiration. Interestingly, we also detected a further decrease of the Rrp6 level by a factor of 2 at the onset of meiosis (SPII 2–6 h) until it approached the threshold level of detection by Western blotting during postmeiotic spore formation (SPII 8–10 h). As opposed to that, protein levels remained essentially constant in all samples in the case of the Pgk1 control (Fig. 4C).
We conclude that RRP6 is constitutively transcribed (showing moderate mRNA fluctuations during the mitotic and meiotic cell cycles) while the Rrp6 protein is down-regulated as cells progress through meiotic development. It is remarkable that Rrp6’s progressive destruction coincides with increasing pervasive accumulation of meiotic ncRNAs (compare Fig. 1 and Fig. 3C with Fig. 4C).
Rrp6 Is Essential for Premeiotic DNA Replication, the Meiotic Divisions, and Spore Formation.
We next asked whether Rrp6 was essential for the transition between vegetative growth and meiotic differentiation. To this end, rrp6 mutant cells were sporulated, and their ability to progress through S-phase to form bi- and tetranucleate cells during M-phase and to package nuclei into four-spored asci was examined. Indeed, the MATa/α rrp6 strain failed to undergo normal premeiotic DNA replication because most cells could not initiate the process, and only a small fraction completed it after a substantial delay. As opposed to that, MATa/α pap2 cells replicated their DNA normally, albeit slightly earlier than the wild-type strain (Fig. 4D). We next determined that cells lacking Rrp6 could not transit through MI, MII, and undergo spore formation at normal levels (Fig. 4E), whereas the MATa/α pap2 mutant initiated M-phase faster than the wild-type background and formed spores as efficiently as the MATa/α PAP2 strain (Fig. S4C).
Taken together, these results confirm that Rrp6 is important for robust mitotic growth (or any growth at all in the SK1 strain background) and suggest that its mitotic and premeiotic presence is required for normal premeiotic DNA replication, execution of the meiotic divisions, and spore formation.
Discussion
The present study addresses the important question of how the mRNA and ncRNA transcriptomes are interleaved during the vegetative and reproductive phases of the budding yeast life cycle. Moreover, it reveals Rrp6—a highly conserved exoribonuclease associated with the nuclear exosome—to be posttranslationally down-regulated during meiotic M-phase and spore formation when pervasive ncRNA accumulation occurs. Finally, it demonstrates Rrp6 to be important for a normal transition from the mitotic to the meiotic cell cycle and efficient gamete formation.
ncRNA Expression and Function During Sporulation.
Exit from mitosis and proper execution of the meiotic developmental pathway requires reprogramming the transcriptome from a cyclical to a linear process. This includes repression of mitotic genes incompatible with meiotic development, activation of genes specifically needed for meiosis and gametogenesis, and adaptation of cell cycle-regulated loci to the meiotic divisions. Although such regulatory functions have typically been attributed to DNA binding transcriptional activators and repressors (27, 28), a number of mitotic ncRNAs were recently shown to positively or negatively influence gene expression via changes of histone modification or transcriptional interference (17, 29–32). It is therefore conceivable that developmentally regulated ncRNAs such as MUTs contribute to the control of genes required for, for example, cytokinesis (CHS2) and the mitotic cell cycle (CLN3, HUG1) through similar mechanisms.
Some of the noncoding RNAs may also regulate protein-coding genes by inhibiting the activity of their promoters, as shown earlier for loci involved in metabolic functions (33; for review see ref. 34). In such a case, it is not the ncRNA itself but its synthesis and the pausing of RNA polymerase during elongation that prevents activator binding (35). For example, the mRNA concentration of CLN2 declines during sporulation, and its 5′-regulatory region is covered by MUT1465; CLN2 is a repressor of IME1 (the inducer of meiosis), and therefore CLN2’s down-regulation is important for the onset of meiosis (36). Another interesting example is CDC6, which is essential for DNA replication (37). The gene's mRNA declines as cells transit through the meiotic divisions, while at least one strongly meiotically induced ncRNA (SUT200) covers its entire upstream region precisely at the onset of meiotic M-phase (www.germonline.org).
Other MUTs could regulate ARS elements during sporulation via transcription interference (38). For example, the mitotically active ARS605 was shown to be inhibited during meiotic prophase by the expression of the early meiotic gene MSH4, which precludes Orc1 binding to its target motif (39). It is thus tempting to speculate that ncRNAs directly contribute to the regulation of ARS activity, either alone or together with mRNAs (see for example ECM23/MUT1498, which cover ARS1621; www.germonline.org), thereby influencing the efficiency of DNA replication at different stages of growth and development.
Meiotic ncRNAs and Rrp6 Show Inverse Patterns of Developmental Regulation.
A striking outcome of our study is that ncRNAs not detected during mitosis in wild-type cells accumulate to high levels during meiosis and spore formation (9, 20, 40). Contrary to this pattern, the Rrp6 protein is stable during mitotic growth (fermentation) but diminishes as cells switch to respiration and sporulation. A simple interpretation of our findings is that Rrp6 directly targets MUTs, CUTs, and rsSUTs during vegetative growth in the presence of glucose and that these ncRNAs are able to exert their functions when Rrp6 levels decline during respiration and sporulation.
Two explanations for the decreasing Rrp6 protein concentrations are conceivable: MUT1312 might inhibit Rrp6 translation via an RNA interference-independent mechanism (Fig. 4 A and B) (41). Alternatively, the Rrp6 protein might be targeted by a protease that is partially induced during respiration (when the Rrp6 protein level drops by 50%) and that reaches its full activity after the induction of meiosis. Intriguingly, this is the case for Ume6, which is destroyed by the anaphase promoting complex/cyclosome during early meiosis (42). Such a mechanism would imply that early meiotic mRNAs and meiotically induced ncRNAs are orchestrated via simultaneous destruction of the DNA-binding repressor Ume6 and the exoribonuclease Rrp6.
Rrp6 Function Is Important for the Transition from Mitosis to Meiosis.
Rrp6 contains three domains required for nuclear localization, exoribonuclease activity, and RNA substrate specificity (43). Although a number of mRNAs are deregulated in the absence of Rrp6 (Dataset S2) it seems that protein-coding transcripts are recognized by the protein as substrates only in mutants that fail to export them from the nucleus (44). The protein is thus predominantly required for efficient 5.8S rRNA 3′-end formation and mitotic processing of snRNAs, snoRNAs, and ncRNAs, the latter in cooperation with Pap2 (9, 16, 20, 40, 45). Importantly, fission yeast Rrp6 was recently shown to target meiotic mRNAs untimely expressed during mitotic growth and the meiotic ncRNA encoded by the sme2 gene, which is essential for Meiosis I (13, 46, 47). This is in keeping with the finding that Rrp6 targets MUTs and other meiotically induced ncRNAs during vegetative growth in the distantly related yeast S. cerevisiae. Given the opposed profiles of meiotic ncRNA and Rrp6 stabilities during growth and development and the phenotypes of rrp6 cells, we speculate that Rrp6 is a negative regulator of meiotic development and that the protein's progressive down-regulation during the transition from fermentation to respiration and sporulation is important for efficient gametogenesis by allowing ncRNAs to accumulate.
The outcome of our analysis, together with reports on meiotic ncRNAs in fission yeast (6, 7), increasing evidence for ncRNAs in the mammalian germline (8), and the finding that fly Rrp6 is important for cell cycle progression (48), raise the intriguing possibility that conserved mechanisms involving noncoding transcripts and RNase D-type ribonucleases contribute to the control of gametogenesis during germ cell development in higher eukaryotes.
Materials and Methods
Yeast Strains, Media, and Sporulation Conditions.
We studied wild-type and mutant derivatives of S. cerevisiae SK1 and JHY222 strains (SI Materials and Methods; Table S1).
Expression Profiling.
Total RNA from diploid cells was quality controlled with a BioAnalyzer (Agilent) and used to enrich for poly-A+ transcripts with the Oligotex kit (Qiagen). Purified polyA+ RNA was reverse transcribed in the presence of actinomycin D to prevent spurious antisense RNA synthesis (49). Proprietary high-density oligonucleotide tiling microarrays (Sc_tlg GeneChips) were used for raw data production using the GCS3OOO TG system (Affymetrix) (17).
Tiling Array Data Analysis.
Data quality control was carried out using R scripts (50). A modified version of the tilingArray Bioconductor package was used for data processing and normalization (18). Segments were identified by first applying the segmentation algorithm to five sample groups, followed by merging similarly expressed segments located within <50 base pairs of each other. Segments corresponding to differentially expressed transcripts identified using AMEN were inspected in a genome-wide manual curation step (50). A detailed description of our approach is available in SI Materials and Methods. Raw data are available via the European Bioinformatics Institute’s repository ArrayExpress (http://www.ebi.ac.uk/arrayexpress) at E-TABM-915 (MATa/α and MATα/α).
Supplementary Material
Acknowledgments
We thank R. Strich for a critical reading of the manuscript; O. Collin, A. Roult, and R. Fabretti for information technology support; P. Demougin, J. Dyczkowski, Z. Xu, and M. Ritchie for technical support; and J. Horecka (Stanford Genome Technology Center, Palo Alto, CA) for JHY222. The Rrp6 antibody was a gift from M. Schmid (University of Aarhus, Aarhus C, Denmark). This work was supported by fellowships from Institut National de la Santé et de la Recherche Médicale (INSERM)/Institut National de l’Environnement Industriel et des Risques (INERIS) and the French Ministry of Education (to A.L. and Y.L., respectively); by INSERM Avenir Grant R07216NS and Fondation pour la Recherche Médicale Grant INE20071111109 (to M.P.); and by National Institutes of Health Grants 5R01 GM068717 and 5P01 HG000205 (to R.W.D. and L.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Raw data are available via the European Bioinformatics Institute’s repository ArrayExpress (http://www.ebi.ac.uk/arrayexpress) [accession no. E-TABM-915 (MATa/α and MATα/α)].
See Commentary on page 891.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016459108/-/DCSupplemental.
References
- 1.Primig M, et al. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 2.Chu S, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 3.Mata J, Lyne R, Burns G, Bähler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 4.Havilio M, Levanon EY, Lerman G, Kupiec M, Eisenberg E. Evidence for abundant transcription of non-coding regions in the Saccharomyces cerevisiae genome. BMC Genomics. 2005;6:93. doi: 10.1186/1471-2164-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miura F, et al. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc Natl Acad Sci USA. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe T, et al. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 2001;29:2327–2337. doi: 10.1093/nar/29.11.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 9.Davis CA, Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 11.Lykke-Andersen S, Brodersen DE, Jensen TH. Origins and activities of the eukaryotic exosome. J Cell Sci. 2009;122:1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- 12.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 2010;29:2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St-André O, et al. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem. 2010;285:27859–27868. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 15.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 17.Granovskaia MV, et al. High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol. 2010;11:R24. doi: 10.1186/gb-2010-11-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber W, Toedling J, Steinmetz LM. Transcript mapping with high-density oligonucleotide tiling arrays. Bioinformatics. 2006;22:1963–1970. doi: 10.1093/bioinformatics/btl289. [DOI] [PubMed] [Google Scholar]
- 20.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gattiker A, Niederhauser-Wiederkehr C, Moore J, Hermida L, Primig M. The GermOnline cross-species systems browser provides comprehensive information on genes and gene products relevant for sexual reproduction. Nucleic Acids Res. 2007;35(Database issue):D457–D462. doi: 10.1093/nar/gkl957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2009;38(Database issue):D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanácová S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castaño IB, Heath-Pagliuso S, Sadoff BU, Fitzhugh DJ, Christman MF. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 1996;24:2404–2410. doi: 10.1093/nar/24.12.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: The link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008;1779:239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Kassir Y, et al. Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol. 2003;224:111–171. doi: 10.1016/s0074-7696(05)24004-4. [DOI] [PubMed] [Google Scholar]
- 28.Raithatha SA, Stuart DT. Meiosis-specific regulation of the Saccharomyces cerevisiae S-phase cyclin CLB5 is dependent on MluI cell cycle box (MCB) elements in its promoter but is independent of MCB-binding factor activity. Genetics. 2005;169:1329–1342. doi: 10.1534/genetics.104.036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci USA. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 33.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 34.Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW. Transcriptional interference: An unexpected layer of complexity in gene regulation. J Cell Sci. 2007;120:2755–2761. doi: 10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- 35.Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol Cell. 2009;34:545–555. doi: 10.1016/j.molcel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purnapatre K, Piccirillo S, Schneider BL, Honigberg SM. The CLN3/SWI6/CLN2 pathway and SNF1 act sequentially to regulate meiotic initiation in Saccharomyces cerevisiae. Genes Cells. 2002;7:675–691. doi: 10.1046/j.1365-2443.2002.00551.x. [DOI] [PubMed] [Google Scholar]
- 37.Boronat S, Campbell JL. Linking mitosis with S-phase: Cdc6 at play. Cell Cycle. 2008;7:597–601. doi: 10.4161/cc.7.5.5519. [DOI] [PubMed] [Google Scholar]
- 38.Snyder M, Sapolsky RJ, Davis RW. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori S, Shirahige K. Perturbation of the activity of replication origin by meiosis-specific transcription. J Biol Chem. 2007;282:4447–4452. doi: 10.1074/jbc.M609671200. [DOI] [PubMed] [Google Scholar]
- 40.Neil H, et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 41.Harrison BR, Yazgan O, Krebs JE. Life without RNAi: Noncoding RNAs and their functions in Saccharomyces cerevisiae. Biochem Cell Biol. 2009;87:767–779. doi: 10.1139/O09-043. [DOI] [PubMed] [Google Scholar]
- 42.Mallory MJ, Cooper KF, Strich R. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol Cell. 2007;27:951–961. doi: 10.1016/j.molcel.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips S, Butler JS. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA. 2003;9:1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuai L, Das B, Sherman F. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:13962–13967. doi: 10.1073/pnas.0506518102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allmang C, et al. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harigaya Y, et al. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 47.Shimada T, Yamashita A, Yamamoto M. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol Biol Cell. 2003;14:2461–2469. doi: 10.1091/mbc.E02-11-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graham AC, Kiss DL, Andrulis ED. Core exosome-independent roles for Rrp6 in cell cycle progression. Mol Biol Cell. 2009;20:2242–2253. doi: 10.1091/mbc.E08-08-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perocchi F, Xu Z, Clauder-Münster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chalmel F, Primig M. The Annotation, Mapping, Expression and Network (AMEN) suite of tools for molecular systems biology. BMC Bioinformatics. 2008;9:86. doi: 10.1186/1471-2105-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.