Abstract
Understanding the nature of interactions between engineered nanomaterials and plants is crucial in comprehending the impact of nanotechnology on the environment and agriculture with a focus on toxicity concerns, plant disease treatment, and genetic engineering. To date, little progress has been made in studying nanoparticle-plant interactions at single nanoparticle and genetic levels. Here, we introduce an advanced platform integrating genetic, Raman, photothermal, and photoacoustic methods. Using this approach, we discovered that multiwall carbon nanotubes induce previously unknown changes in gene expression in tomato leaves and roots, particularly, up-regulation of the stress-related genes, including those induced by pathogens and the water-channel LeAqp2 gene. A nano-bubble amplified photothermal/photoacoustic imaging, spectroscopy, and burning technique demonstrated the detection of multiwall carbon nanotubes in roots, leaves, and fruits down to the single nanoparticle and cell level. Thus, our integrated platform allows the study of nanoparticles’ impact on plants with higher sensitivity and specificity, compared to existing assays.
Keywords: microarray, laser spectroscopy, tomato plants, aquaporins, carbon nanomaterials
The use of nanostructures in biomedicine (1) and, recently, in agriculture (2) is one of the most intensely studied areas in nanotechnology. Nanoscale materials have been shown to be uptaken by tumor cells (3), bacteria (4), plant cells (5), and animal tissues (6). In particular, carbon nanotubes (CNTs) with their unique structural and dimensional properties have been intensively studied for drug and gene delivery, tissue engineering, and other biomedical applications (7–9). It has also been shown that carbon nanotubes have the ability to penetrate plant cells (5) and induce phytotoxicity at high doses (10). We have demonstrated that single-wall CNTs at relatively low doses can penetrate even thick seed coats, stimulate germination, and activate enhanced growth of tomato plants (11). However, a thorough understanding of the effects induced by the nano-sized engineered materials on plant physiology at the molecular level is still lacking. In addition, the methods used for detecting such nanostructures in plant tissues are not well established and most of them are time consuming and labor intensive. Moreover, existing nanoparticle detection techniques usually decompose and destroy samples to prove the presence of nanomaterials; as a result, the same plant samples cannot be assessed for genomic/proteomic analysis. For example, the detection of magnetic nanoparticles in pumpkin plants by vibrating sample magnetometer requires drying and digestion of tissue samples with HNO3 (12). Transmission electron microscopy (TEM) has been used to monitor the uptake and transportation of CNTs in rice (13), but it has few quantitative capabilities and may result in false positive interpretation because of considerable similarity in TEM images of CNTs and natural plant structures. Consequently, the analysis has to be combined with spectroscopic studies for the exact identification and assessment of the CNTs in the host plant tissue, and this requires the total destruction of the samples (13). Moreover, the majority of the detection methods require additional nanoparticle labeling (14) that may unpredictably modify both the nanoparticles’ biodistribution properties and plant responses. It is obvious that a comprehensive study of the nature of nanoparticles-plant cell interactions will require the development of ultrasensitive methods for in situ real-time monitoring of nanoparticle transportation in plants.
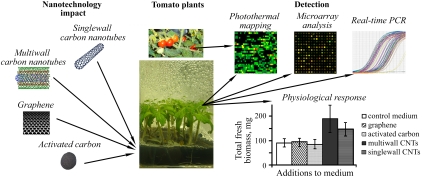
The main goal of this work is to demonstrate an integrated approach for studying the processes that take place during the exposure of tomato plants to carbon nanotubes, e.g., changes in gene expression, and correlate these findings with the presence of nanoparticles from roots to leaves. To perform this task, we linked the observed physiological responses of tomato plants with the complex sets of information provided by a unique combination of microarray analysis and nano-bubble amplified photothermal and photoacoustic imaging of nanomaterials in 10-day-old tomato seedlings grown on a medium containing various carbon nanostructural species (Fig. 1).
Fig. 1.
Schematics of integrated genomic and photothermal-based analysis of nanoparticle-plant interaction. The right bottom shows the effect of carbon materials (activated carbon, few-layer graphene structures, and single- and multiwall CNTs) on biomass accumulation of tomato plants. Pseudocolor in example of photothermal leaf map on right top indicates signals from small CNT clusters (red- maximum signal, green low signals).
Results
To understand the role of the structural shapes of the carbonaceous materials on plant physiology and to select those with the highest impact, four carbon-based structures—activated carbon (AC), few-layer graphene structures, multiwall and single-wall CNTs—were added separately to the Murashige and Skoog (MS) growth medium at an identical concentration of 50 μg/mL. The maximum physiological responses, such as the biomass enhancement (both for fresh and dried plants), were observed for the single- and multiwall CNTs only (see Fig. 1, bottom-right and Fig. S1A). The plants exposed to multiwall CNTs were then chosen for further detailed examination (multiwall carbon nanotubes are hereinafter referred to simply as “CNTs”). In this study, samples from the same plant seedlings exposed to CNTs were divided into two groups that were used in parallel for photoacoustic (PA)/photothermal (PT) detection and genetic analysis. Specifically, different leaf/root tissues from the same plants were used for the PA/PT detection and the genetic assays. We did not use the same identical tissues for both laser exposure and genetic analysis, in order to avoid additional stress that could be induced by the laser irradiation and that could impact the genetic results. For this reason, the changes in gene expression that we report could only be related to the presence of nanoparticles in plant tissues.
Photothermal and Photoacoustic Mapping of Multiwall CNTs in Tomato Roots, Leaves, and Fruits.
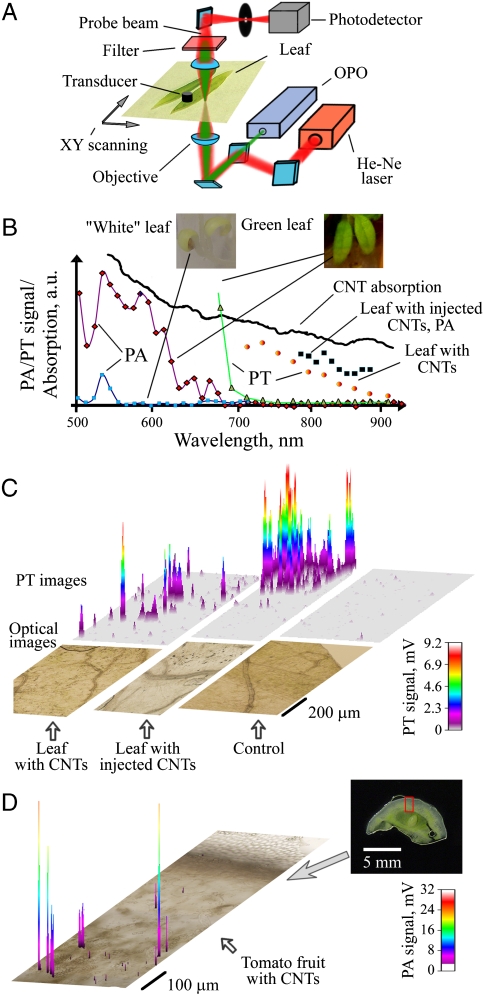
Among different imaging methods, PT and especially PA techniques based on the nonirradiative conversion of absorbed laser energy into heat and acoustic phenomena demonstrated higher sensitivity and resolution in deeper tissues in vivo compared to other optical imaging approaches (15, 16). However, the capability of these techniques to detect nanoparticles, specifically CNTs, in plants has not yet been demonstrated. Based on our previous experience in this field (17–19), we developed a PT/PA scanning cytometry platform on the basis of an invert microscope, a spectrally tunable optical parametric oscillator (OPO) with increased pulse rate of up to 100 Hz, and automated scanning (Fig. 2A and Fig. S2). Laser-induced generation of nanobubbles around the overheated individual CNTs, especially their clusters, was used as the selective and significant (5–15 fold compared to control) nonlinear PT and PA signal amplifier. The near-infrared spectral range 800–900 nm was the most promising for the detection of CNTs as its absorption at 900 nm is only two times lower than that at 600 nm, while PA spectra indicated that background light absorbance of the plants was dramatically lower than that at 600 nm, especially for tomato seedlings grown under dark conditions (Fig. 2B and Fig. S3). From the control plants analyzed at 903 nm, we observed only low PA signals (signal-to-noise up to 3), and low positive PT signals (signal-to-noise ∼2–4) defined by chlorophyll and water absorption (Fig. S3). Negative PT signal amplitudes (nonlinear effects) from the control plant corresponded to the photodetector electronic noise. For plants grown on MS medium supplemented with CNTs (50 μg/mL), we observed numerous randomly distributed and intense PA and PT signals in roots (Fig. S4) and in the leaves (Fig. 2C and Fig. S5 A–C). These negative PT and strong PA signals (signal-to-noise ratio up to 20 and 8, respectively) were associated with the localized CNT light absorbance in plant tissues. To verify the source of these signals, we locally injected CNT solution containing a mixture of individually dispersed and clustered 10 μm nanoparticles into leaves. For this calibration model, we also observed strong PT and PA signals above background level (signal-to-noise up to 100 and 12, respectively). The spatial distribution of these signals correlated well with the injection site and the position of relatively large CNT clusters visible in the optical transmission images (Fig. 2C, center, and Fig. S5G). PT/PA spectra were obtained from tomatoes germinated on CNT-containing medium, as well as from the leaves with injected CNTs. PT/PA spectra taken from the areas with high PT/PA signal were similar to the absorption spectra of the original CNTs (Fig. 2B). The majority of the detected CNTs were randomly distributed outside the leaves’ vascular system, among individual cells; only a few were found in close proximity to the leaf vasculatures (Fig. S5 D and E). This finding suggested that, at the time of examination, the CNTs were already cleared from the plant vascular system. In roots (Fig. S4), relatively large CNT aggregates (a few of them were also visible with optical transmission microscope) were located in the bulk of the root’s wall, and, probably, were filtered out from the medium. Because there were no large clusters found in the leaves by conventional microscopy, the transportation of smaller clusters and likely individual CNTs in the roots was more efficient and most probably regulated the further accumulation of the nanomaterial in the leaves (Fig. 2C and Fig. S5 A–E). Additionally, we were able to detect CNTs in tomato fruits (Fig. 2D and Fig. S6). In this case, PA detection proved to be more sensitive in 1 mm thick slices of tomato fruits compared to the PT mode due to a significant scattering of the probe beam by tomato tissues. These results showing the presence of CNTs in tomato fruits are in good correlation with our Raman spectroscopy analysis of the homogenized fruit samples (Fig. S6).
Fig. 2.
Photothermal and photoacoustic detection of multiwall CNTs in tomato leaves. Schematic of integrated PA/PT scanning cytometer (A). Spectral PA and PT identification of CNTs (B); given are images of tomato leaves grown in darkness (white) and under light (green). Two-dimensional PT maps (with three-dimensional simulation) of CNT distribution in tomato leaves compared to conventional optical images (C). Calibration model was constructed by injection of CNTs into leaf. PA detection of CNTs in 1 mm thick section of tomato fruit (D).
The two-dimensional scanning of the tomato leaves with relatively high energy laser radiation (740 nm) led to the clearing of the optical imaging from dark structures without influencing the CNT distribution, which allowed better distinction of leaf vasculatures and improved CNT localization (Fig. S5F). This optical contrast improvement was associated with “laser burning” (i.e., photomodification) of natural leaf structures that had absorbed laser radiation.
Analysis of Total Gene Expression in Tomato Plants Exposed to Multiwall CNTs.
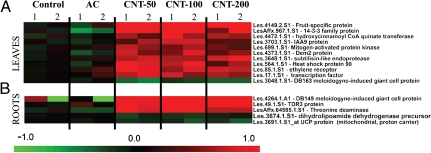
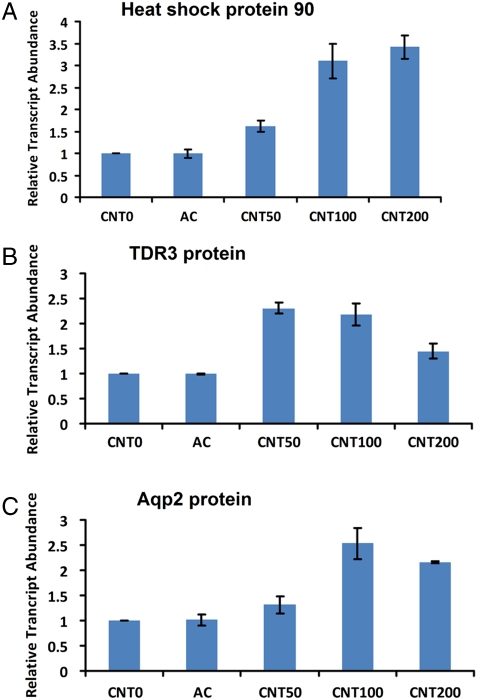
Because we had demonstrated the presence of CNTs in both roots and leaves by PA and PT scanning cytometry, genomic analysis was performed next in order to determine the possible impact of CNTs on plant tissues at the molecular and genetic levels. Indeed, plant genes are the units fundamentally responsible for controlling protein synthesis and, ultimately, plant functions underlying physiological processes. The microarray technology is a powerful profiling tool for studying altered gene expression induced by external stress factors (20) that could be linked to the CNT uptake by the plants as demonstrated by PA/PT cytometry. Our hypothesis was that monitoring the mRNA abundance changes for large sets of genes would enable us to identify gene clusters that are coregulated or show temporal interdependence to the presence of CNTs in tomato plants. Here, we carried out a comparative transcriptome analysis using Affymetrix Microarray Technology. Differences in gene expression in the roots and first two leaves of 10-day-old seedlings exposed to multiwall CNTs at different concentration levels (50; 100; 200 μg/mL), activated carbon (50 μg/mL), and the control tomato seedlings grown on regular MS medium were quantified by Affymetrix Tomato GeneChips. Based on these studies and after statistical analysis (T-test, hierachical clustering), we identified 91 transcripts in leaves and 49 transcripts in roots that showed significant differences in transcript abundance between the CNT-exposed seedlings and two controls (seedlings exposed to activated carbon and seedlings unexposed to any carbon structures) in all biological replicates that were analyzed (Fig. 3 for the 16 genes with known functions; Figs. S7 and S8 presented full datasets). The differently expressed profiles of nanoparticle-regulated genes were categorized according to their involvement in specific biological processes as depicted in the Tomato Database (Table S1). In leaves, the majority of CNT-regulated genes were involved in cellular responses (29 genes), stress responses (39 genes), transport (14 genes), signal transduction (13 genes), and metabolic and biosynthetic processes (25 genes). In roots, the largest numbers of CNT-regulated genes were involved in stress responses (10 genes), cellular processes (9 genes), transport (6 genes), and catabolic, metabolic, and biosynthetic processes (22 genes). The particular functions of several genes that showed altered expressions in response to CNTs are known. Thus, several tomato genes (Les.17.1.S1—transcription factor; LesAffx.967.1.S1—14-3-3 family protein; Les.49.1.S1—TDR3 protein) involved in the regulation of transcription and hormone pathways (Les.85.1.S1—ethylene receptor; Les.3703.1.S1—IAA9 protein) were up-regulated in seedlings in response to the exposure to CNTs. Expression of mitogen-activated protein kinase (Les.699.1.S1) was also significantly up-regulated in leaves exposed to CNTs (Fig. 3). It was documented earlier that mitogen-activated protein kinases play a positive role in the control of plant cell division and growth (21) and have important functions in stress signal transduction pathways in plants (22). A number of stress-related genes (Les.564.1.S1—heat shock protein 90; Les.3048.1.S1—DB163 meloidogyne-induced giant cell protein; Les.3648.1.S1—subtilisin-like endoprotease; LesAffx.64585.1.S1—threonine deaminase) were differently expressed in tissues exposed to CNTs. To validate the microarray data, we generated sequence-specific primers and performed quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR) using three independent biological replicates for four identified up-regulated genes in tomato tissues (Les.4264.1.A1—DB149 meloidogyne-induced giant cell protein; Les.49.1.S1—TDR3 protein; Les.564.1.S1—heat shock protein 90; Les.4373.1.S1—Dem2 protein). Data for two genes (Les.564.1.S1—heat shock protein 90 and Les.49.1.S1—TDR3 protein) are shown in Fig. 4 A and B. This analysis confirmed above presented data of microarray assay and revealed that the expression of all analyzed genes was dramatically higher in tissues of seedlings exposed to CNTs but not in control tissues or seedlings exposed to activated carbon. The intriguing finding of the microarray analysis is that several up-regulated genes (Les.3648.1.S1—subtilisin-like endoprotease; Les.3048.1.S1—DB163 meloidogyne-induced giant cell protein; LesAffx.64585.1.S1—threonine deaminase) in response to CNTs can also be activated in response to specific biotic stress factors (pathogens or herbivores). For example, expression of threonine deaminase was activated in tomato leaves in response to herbivore attack (23). In our experiment, expression of the same gene was up-regulated in tomato roots exposed to CNTs (Fig. 3). This observation suggests that the penetration of nano-sized material into plant tissues can be sensed by plants as a stress factor similar to pathogens or herbivore attack. In this case, important stress-signaling pathways and cascades could be modified/activated in response to the uptake of nanoparticles. Such possible changes might have a significant impact on most of the major physiological processes in planta. Our hypothesis was verified in a separate experiment. The expression of the gene for the tomato water-channel protein (LeAqp2) in the control tissues and tissues exposed to CNTs (50; 100; 200 μg/mL) was monitored by real-time RT-qPCR (Fig. 4C). As shown before, water channels (aquaporins) play a key role in germination and plant growth by regulating the water permeability at the level of individual plant cells (24, 25). The activity of aquaporins was previously shown to be influenced by a number of stress factors such as heavy metals, salinity, pH, and anoxia (24). Moreover, the expression of tomato aquaporin (LeAqp2) can be activated in response to the parasite plant Cuscuta reflexa (26). Here, we demonstrated that expression of LeAqp2 gene can be significantly activated in tomato roots by exposure to CNTs as compared to roots from plants grown on regular MS medium or a medium supplemented with regular carbon (Fig. 4C).
Fig. 3.
Microarray data of transcripts showing significant quantitative differences between the tomato seedlings exposed to CNTs in concentrations of 50; 100; and 200 μg/mL and two groups of control seedlings (exposed to activated carbon and unexposed to any carbon material) in two tissues of 10-day-old plants [roots (A) and first two leaves (B)]. Statistical analysis was performed by T-test (P < 10-3) based on log (2) fold changes of mRNA abundance over the average abundance of the specific transcript in the seedlings not exposed to carbon material. Only genes with known functions are presented here.
Fig. 4.
Relative transcript abundances of Les.564.1.S1- heat shock protein 90 in leaves (A), Les.49.1.S1- TDR3 protein in roots (B), and LeAqp2 protein in roots (C) of tomato seedlings exposed to AC, CNTs in concentrations 50, 100, and 200 μg/mL), and seedlings not exposed to carbon materials (CNT 0). Expression of genes was analyzed by real-time RT-qPCR. Results are shown as the average of three independent biological replicates. Relative expression levels were normalized to an internal standard (actin transcript) for each treatment. Bars represent standard error (SE) of means.
Discussion
In this multidisciplinary work, we proved that the interaction of multiwall CNTs with the cells of tomato seedlings resulted in significant changes in total gene expression. The groups of genes with altered expression were identified in the roots and leaves of tomato plants growing on a medium supplemented with CNTs. The exposure of tomato cells to CNTs can lead to the activation of many stress-related genes including the gene for tomato water-channel protein (LeAqp2). In addition, our data demonstrated that the up-regulation of the expression of the water channel’s gene in CNT-exposed roots and leaves can have a significant impact on the observed phenomena of activation, enhanced germination, and growth of tomato seedlings on a medium supplemented with CNTs. (See Fig. 1, the inset in right bottom; Fig. S1A.) It is interesting that the few-layer graphene carbon structures, although being at the nano-scale, did not significantly affect plant growth rates, probably because of their inability to penetrate plant tissues in a manner similar to that of CNTs.
The association of the observed molecular changes with the presence of CNTs in tomato roots and leaves was validated by performing advanced PA and PT scanning cytometry. PA/PT imaging revealed no distinguishable CNT-specific signals in the control plant tissues, while providing detectable and spectrally identified signals in the exposed plants. The sensitivity of this technique is sufficiently high for the detection of CNT clusters, if not individual CNTs, with sizes below the diffraction limit of conventional optical microscopy (250–300 nm). The sensitivity of PA method was comparable to that of PT method (Fig. S5 B and C); however PA demonstrated much higher sensitivity in detecting CNTs in relatively thick samples (Fig. 2D and Fig. S6D). In addition, PA backward schematics (i.e., laser and ultrasound transducer on one side) are better suited for assessing intact plant leaves in situ by delivering the laser radiation to the sample by optical fibers (27). For transparent plant cells, PT detection sensitivity is much higher than that of the PA mode and is sufficient for detection of submicrometer CNT clusters (Fig. 2 C and D). Laser-induced nano- and micro-bubbles around the overheated CNTs provided high specificity of PT mode through the appearance of a negative PT signals (17, 18). A combination of both PT and PA methods provides signal amplification and the additional identification of strongly absorbing CNTs that are present among natural biological backgrounds (Fig. 2C). PA and PT methods could beneficially supplement each other and, in combination, provide a powerful unique biological tool to study the nanomaterials’ impact on plants at the single nanoparticle and cell level including real-time monitoring of nanoparticle transport in plant vasculature analogous to the detection of nanoparticles and cells in blood and lymph systems (18, 19). We found that tomato plants grown in soil containing CNTs are able to uptake nanoparticles from soil and biodistribute them in various tissues, including fruits (Fig. S6). Based on this observation, we can predict that nanoparticles can be found in various parts of the plants, including the reproductive organs, grown in field conditions on soil containing nanoparticles either as fertilizers or contaminants. Thus, spectroscopic laser-based techniques such as PA/PT or Raman scattering can be used for highly sensitive detection of nanoparticles in the seeds or fruits of such plants.
The integration of the genomic and PT/PA detection methods can provide an advanced platform for the identification of genes affected by the documented presence of nanoparticles in certain plant tissues or individual plant cells and shed light on the possible mechanisms of the positive effects of nanoparticles on plant growth and development. Further studies involving various plant species and the possible proteomic and hormonal changes induced by CNTs need to be performed in order to fully understand the complex interaction between nanoparticles and plant systems. The application of nanoparticles to plant cells or young plants could be a possible alternative to classical genetic engineering techniques (28, 29) to create more productive and stress-tolerant plants for various applications ranging from biofuels crops to space-grown plants.
Methods
Nanoparticles and Plant Growth Conditions.
The carbonaceous nanomaterials (single-, multiwall CNTs, and few-layer graphene nanomaterials) that were used in this study were synthesized and analyzed according to published procedures (30, 31). TEM analysis indicated that the multiwall CNTs described in this manuscript have outer diameters varying between 10–35 nm, an average length of 6 μm, and purity of 98.5% ± 0.5%. The single-wall CNTs with a purity of 98% ± 0.7% were composed of tubes with diameters ranging from 0.86 nm to 2.22 nm; the corresponding bundles were found to be a few microns long. The diameter of the few-layer graphene structures (with a thickness of 2–5 nm) varies between 100–120 nm and had a purity of 98% ± 0.5%. The carbon structures utilized in this experiment were slightly functionalized to enhance their water solubility through a mild nitric acid treatment (gentle sonication for 30 min). In this way, carboxylic functional groups were introduced onto the surface of the nanostructures enhancing their water dispersion and creating a homogeneous solution. The representative TEM analysis of the nanomaterials used in this study is provided in Fig. S1B.
For assessment of physiological response (biomass accumulation), the single-wall CNTs, multiwall CNTs, few-layer graphene materials, and activated carbon were introduced into MS media in identical concentrations (50 μg/mL). For PT/PA analysis, plants grown on medium supplemented with 50 μg/mL multiwall CNTs were used. Standard MS medium was used as the control. Tomato seeds were surface-sterilized, germinated, and grown on all experimental variations of sterile MS medium as previously reported (11). Leaves and roots of 10-day-old tomato seedlings were used for subsequent experiments.
Microarray Analysis and Real-Time Quantitative Polymerase Chain Reaction.
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen). Microarray analysis was performed in two independent biological replicates using mRNA of the first two leaves and root tips of 10-day-old tomato seedlings that were exposed to multiwall CNTs (50 μg/mL; 100 μg/mL; and 200 μg/mL), nonexposed to multiwall CNTs (control 1), and exposed to 50 μg/mL activated carbon (control 2). Each sample for each replicate contained combined material from six individual plants. Affymetrix Tomato Genome Arrays methodology was used for our experiment. Biotinylated cRNA targets were synthesized according to Affymetrix IVT Express target labeling assay as specified in the Affymetrix GeneChip Expression Analysis Technical Manual. Hybridization reactions to the Affymetrix Tomato GeneChips were carried out by Expression Analysis, Inc. The differences in transcript abundance of several genes identified by microarray analysis were validated by using real-time RT-qPCR (SYBR Green detection). Three independent biological replicates were used. Complimentary DNA (cDNA) was synthesized for each sample using the SuperScript™ III First-Strand Synthesis System (Invitrogen). RT-qPCR assay was performed and analyzed as described earlier (28).
Statistical Analysis of Microarray.
Microarray raw data were analyzed using MAS5 (Affymetrix). Scanned arrays were normalized to a baseline array with median overall expression. Statistical analysis by Student T-test and hierarchical clustering (Euclidean distance) of log (2) fold changes were performed using TM4 Microarray Suite from TIGR (32). The functional characterization of up- and down-regulated genes in tomato seedlings exposed to CNTs was carried out by using Tomato Functional Genomics Database (http://ted.bti.cornell.edu/cgi-bin/TFGD/array/funcat.cgi). All generated microarray data were deposited in public database GEO under accession number GSE22803.
Integrated PA/PT Scanning Cytometry.
PA/PT cytometer was based on the technical platform of an Olympus invert IX81 microscope (Olympus America, Inc.). PT and PA effects occur in the sample upon exposure to irradiation of a tunable OPO (Opolette HR 355 LD, OPOTEK, Inc.) with 5 ns pulsewidth, 100 Hz repetition rate, a wavelength range of 410–2,200 nm, and a fluence range of 1–104 mJ/cm2 (Fig. 2A and Fig. S2). The energy of each excitation laser pulse was controlled by energy meter (PE10-SH, OPHIR). PT/PA mapping of the sample was realized by raster scanning with XY translation stage (H117 ProScan II, Prior Scientific, Inc.); stage positioning accuracy is up to 50 nm; laser spot size ∼1 μm at 20× magnification. PT thermal-lens effect was assessed by recording intensity modulations in the center of a stabilized continuous wave He-Ne laser beam coaxial to excitation beam (wavelength 633 nm, power 1.4 mW, frequency stabilized mode, model 117A, Spectra-Physics Inc.) by the pinholed photodetector (PDA36A, 40 dB amplification, ThorLabs Inc.). PT signal showed the linear positive asymmetric component associated with fast heating and slower cooling effects and a nonlinear sharp negative peak associated with nano-bubble formation (Fig. S4). PA effect and corresponding ultrasonic waves were detected by ultrasound transducer (model 6528101, 3.5 MHz, 4.5 mm in diameter; Imasonic Inc.) and amplified (preamplifier model 5662B; bandwidth, 50 kHz–5 MHz; gain 54 dB; Panametrics NDT, Olympus NDT Inc.). The PA signal had a classic bipolar shape that transformed into a pulse train due to reflections and diffraction effects (Fig. S6, top right). PC (Dell Precision 690) equipped with a high-speed (200 MHz) analog-to-digital converter board PCI-5124, 12-bit card, 128 MB of memory (National Instruments, Inc.) was used to acquire signals from the transducer, photodiode, and energy meter. Synchronization of the excitation laser, signal acquisition/procession, and control over translation stage was implemented in a single software module (customized LabView 8.5 complex, National Instruments, Inc.). Optical transmission images were obtained by color CCD camera (DP72, Olympus Inc.). The sample preparation and signal processing details are given in SI Text.
Supplementary Material
Acknowledgments.
This work was supported in part by the National Institute of Health Grants R01EB000873, R01CA131164, R01 EB009230, and R21CA139373 (V.P.Z), the National Science Foundation Grant DBI-0852737 (V.P.Z). The financial support from Arkansas Science and Technology Authority (ASTA) Grant 08-CAT-03 (for A.S.B.) and EPSCoR-NSF-P3 Center (Grant P3-202 for M.V.K.) is highly appreciated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008856108/-/DCSupplemental.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE22803).
References
- 1.Zhang L, et al. Nanoparticles in medicine: therapeutic applications and developments. Clin Pharmacol Ther. 2008;83:761–769. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 2.Joseph T, Morrison M. Nanotechnology in agriculture and food. 2006. www.nanoforum.org.
- 3.Kam NWS, O’Connell M, Wisdom JA, Dai H. Carbon nanotubes as multifunctional biological transporters and near-infraredagents for selective cancer cell destruction. Proc Nat Acad Sci USA. 2005;102:11600–11605. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu SB, et al. Sharper and faster “Nano Darts” kill more bacteria: a study of antibacterial activity of individually dispersed pristine single-walled carbon nanotubes. ACS Nano. 2009;3:3891–3902. doi: 10.1021/nn901252r. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, et al. Carbon nanotubes as molecular transporters for walled plant cells. Nano Lett. 2009;9:1007–1010. doi: 10.1021/nl803083u. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, et al. Circulation and long-term fate of functionalized, biocompatible single walled carbon nanotubes in mice probed by Raman spectroscopy. Proc Natl Acad Sci USA. 2008;105:1410–1415. doi: 10.1073/pnas.0707654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polizu S, Savadogo O, Poulin P, Yahia L. Applications of carbon nanotubes- based biomaterials in biomedical nanotechnology. J Nanosci Nanotechno. 2006;6:1883–1904. doi: 10.1166/jnn.2006.197. [DOI] [PubMed] [Google Scholar]
- 8.Harrison B, Atala A. Carbon nanotube applications for tissue engineering. Biomaterials. 2007;28:344–353. doi: 10.1016/j.biomaterials.2006.07.044. [DOI] [PubMed] [Google Scholar]
- 9.Saito N, et al. Carbon nanotubes: biomaterial applications. Chem Soc Rev. 2009;38:1897–1903. doi: 10.1039/b804822n. [DOI] [PubMed] [Google Scholar]
- 10.Stampoulis D, Sinha SK, White JC. Assay-dependent phytotoxicity of nanoparticles to plants. Environ Sci Technol. 2009;43:9473–9479. doi: 10.1021/es901695c. [DOI] [PubMed] [Google Scholar]
- 11.Khodakovskaya M, et al. Carbon nanotubes are able to penetrate plant seed coat and dramatically affect seed germination and plant growth. ACS Nano. 2009;3:3221–3227. doi: 10.1021/nn900887m. [DOI] [PubMed] [Google Scholar]
- 12.Zhu H, Han J, Xiao JQ, Jin Y. Uptake, translocation, and accumulation of manufactured iron oxide nanoparticles by pumpkin plants. J Environ Monitor. 2008;10:713–717. doi: 10.1039/b805998e. [DOI] [PubMed] [Google Scholar]
- 13.Lin S, et al. Uptake, translocation, and transmission of carbon nanomaterials in rice plants. Small. 2009;5:1128–1132. doi: 10.1002/smll.200801556. [DOI] [PubMed] [Google Scholar]
- 14.Singh R, et al. Tissue biodistribution and blood clearance rates of intravenously administered carbon nanotube radiotracers. Proc Natl Acad Sci USA. 2006;103:3357–3362. doi: 10.1073/pnas.0509009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zharov VP, Letokhov VS. Laser optoacoustic spectroscopy. Berlin Heidelberg: Springer- Verlag; 1986. [Google Scholar]
- 16.Wang LV, editor. Photoacoustic imaging and spectroscopy. Boca Raton, FL: CRC; 2009. [Google Scholar]
- 17.Zharov VP, Lapotko DO. Photothermal imaging of nanoparticles and cells. IEEE J Sel Topics Quant. 2005;11:733–751. [Google Scholar]
- 18.Kim J-W, Galanzha EI, Shashkov EV, Moon H-M, Zharov VP. Golden carbon nanotubes as multimodal photoacoustic and photothermal high-contrast molecular agents. Nature Nanotechnol. 2009;4:688–694. doi: 10.1038/nnano.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galanzha EI, et al. In vivo magnetic enrichment and multiplex photoacoustic detection of circulating tumor cells. Nature Nanotechnol. 2009;12:855–860. doi: 10.1038/nnano.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rensink WA, Buell CR. Microarray expression profiling resources for plant genomics. TRENDS Plant Sci. 2005;10:603–609. doi: 10.1016/j.tplants.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Krysan PJ, Jester PJ, Gottward JR, Sussman MR. An Arabidopsis mitogen- activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell. 2002;14:1109–1220. doi: 10.1105/tpc.001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menke FLH, van Pelt JA, Pieterse CMJ, Klessig DF. Silencing of the mitogene-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell. 2004;16:897–907. doi: 10.1105/tpc.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Gonzales-Vigil E, Wilkerson CG, Howe GA. Stability of plant defense proteins in the gut of insect herbivores. Plant Physiol. 2007;143:1954–1967. doi: 10.1104/pp.106.095588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyerman SD, Bohnert HJ, Maurel C, Steudle E, Smith JAC. Plant aquaporins: their molecular biology, biophysics and significance for plant water relations. J Exp Bot. 1999;50:1055–1071. [Google Scholar]
- 25.Heinen RB, Ye Q, Chaumont F. Role aquaporins in leaf physiology. J Exp Bot. 2009;11:2971–2985. doi: 10.1093/jxb/erp171. [DOI] [PubMed] [Google Scholar]
- 26.Werner M, Uehlein N, Proksch P, Kaldenhoff R. Characterization of two tomato aquaporins and expression during incompatible interaction of tomato with the plant parasite Cuscuta reflexa. Planta. 2001;213:550–555. doi: 10.1007/s004250100533. [DOI] [PubMed] [Google Scholar]
- 27.Galanzha EI, et al. In vivo fiber-based photoacoustic detection and photothermal purging of metastasis in sentinel lymph nodes targeted by nanoparticles. J Biophotonics. 2009;2:528–539. doi: 10.1002/jbio.200910046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khodakovskaya M, et al. Increasing inositol (1,4,5)-triphosphate metaboilism affects drought tolerance, carbohydrate metabolism and phosphate-sensitive biomass increases in tomato. Plant Biotechnol J. 2010;8:170–183. doi: 10.1111/j.1467-7652.2009.00472.x. [DOI] [PubMed] [Google Scholar]
- 29.Li J, et al. Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science. 2005;310:121–125. doi: 10.1126/science.1115711. [DOI] [PubMed] [Google Scholar]
- 30.Dervishi E, et al. Morphology of multiwall carbon nanotubes affected by the thermal stability of the catalyst system. Chem Mater. 2007;19:179–184. [Google Scholar]
- 31.Dervishi E, et al. Versatile catalytic system for the large-scale and controlled synthesis of single-wall, double-wall, multiwall, and graphene carbon nanostructures. Chem Mater. 2009;21:5491–5498. [Google Scholar]
- 32.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.