Abstract
The epithelial Na+ channel ENaC is a key player in the maintenance of whole body Na+ balance, and consequently of blood pressure. It is tightly regulated by numerous signaling pathways including ubiquitylation via the ubiquitin-protein ligase Nedd4-2. This mechanism is itself under the control of several kinases, which phosphorylate Nedd4-2, thereby interfering with ENaC/Nedd4-2 interaction, or by Usp2-45, which binds to and deubiquitylates ENaC. Another, different regulatory mechanism concerns the proteolytic activation of ENaC, during which the channel is cleaved on its luminal side by intracellular convertases such as furin, and further activated by extracellular proteases such as CAP-1. This process is regulated as well but the underlying mechanisms are not understood. Previously, evidence was provided that the ubiquitylation status of ENaC may affect the cleavage of the channel. When ubiquitylation of ENaC was reduced, either by co-expressing Usp2–45, or mutating either the ENaC PY-motifs (i.e. the binding sites for Nedd4-2) or intracellular lysines (i.e. ubiquitylation sites), the level of channel cleavage was increased. Here we demonstrate that lysine-mutated ENaC channels are not ubiquitylated at the cell surface, are preferentially cleaved, and Usp2-45 does not affect their cleavage efficiency. We further show by limited proteolysis that the intracellular ubiquitylation status of ENaC affects the extracellular conformation of αENaC, by demonstrating that non-ubiquitylated channels are more efficiently cleaved when treated with extracellularly added trypsin or chymotrypsin. These results present a new paradigm in which an intracellular, post-translational modification (e.g. ubiquitylation) of a transmembrane protein can affect its extracellular conformation.
Keywords: Membrane Proteins, Post-translational Modification, Proteolytic Enzymes, Sodium Channels, Ubiquitin
Introduction
Na+ homeostasis and blood pressure control are tightly linked and primarily regulated in the kidney, as evidenced by the genetic association of mutations in the genes encoding the epithelial Na+ channel (ENaC)2 with early onset of salt-sensitive hypertension in Liddle's syndrome (1, 2). ENaC is composed of three homologous subunits (αβγ), each comprising short cytosolic N and C termini, 2 transmembrane domains, and a large cytoplasmic loop (3); it is expressed in the apical membrane of high resistant epithelia in various tissues including the kidney, colon, and lung (3). ENaC is strongly regulated by numerous hormonally controlled signaling pathways that act via ubiquitylation and proteolytic cleavage. Indeed, in Liddle's syndrome, mutations in the genes encoding β- or γ-ENaC lead to the inactivation of cytosolic PY-motifs that serve as binding sites for the ubiquitin-protein ligase Nedd4-2 (4, 5), suggesting that 1) ENaC is regulated by Nedd4-2-dependent ubiquitylation, and 2) this regulation is impaired in Liddle's syndrome. This concept was confirmed recently in a Nedd4-2 KO model, which displays salt-sensitive hypertension (6), and by a number of studies linking Nedd4-2 polymorphisms with essential hypertension (7–10). Regulation of ENaC by proteolytic cleavage is the other pathway that has drawn a lot of attention in recent years (for a review see Ref. 11). The idea that ENaC is activated by proteases was first proposed by Rossier and co-workers (12) who identified an extracellular channel activating protease (CAP-1 or human prostasin) that enhanced ENaC activity when co-expressed with ENaC in oocytes. Since this pioneering work, and the demonstration that endogenous ENaC subunits are indeed cleaved in a regulated fashion that is stimulated by aldosterone (13), a number of different proteases have been shown to be involved in ENaC regulation, including CAP 1 to 3 (14), plasmin (15, 16), elastase (17), or furin (18), and it was shown that these proteases cleave the α- and γ-subunit. The importance of these proteolytic mechanisms for Na+ transport was underlined by the demonstration that plasmin plays a key role in nephrotic syndrome (15), whereas elastase is relevant in cystic fibrosis (17). Recent data suggests that the two mechanisms (ubiquitylation, proteolysis) are connected to each other. Indeed, Snyder and co-workers (19) reported that ENaC Liddle's syndrome mutations (with channels missing the binding sites for Nedd4-2), are more efficiently cleaved than wild-type channels, and suggested that this depends on the ubiquitylation status of ENaC. Consistently, the balance of ubiquitylation/deubiquitylation (i.e. the ratio of Nedd4-2 to Usp2-45 activity) controls the cleavage of α- and γ-ENaC via 2 different mechanisms (20): 1) Deubiquitylation directly promotes αENaC proteolysis independently of its turnover at the cell surface, whereas 2) γENaC cleavage can be induced by inhibition of endocytosis with dominant-negative dynamin, indicating that prolonged length of time at the cell surface stimulates proteolysis.
The observation that an intracellular process (ubiquitylation/deubiquitylation) controls extracellular events is intriguing and suggests that the modification by ubiquitin, either directly or indirectly via interaction with other proteins, changes the accessibility of the extracellular proteolytic sites. We have investigated this question by applying limited proteolysis on ENaC channels that are either ubiquitylated or deubiquitylated and confirm that deubiquitylation causes a conformational change in ENaC, facilitating its cleavage.
EXPERIMENTAL PROCEDURES
DNA Constructs
All ENaC constructs were based on rat ENaC and most of them described previously (20). Briefly, αENaC-(HA)3 was subcloned into pCMV4 (21), βENaC into pCDNA3, and γENaC into pcDNA3.1(+)Zeo. βENaC was tagged with a C-terminal Myc tag, and γENaC with a C-terminal VSV tag. The ENaC lysine to arginine mutants were generated by PCR as described (22). Furin sites on αENaC (R205A, R231A) or γENaC (R138A) were mutated either in wild-type ENaC or in the lysine mutant. Usp2-45 WT and the inactive mutant (C67A) were generated by PCR as described before and cloned into pcDNA3 (20). The dynamin constructs were previously described (20).
Cell Culture and Transfection
Hek293 (Human Embryonic Kidney) cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% of fetal bovine serum (Invitrogen) and 0.05 units/ml of penicillin/streptomycin (Invitrogen) and incubated at 37 °C/5%CO2. Cells were transiently transfected in 10-cm dishes at 60% confluence, using the Ca2+ phosphate method.
Cell Surface Biotinylation
24 h after transfection, cells were washed twice with cold PBS(1X), then treated 30 min with 4 ml of biotin (0.3 mg/ml) (EZ link Sulfo-NHS-SS-Biotin from Perbio) per 10-cm dish at 4 °C. Cells were washed twice with cold TBS(1×) and lysed in 1 ml/dish lysis buffer (50 mm Hepes pH 7.4, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA pH 8.0, 10% glycerol, 1% Triton X-100, 1.2 mg/ml N-ethylmaleimide and Complete protease inhibitor mixture (1 tablet/20 ml; Roche)). The cells were solubilized 3 h on a wheel at 4 °C and centrifuged 30 min at 20,000 × g at 4 °C. Supernatants were recovered, and protein concentrations were quantified by the Bradford method (Coo protein dosage kit; Interchim). 30 μl of streptavidin Sepharose beads were added to 0.7 to 2 mg of total proteins and incubated overnight on a wheel at 4 °C. A small part of the supernatants were recovered to have the non biotinylated fraction. The beads were washed five times with lysis buffer, and dried beads were resuspended in 30 μl of 100 mm dithiothreitol. They were incubated 1 h at 37 °C with shaking. Then they were centrifuged 2 min at 11′000 × g to recover the supernatant. 5× sample buffer (1.47 m sucrose, 10% SDS, 5 mm EDTA, 300 mm Tris pH 8.8, 0.25% bromphenol blue, and 130 mm dithiothreitol) was added, and the proteins were run on SDS-PAGE, transferred on nitrocellulose, and analyzed by Western blotting. All the experiments were carried out at least twice or three times.
Antibodies/Western Blotting
Anti-αENaC antibodies were raised against the N terminus, and the β,γ antibodies against the C terminus, as described previously (23) and used at a dilution of 1:500. Anti-Usp2-45 antibodies were used as described before at a dilution of 1:500 (20). Anti-actin (Sigma) is used at a dilution of 1:2,000. Secondary antibodies were coupled with horseradish peroxidase (GE Healthcare), and Westerns revealed with ECL (GE Healthcare or Pierce). Anti-ubiquitin antibodies (FK2, Enzo Life Sciences) were used at a dilution of 1:500.
Immunoprecipitations
Cells were washed two times in cold PBS 1×. They were lysed in 1 ml of lysis buffer (50 mm Hepes, 150 mm NaCl, 1 mm EGTA, 10% glycerol, and 1% Triton X-100; containing proteases inhibitors (protease inhibitor mixture complete; Roche), 0.2 mm N-ethylmaleimide (Fluka), 1 mm dithiothreitol (Sigma), and phosphatases inhibitors (100 mm NaF and 10 mm di-Na-pyrophosphate). Cells were vortexed 15 s and then centrifuged 15 min at 20,000 × g. Supernatants were recovered and protein concentrations were quantified by the Bradford method (Coo protein dosage kit; Interchim). Immunoprecipitations were performed with a minimum of 700 μg of total proteins.1:250 of antibody (HA (Santa Cruz Biotechnology) for αENaC, c-Myc (Sigma) for βENaC) were added to the lysate and incubated 2 h on a rotating wheel at 4 °C. Then 20 μl of protein G-Sepharose (GE Healthcare) were added to the mix protein-antibody and incubated in the same conditions over 1 h. Four washes were done with 1 ml of wash buffer (50 mm Hepes pH 7.4, 150 mm NaCl, 1 mm EGTA, 10% glycerol, 0.2% Triton X-100, and 1.5 mm MgCl2), the centrifugations were done at 3,000 × g. Dried beads were resuspended in 40 μl of sample buffer 2×. Then the samples were run on SDS-PAGE, transferred on nitrocellulose, and analyzed by Western blot.
Ubiquitylation of Cell Surface Proteins
Cells were biotinylated as described below. Then they were lysed in 500 μl of lysis buffer (50 mm Hepes, pH 7.5 150 mm NaCl, 1 mm EGTA, 10% glycerol, and 1% Triton X-100 in H2O) containing proteases inhibitors (protease inhibitor mixture (Complete; Roche) and 0.2 mm N-ethylmaleimide (Fluka) in 20 ml of lysis buffer). Cells were vortexed for 15 s and then centrifuged 15 min at 20,000 × g at 4 °C. Then we followed the protocol of immunoprecipitation until the washes (see above). Dried beads were incubated with 100 μl of PBS/1% SDS 15 min at 99 °C under agitation. Supernatants were recovered, and we added 900 μl of PBS and 30 μl streptavidin-Sepharose beads (GE Healthcare). They were incubated 1 h on a rotating wheel at 4 °C. Beads were then washed two times with PBS 1% Triton X-100. Dried beads were resuspended in 30 μl of 2× sample buffer. The samples were run on SDS-PAGE, transferred on nitrocellulose, and analyzed by Western blot using either anti-ubiquitin, or anti ENaC antibodies.
Trypsin and Chymotrypsin Treatment
The cells were treated 24 h after transfection. Cells were washed two times with PBS. 1 μg/ml of trypsin (Sigma) or chymotrypsin (Sigma) was added to the medium (without FBS), and cells were incubated 5min at 37 °C. After extensive washing with PBS, cells were then biotinylated and recovered as described above.
Quantification of Blots
The levels of full-length and cleaved αENaC were detected by Western blot fluorography and quantified on a molecular imager FX. The ratios between cleaved and full-length αENaC were calculated, and values were normalized to the control (ENaC alone) and displayed as mean ± S.E. (n ≥ three independent experiments). Bilateral Student t test for unpaired data were performed to determine statistical significance.
RESULTS
ENaC Gets Preferentially Cleaved in Its Non-ubiquitylated State
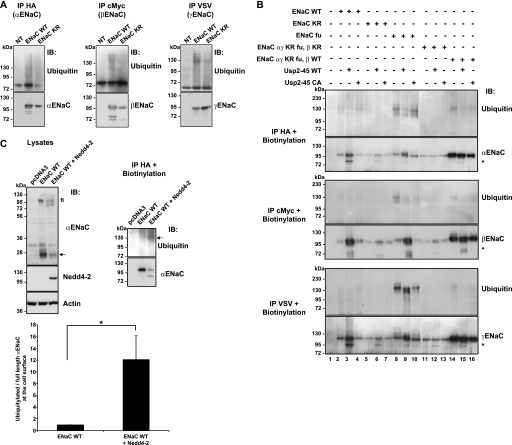
Previous findings suggest that ubiquitylation and deubiquitylation of ENaC (involving Nedd4-2 and/or Usp2-45) affect the proteolytic cleavage of α- and γ-ENaC (19, 20). To obtain additional evidence that it is indeed the ubiquitylation of ENaC, which affects the proteolytic cleavage, we expressed in Hek293 cells either wild-type ENaC, or a previously described ENaC mutant (22), in which all cytoplasmic lysines of the 3 subunits were mutated to arginine (ENaC-KR), hence representing a channel complex that cannot become modified by ubiquitin on these sites. We immunoprecipitated the ENaC subunits individually, and blotted the precipitated material with either anti-ENaC or anti-ubiquitin antibodies (Fig. 1A). As shown previously by many laboratories the wild-type ENaC subunits were poly-ubiquitylated (upper panels). Intriguingly, however, the KR mutant channels still displayed polyubiquitylated chains, albeit weaker. This may be due to associated proteins that are ubiquitylated, or, alternatively, as we are dealing with a powerful overexpression system (Hek293 cells), misfolded or improperly inserted channel subunits, in which lysines of the normally extracellular loop are exposed to the cytosolic side. To study only properly assembled channels, we decided to monitor ubiquitylation of ENaC at the cells surface. We surface biotinylated cells 24 h after transfection, lysed them, and recovered the ENaC subunits by immunoprecipitation using either anti-HA (αENaC), anti-c-Myc (βENaC), or anti-VSV (γENaC) antibodies. The recovered subunits were then precipitated with streptavidin-Sepharose, which binds the cell surface biotinylated ENaC pool and analyzed this material by SDS-PAGE/Western blotting using either anti-ubiquitin or anti-ENaC subunit antibodies (Fig. 1B). As shown previously (20), cell surface wild-type ENaC subunits (lanes 2–4) detected by anti-ubiquitin antibodies (upper panels) migrated as several discrete bands at an apparent molecular mass of ∼130 kDa, whereas the non-modified forms (recognized by the ENaC antibodies), migrated at a lower apparent molecular weight (95 to 110 kDa). The lysine-free ENaC subunits (lanes 5–7 or 11–13) were observable in the ENaC blots (lower panels), but hardly in the ubiquitin blots, demonstrating the importance of these lysines for ubiquitylation of the channel at the cell surface. A weak signal was observable for αENaC-KR mutant (Fig. 1B, lane 5). Currently, we do not know its nature but it may be caused by the polyubiquitylation seen in Fig. 1A. In order to provide further evidence that αENaC is ubiquitylated at the cell surface, we co-expressed αβγENaC with the ubiquitin-protein ligase Nedd4-2, and observed that the relative ubiquitylation level of αENaC (ubiquitylated to full-length αENaC) was increased more than 10-fold when Nedd4-2 was co-expressed (Fig. 1C), supporting the notion that this subunit is ubiquitylated at the cell surface. We noticed that ENaC subunits mutated on the furin sites described previously (18, 20) were stronger ubiquitylated (lanes 8–10) than wild-type channels. We also observed that ENaC channels composed of lysine-free αγ combined with wt β (lanes 14–16) were stronger expressed at the cell surface than wild-type channels or channels mutated on the lysines of all three subunits (lanes 11–13). This suggests that 1) ubiquitylation of α and γENaC has an impact on ENaC cell surface expression, and 2) some of the cytoplasmic lysine residues in β are important for proper assembly and export of the channel to the plasma membrane consistent with our previous findings (22, 24).
FIGURE 1.
ENaC ubiquitylation in whole cell lysates and at the cell surface. A, Hek293 cells were transiently transfected with either wild-type or KR mutant ENaC channels. The ubiquitylated and the total amount of ENaC expressed in Hek293 cells were visualized by Western blotting of immunoprecipitated α-(HA), β-(c-Myc), and γ-(VSV)-ENaC, with either anti ubiquitin or anti ENaC antibodies. B, cells were biotinylated, lysed and then immunoprecipitated with HA tag (for α), c-Myc tag (for β), and VSV tag (for γ). The channels present at the cells surface were recovered using streptavidin-Sepharose beads. Biotinylated proteins were analyzed by SDS-PAGE/Western blotting as indicated. ENaC KR: K to R mutation on all cytoplasmic lysines. fu: mutation on furin sites. The nature of a fragment seen occasionally at ∼70 kDa in the ENaC blots is not known (*). C, Hek293 cells were transiently transfected with ENaC WT +/− Nedd4-2. Cells were biotinylated 24 h after transfection. Then they were lysed and an immunoprecipitation was performed with anti-HA to recover αENaC. The cell surface channels were recovered using streptavidin beads. Then proteins were run on SDS/PAGE and Western blot were performed as describe. Quantification of three individual experiments of the ratio ubiquitylated/full-length αENaC was calculated. They were normalized to αENaC full-length and displayed as mean ± S.E. (n = 3 experiments, *, p < 0.05). All the blots are representative of three independent experiments.
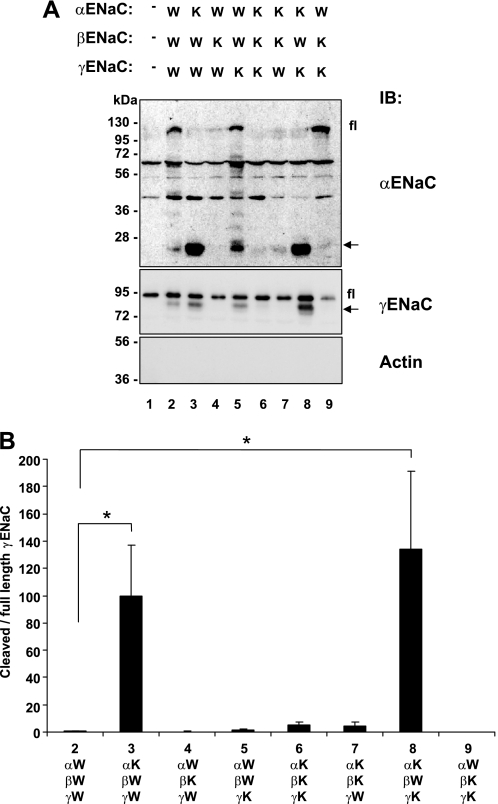
Having established that the lysine-free channels were not ubiquitylated at the cell surface, we followed the cleavage of ENaC in either wild-type channels or channels comprising various combinations of lysine-free and wild-type subunits at the cell surface (Fig. 2; W: wild-type; K: lysine mutated). As shown in lane 1, occasionally endogenous crossreacting bands were detected with the ENaC antibodies, which represent endogenous proteins unrelated to ENaC (20). We confirmed previous findings showing that wild-type ENaC was cleaved on the α- and γ-subunit (Fig. 2, lane 2). Importantly, when α-ENaC was mutated, the cleavage of α and γ was increased, (Fig. 2A, conditions 3 and 8 and quantification in Fig. 2B of the ratio of cleaved to full-length αENaC), whereas mutation of the other subunits had no effect. This supports the idea that 1) it is direct ubiquitylation of αENaC that controls the process and 2) the α subunit needs to be in a non-ubiquitylated state to get efficient ENaC cleavage by endogenous convertases such as furin.
FIGURE 2.
Proteolytic cleavage of wild-type and mutant channels mutated on cytoplasmic lysines. A, Hek293 cells were transiently transfected with either wild-type (W) or cytoplasmic lysine mutant (K) ENaC. 24 h after transfection, cells were biotinylated, recovered with streptavidin-Sepharose and analyzed by SDS-PAGE/Western blotting using anti α-, or γ-ENaC antibodies as indicated. fl: full-length α- or γ-ENaC; arrow: cleaved α- or γ-ENaC. α- and γENaC antibodies crossreact with endogenous proteins, as shown in lane 1. In our previous work we have provided evidence that these do not represent endogenous ENaC. B, quantification of the ratio of cleaved to full-length αENaC (as described under “Experimental Procedures”), normalized to wild-type ENaC (condition 2), and displayed as mean ± S.E. (n = 3 experiments; *, p < 0.05 versus wt ENaC (2) as determined by Student's t test).
Usp2–45 Does Not Affect the Cleavage of Lysine Channel Mutants
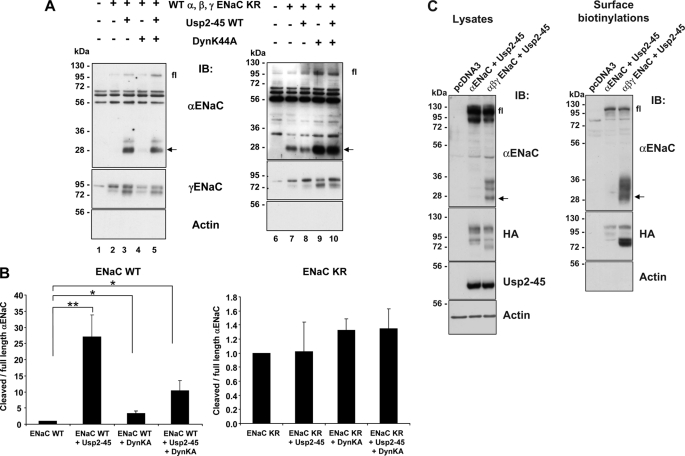
If ENaC subunits are in a non-ubiquitylated state, Usp2-45 should not affect their cleavage. We confirmed this assumption by expressing either wild-type or lysine-free ENaC in Hek293 cells together with either Usp2–45, or dominant-negative dynamin (DynK44A) and followed α- and γENaC in the biotinylated fraction. Similarly to what was described previously (20), both Usp2-45 and DynK44A increased the cleavage of wild-type γ-ENaC at the cell surface to a similar extent (Fig. 3, compare lanes 3, 4, or 5 with lane 2), whereas the cleavage of αENaC was strongly promoted by Usp2-45 (appearance of a 28 kDa and a weaker ∼32-kDa fragment) and hardly by DynK44A (lanes 3 and 5 and quantification in Fig. 3B). The lysine-free ENaC mutant behaved differently as both α- and γENaC-subunits were cleaved at the plasma membrane in the absence of Usp2-45, and co-expression of Usp2-45 did not affect cleavage (Fig. 3A, compare lanes 7 and 8 and Fig. 3B). Interestingly, when expressing ENaC-KR, only a 28-kDa fragment was seen. Interfering with clathrin-mediated endocytosis by DynK44A increased the density of cleaved ENaC fragments at the plasma membrane without significantly changing the ratio of cleaved to full-length αENaC (lanes 9 and 10 and Fig. 3B). If ubiquitylation is impaired or reduced by either mutating the cytoplasmic lysines, or by co-expressing Usp2-45, one could also expect that the ER-associated degradation is impaired and that more non-assembled channels may escape to the plasma membrane, hence subunits that may be more susceptible for cleavage. We therefore expressed αENaC alone, or αβγENaC, together with or without Usp2–45 (Fig. 3C). Western blotting of the biotinylated fraction revealed that when αENaC was not assembled with the other subunits, it was less cleaved than the fully assembled channel, suggesting that the assembled channel is more prone for such proteolysis. Our findings therefore further support the idea that αENaC cleavage is efficient only if the channel is deubiquitylated, and that Usp2-45 directly acts on ubiquitylated ENaC channels.
FIGURE 3.
Effect of Usp2-45 and DynK44A on proteolytic cleavage of either wild-type or KR channel. A, Hek293 cells were transiently transfected with either wild-type or cytoplasmic lysine mutant ENaC together with Usp2-45, with dominant negative dynamin (DynK44A) or both. 24 h after transfection, cells were biotinylated and biotinylated proteins analyzed as in Fig. 2 with either anti α-, γENaC, or anti-actin antibodies. Fl: full-length αENaC; arrow: cleaved αENaC. B, full-length αENaC and its cleaved fragment were quantified and the ratio of cleaved to full-length is displayed (n = 3 experiments; *, p < 0.05; **, p < 0.01 versus wt ENaC). C, Hek293 cells were transiently transfected with αENaC WT + Usp2-45 or αβγENaC WT + Usp2-45. 24 h after transfection, cells were biotinylated then lysed. Surface proteins were recovered, run on SDS/PAGE, and analyzed by Western blot as indicated (n = 3).
αENaC Undergoes Conformational Changes upon Ubiquitylation or Deubiquitylation
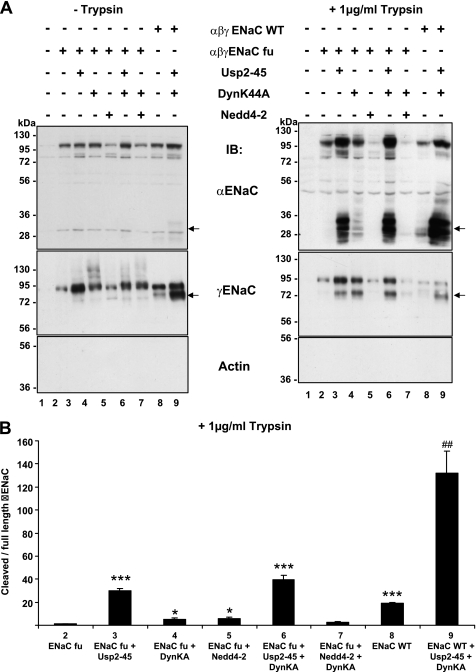
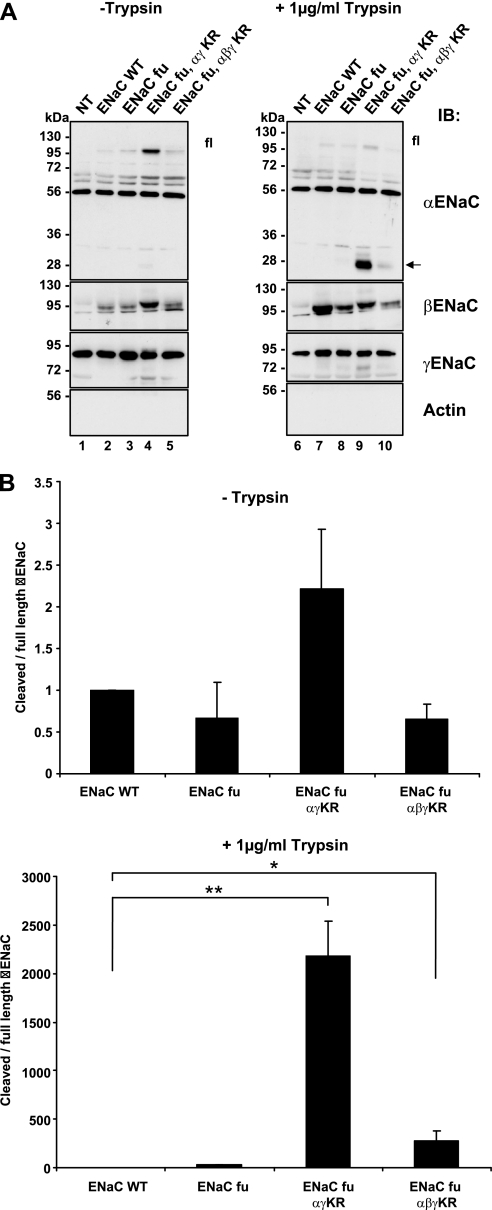
Assembled ENaC at the plasma membrane is ubiquitylated at the cytosolic side (Fig. 1B), whereas proteolytic cleavage takes place on the extracellular, or luminal side. Consequently, deubiquitylation may control ENaC cleavage either by prolonging the time period that the channel remains at the cell surface, or alternatively, by inducing a conformational change that exposes the proteolytic site(s). Our previous findings suggest that cleavage of γENaC by endogenous proteases is primarily controlled by the turnover of the channel at the cell surface, whereas αENaC is regulated by other means (20). To detect possible conformational changes we decided to carry out experiments of limited proteolysis using extracellularly added trypsin. With such a classical approach we expected to detect different cleavage patterns if conformational changes at the extracellular side take place. Importantly, the short addition (5 min) of a protease to the extracellular side will provide a snapshot of the channels at the cell surface and not be affected by exocytosis or endocytosis rates of the channel. Because wild-type ENaC undergoes cleavage by endogenous convertases such as furin (20), we used furin-site mutants of α- and γ-ENaC that were described previously (18). These mutant channels were not or hardly cleaved on α- or γ-ENaC by endogenous proteases as shown in Fig. 4 (left, lanes 2–7, α- or γ-ENaC, arrow). As comparison we loaded wt channels that were strongly cleaved as shown above (lanes 8–9). When the cells were treated with limiting amounts of trypsin (1 μg/ml, 5 min, 37 °C), hardly any cleavage product of α- or γENaC was seen (right panel, lane 2, and quantification in B). When Usp2–45 was co-expressed, both α- and γ-ENaC were cleaved by trypsin, as evidenced by the presence of 2 major fragments of 28 and 32 kDa for α, and an enhanced 75 kDa product for γENaC (Fig. 4, right side). Importantly, the ratio of cleaved to full-length αENaC was strongly increased, suggesting a conformational change when Usp2–45 was co-expressed (Fig. 4B). Interfering with endocytosis with DynK44A did only very faintly reveal the cleaved bands, corroborating the idea that the effect of Usp2-45 on αENaC was not caused by inhibition of endocytosis, but by another process, possibly a conformational change. To confirm that it was the ubiquitylation state of ENaC that was affected by Usp2-45, we expressed lysine-free channels, which comprised also the furin site mutations, and carried out another set of limited proteolysis experiments with trypsin (Fig. 5). Wild-type, furin sites mutant (fu), furin-sites (fu) + αγ KR mutant, and furin sites (fu) + αβγ KR ENaC mutant ENaC channels were expressed. In lanes 1–5 (biotinylated fraction, no trypsin addition) hardly any proteolytic fragment either for α or γENaC was observed. When trypsin was added extracellularly (lanes 6–10), proteolytic fragments were hardly visible in the wild-type channel (arrow). When KR mutated ENaC subunits were expressed the cleavage of αENaC was strongly enhanced (appearance of only the 28-kDa fragment) as compared with the furin mutant (compare lanes 9 and 10 with 8, arrow and quantification in B), and also cleavage of γENaC was stronger when α and γ were KR mutated (compare lanes 8 and 9). Quantification of the ratio between cleaved and full-length αENaC confirmed the very strong increase in cleavage of the αENaC KR mutants (Fig. 5B). These findings suggest that non-ubiquitylated ENaC is prone to be cleaved by limiting amounts of trypsin, likely due to a conformational change.
FIGURE 4.
Limited proteolysis using trypsin on ENaC channels co-expressed with Usp2-45 and/or DynK44A. A, Hek293 cells were transiently transfected with either wild-type or furin mutant (fu) ENaC together with Usp2-45 WT or with DynK44A, or with Nedd4-2 WT. 24 h after transfection, cells were treated with 1 μg/ml trypsin for 5 min at 37 °C, and biotinylation was performed to recover proteins present at the cell surface. Recovered proteins were run on a SDS/PAGE and analyzed by Western blot as indicated. B, in 4 individual experiments the cleaved 30-kDa α fragment and full-length αENaC was quantified. The ratio of cleaved to full-length α is displayed. *, p < 0.05 versus ENaC fu; ***, p < 0.001, ##, p < 0.01 versus wt ENaC (8) (n = 4).
FIGURE 5.
Limited proteolysis using trypsin on wild-type or lysine-mutated ENaC channels. A, Hek293 cells were transiently transfected with wild-type ENaC (WT), or on ENaC mutated on the furin sites (fu), or on ENaC mutated both on the furin sites and on the cytoplasmic lysines (fu, αγKR, αβγKR). 24 h after transfection, cells were treated or not with 1 μg/ml trypsin, and then biotinylated. Biotinylated proteins were recovered and analyzed on SDS/PAGE/Western blotting as indicated. B, quantification as described in Figs. 2, 3, and 4 (n = 4).
Sensitivity of ENaC to Both Trypsin and Chymotrypsin Cleavage Depends on the Ubiquitylation Status of ENaC
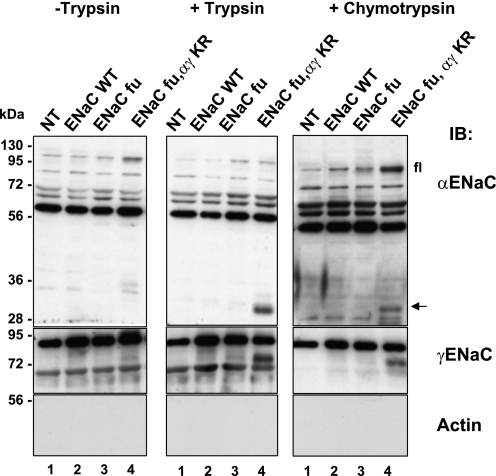
To acquire further evidence that there may be a conformational change on the extracellular side of ENaC, we carried out similar proteolysis experiments using chymotrypsin. Chymotrypsin has been shown to stimulate ENaC activity as well (25, 26). In Fig. 6, extracellularly added chymotrypsin, as well as trypsin, cleaved preferentially ENaC containing the KR mutations on α and γENaC (compare lane 4 with the other lanes). Hence, our results suggest that ubiquitylation or deubiquitylation of ENaC influence the efficiency by which either endogenous convertases such as furin, or extracellularly added proteases such as trypsin or chymotrypsin cleave proteolytically the epithelial Na+ channel ENaC.
FIGURE 6.
Non-ubiquitylated ENaC channels are more efficiently cleaved by exogenously added trypsin and/or chymotrypsin. Hek293 cells were transiently transfected with ENaC WT or fu mutant or fu/αγKR mutant. 24 h after transfection, cells were treated with 1 μg/ml trypsin or with 1 μg/ml chymotrypsin, and then biotinylated. Proteins were run on a SDS/PAGE and analyzed by Western blot as indicated in the previous figures. Shown are representative blots of two individual experiments.
DISCUSSION
Data presented in this work show that the ubiquitylation status of ENaC influences the sensitivity to proteolysis of α and γ ENaC subunits on their extracellular loop. For αENaC, the data are most compatible with a model in which either ubiquitylation or deubiquitylation induce a change of conformation, rendering proteolytic sites inassessable or assessable to relevant proteases. To our knowledge this is the first demonstration that ubiquitylation regulates the conformation of a transmembrane protein across the membrane.
These conclusions are based on the following observations: 1) When Usp2–45 is co-expressed with ENaC, the channel becomes deubiquitylated, and preferentially cleaved on α and γENaC subunits by endogenous proteases/convertases, likely by furin (Fig. 3 and Ref. 20). This cleavage requires the presence (and likely the assembly) of the three subunits, as αENaC alone is not cleaved. 2) Ubiquitylation-deficient ENaC mutants missing all the cytoplasmic lysines are constitutively cleaved (particularly when mutated on αENaC), and the cleavage is not further affected by Usp2-45, demonstrating that this enzyme controls ENaC cleavage by acting directly on the channel (Figs. 2 and 3). 3) Further in support are previous published data showing that Liddle's channels, missing bindings sites for the ubiquitin-protein ligase Nedd4-2, are preferentially cleaved (19). 4) Channels that are not ubiquitylated, either because they are devoid of cytosolic lysines or because they are deubiquitylated by Usp2-45, show different proteolysis pattern by limited treatment with 2 different extracellularly added proteases (trypsin or chymotrypsin) (Figs. 5 and 6). These treatments are short (5 min); therefore these experiments likely show the steady state level of proteolytically accessible channels at the cell surface. Hence, changes in the ratios of cleaved fragments to uncleaved, full-length channels cannot be explained by alterations in expression of the channels (or their proteolytic fragments) at the cell surface, and therefore likely represent changes in the conformation of the channel. However, eliminating the ubiquitylation sites by mutating cytoplasmic lysines to arginines, or deubiquitylating ENaC subunits by Usp2-45, may affect quality control in the ER and consequently ERAD (ER-associated degradation). We can therefore not firmly exclude that under such conditions αβγ channels form, which are not properly folded in the protease-sensitive region and escape to the plasma membrane, where they may exhibit enhanced sensitivity to proteases such as trypsin. Future studies will aim to address this issue in further detail.
At this stage, we can only speculate on how the ubiquitin modification of ENaC affects the extracellular conformation of ENaC. Judged from the molecular weight shift from non-ubiquitylated (lower panels in Fig. 1) to ubiquitylated (upper panels in Fig. 1) ENaC subunits, which is ∼30 to 40 kDa, we estimate that there are 4 to 5 ubiquitins of ∼8 kDa, which are attached to each subunit, representing an increase in molecular weight of roughly 30%. These modifications may represent either multiple mono-ubiquitylations or short poly-ubiquitylated chains (as is often observed for membrane proteins) (27). These bulky modifications occur on the short N termini (24) and likely change their conformational arrangement sufficiently to affect the whole transmembrane channel complex (including the extracellular loops). Moreover, it is well recognized that ubiquitin acts as a module that binds to numerous cellular proteins. The number of known ubiquitin-binding proteins is constantly growing and estimated to be larger than 150 (28). Such proteins interact via so-called ubiquitin-binding motifs, or domains (more than 20 different ones have been identified so far), and translate an ubiquitin signal into a cellular response. It is likely that the attached ubiquitins on ENaC bind to other cellular proteins, which may influence the conformation of the channel complex. Although ubiquitylation is known to lead to the formation of both intra- or intermolecular interactions in cytoplasmic proteins (29), the case of ENaC is, to our knowledge, the first one to demonstrate that ubiquitylation changes the conformation at the extracellular side of a transmembrane protein complex.
With respect to the conformational change on the extracellular loops of ENaC, it is currently difficult to predict its nature. We have relatively sparse information on the conformation of the extracellular loops of ENaC, although the structure of a closely related ion channel (ASIC) has been resolved recently (30). It will be necessary to solve the structure of active and inactive ENaC, or ubiquitylated and non-ubiquitylated channels, to understand the nature of the changes that occur within ENaC.
We know that ENaC can be cleaved on “furin-sites” that have been defined by Hughey and collaborators on the extracellular loops of α- and γ-ENaC (18), and which correspond to a consensus of RXXR. There are 2 such sites on αENaC, and one on γENaC. In the region around these cleavage sites are numerous additional positively charged amino acids (lysines or arginines), which may serve as potential cleavage sites for trypsin; there are also many large aliphatic residues (phenylalanines, tyrosines, or tryptophans), which represent putative cleavage sites for chymotrypsin. Consequently, this is likely the region which is affected by the ubiquitylation/deubiquitylation of the complex, thereby masking or unmasking the proteolytic sites. To get further insides into these conformational arrangements we will need a better understanding of the structure of ENaC.
Although it is now well established that ENaC is activated by proteolysis, we still do not know how this process is regulated (for a review see Ref. 11). It has been shown over a decade ago, that low Na+ diet or aldosterone promotes the cleavage of γENaC (13), an observation that was further supported by Ergonul et al. (31), who demonstrated that a 7–9-day-long low Na+ diet increased the cleavage of α- and γ-ENaC. Recently it was shown that several mechanisms could be envisioned to explain the phenomena. It is possible that aldosterone stimulates the expression of a protease, which may then cleave ENaC, or alternatively, that endogenous protease inhibitors are suppressed by aldosterone. Such a mechanism has been proposed by Wakida et al. (32). Alternatively, the link to ubiquitylation described in the present work may be important as well, and there is considerable, albeit indirect, evidence in favor of such a regulatory pathway. ENaC channels containing Liddle's mutations, and hence lacking the binding sites for Nedd4-2 have been shown to be preferentially cleaved (19) and to display increased open probability Po (33), compatible with the idea that cleavage of ENaC increases Po (12). Aldosterone induces the expression of GILZ, Sgk1, and Usp2–45, which all are involved in the suppression of Nedd4-2-dependent ENaC ubiquitylation (22, 34, 35); accordingly they are also expected to induce ENaC cleavage, as shown for Usp2–45 in this paper. Indeed, Fejes-Toth et al. (36) presented recently evidence that the proteolytic processing of γENaC is reduced in mice lacking Sgk1. On the other hand the in vivo role of GILZ or Usp2-45 in ENaC regulation and processing has not been studied so far, nor has proteolytic processing of ENaC been observed in Nedd4-2 KO mice (6). Importantly, the relevance of ENaC proteolysis in regulation and activity of ENaC, as it is seen in vivo in the nephron, remains to be established, as there is not always a strict correlation between cleavage and function of ENaC (36).
In conclusion we have shown that the ubiquitylation status of ENaC plays an important role for the susceptibility of ENaC to luminal proteolysis, either by intracellular convertases such as furin, but also by extracellularly added proteases such as trypsin or chymotrypsin. The most likely explanation for these findings is that addition (or removal of ubiquitin) from the ENaC subunits causes important conformational changes that either expose (or mask) the proteolytic sites present on the loops of either α- and/or γENaC.
This work was supported by the Swiss National Science Foundation (31003A_125422/1), by the Leducq Foundation (Transatlantic Network on Hypertension), and by the Swiss Kidney Foundation.
- ENaC
- epithelial Na+ channel
- KO
- knockout.
REFERENCES
- 1. Shimkets R. A., Warnock D. G., Bositis C. M., Nelson-Williams C., Hansson J. H., Schambelan M., Gill J. R., Jr., Ulick S., Milora R. V., Findling J. W. (1994) Cell 79, 407–414 [DOI] [PubMed] [Google Scholar]
- 2. Hansson J. H., Nelson-Williams C., Suzuki H., Schild L., Shimkets R., Lu Y., Canessa C., Iwasaki T., Rossier B., Lifton R. P. (1995) Nat. Genet. 11, 76–82 [DOI] [PubMed] [Google Scholar]
- 3. Kellenberger S., Schild L. (2002) Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 4. Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., Rotin D. (1996) EMBO J. 15, 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- 5. Kamynina E., Debonneville C., Bens M., Vandewalle A., Staub O. (2001) FASEB J. 15, 204–214 [DOI] [PubMed] [Google Scholar]
- 6. Shi P. P., Cao X. R., Sweezer E. M., Kinney T. S., Williams N. R., Husted R. F., Nair R., Weiss R. M., Williamson R. A., Sigmund C. D., Snyder P. M., Staub O., Stokes J. B., Yang B. (2008) Am. J. Physiol. Renal Physiol 295, F462–F470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Russo C. J., Melista E., Cui J., DeStefano A. L., Bakris G. L., Manolis A. J., Gavras H., Baldwin C. T. (2005) Hypertension 46, 488–491 [DOI] [PubMed] [Google Scholar]
- 8. Fava C., von Wowern F., Berglund G., Carlson J., Hedblad B., Rosberg L., Burri P., Almgren P., Melander O. (2006) Kidney Int. 70, 562–569 [DOI] [PubMed] [Google Scholar]
- 9. Dahlberg J., Nilsson L. O., von Wowern F., Melander O. (2007) PLoS ONE 2, e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wen H., Lin R., Jiao Y., Wang F., Wang S., Lu D., Qian J., Jin L., Wang X. (2008) Clin. Exp. Hypertens. 30, 87–94 [DOI] [PubMed] [Google Scholar]
- 11. Rossier B. C., Stutts M. J. (2009) Annu. Rev. Physiol. 71, 361–379 [DOI] [PubMed] [Google Scholar]
- 12. Vallet V., Chraibi A., Gaeggeler H. P., Horisberger J. D., Rossier B. C. (1997) Nature 389, 607–610 [DOI] [PubMed] [Google Scholar]
- 13. Masilamani S., Kim G. H., Mitchell C., Wade J. B., Knepper M. A. (1999) J. Clin. Invest. 104, R19–R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vuagniaux G., Vallet V., Jaeger N. F., Hummler E., Rossier B. C. (2002) J. Gen. Physiol. 120, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Svenningsen P., Bistrup C., Friis U. G., Bertog M., Haerteis S., Krueger B., Stubbe J., Jensen O. N., Thiesson H. C., Uhrenholt T. R., Jespersen B., Jensen B. L., Korbmacher C., Skøtt O. (2009) J. Am. Soc. Nephrol. 20, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Passero C. J., Mueller G. M., Rondon-Berrios H., Tofovic S. P., Hughey R. P., Kleyman T. R. (2008) J. Biol. Chem. 283, 36586–36591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris M., Firsov D., Vuagniaux G., Stutts M. J., Rossier B. C. (2007) J. Biol. Chem. 282, 58–64 [DOI] [PubMed] [Google Scholar]
- 18. Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., Kleyman T. R. (2004) J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 19. Knight K. K., Olson D. R., Zhou R., Snyder P. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2805–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruffieux-Daidié D., Poirot O., Boulkroun S., Verrey F., Kellenberger S., Staub O. (2008) J. Am. Soc. Nephrol. 19, 2170–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanswell A. K., Staub O., Isles R., Belcastro R., Cabacungan J., Sedlackova L., Steer B., Wen Y., Hu J., O'Brodovich H. (1998) Am. J. Physiol. 275, L452–L460 [DOI] [PubMed] [Google Scholar]
- 22. Fakitsas P., Adam G., Daidié D., van Bemmelen M. X., Fouladkou F., Patrignani A., Wagner U., Warth R., Camargo S. M., Staub O., Verrey F. (2007) J. Am. Soc. Nephrol. 18, 1084–1092 [DOI] [PubMed] [Google Scholar]
- 23. Duc C., Farman N., Canessa C. M., Bonvalet J. P., Rossier B. C. (1994) J. Cell Biol. 127, 1907–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., Rotin D. (1997) EMBO J. 16, 6325–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chraïbi A., Vallet V., Firsov D., Hess S. K., Horisberger J. D. (1998) J. Gen. Physiol. 111, 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diakov A., Bera K., Mokrushina M., Krueger B., Korbmacher C. (2008) J. Physiol. 586, 4587–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galan J. M., Haguenauer-Tsapis R. (1997) EMBO J. 16, 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dikic I., Wakatsuki S., Walters K. J. (2009) Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Woelk T., Oldrini B., Maspero E., Confalonieri S., Cavallaro E., Di Fiore P. P., Polo S. (2006) Nat. Cell Biol. 8, 1246–1254 [DOI] [PubMed] [Google Scholar]
- 30. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 31. Ergonul Z., Frindt G., Palmer L. G. (2006) Am. J. Physiol. Renal Physiol. 291, F683–693 [DOI] [PubMed] [Google Scholar]
- 32. Wakida N., Kitamura K., Tuyen D. G., Maekawa A., Miyoshi T., Adachi M., Shiraishi N., Ko T., Ha V., Nonoguchi H., Tomita K. (2006) Kidney Int. 70, 1432–1438 [DOI] [PubMed] [Google Scholar]
- 33. Firsov D., Schild L., Gautschi I., Mérillat A. M., Schneeberger E., Rossier B. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soundararajan R., Melters D., Shih I. C., Wang J., Pearce D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7804–7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Debonneville C., Flores S. Y., Kamynina E., Plant P. J., Tauxe C., Thomas M. A., Münster C., Chraïbi A., Pratt J. H., Horisberger J. D., Pearce D., Loffing J., Staub O. (2001) EMBO J. 20, 7052–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fejes-Tóth G., Frindt G., Náray-Fejes-Tóth A., Palmer L. G. (2008) Am. J. Physiol. Renal Physiol. 294, F1298–1305 [DOI] [PubMed] [Google Scholar]