Abstract
The SLC36 family of transporters consists of four genes, two of which, SLC36A1 and SLC36A2, have been demonstrated to code for human proton-coupled amino acid transporters or hPATs. Here we report the characterization of the fourth member of the family, SLC36A4 or hPAT4, which when expressed in Xenopus laevis oocytes also encodes a plasma membrane amino acid transporter, but one that is not proton-coupled and has a very high substrate affinity for the amino acids proline and tryptophan. hPAT4 in Xenopus oocytes mediated sodium-independent, electroneutral uptake of [3H]proline, with the highest rate of uptake when the uptake medium pH was 7.4 and an affinity of 3.13 μm. Tryptophan was also an excellently transported substrate with a similarly high affinity (1.72 μm). Other amino acids that inhibited [3H]proline were isoleucine (Ki 0.23 mm), glutamine (0.43 mm), methionine (0.44 mm), and alanine (1.48 mm), and with lower affinity, glycine, threonine, and cysteine (Ki >5 mm for all). Of the amino acids directly tested for transport, only proline, tryptophan, and alanine showed significant uptake, whereas glycine and cysteine did not. Of the non-proteogenic amino acids and drugs tested, only sarcosine produced inhibition (Ki 1.09 mm), whereas γ-aminobutyric acid (GABA), β-alanine, l-Dopa, d-serine, and δ-aminolevulinic acid were without effect on [3H]proline uptake. This characterization of hPAT4 as a very high affinity/low capacity non-proton-coupled amino acid transporter raises questions about its physiological role, especially as the transport characteristics of hPAT4 are very similar to the Drosophila orthologue PATH, an amino acid “transceptor” that plays a role in nutrient sensing.
Keywords: Amino Acid Transport, Membrane Proteins, Plasma Membrane, Transport Amino Acids, Xenopus, SLC36, Membrane Transport, Xenopus Oocyte Expression
Introduction
The solute-linked carriers (SLCs)2 consist of 48 families containing over 300 genes that encode transporters (1). Members present within the same SLC family have at least 20% identity to other members within the same family. One of these transporter families is the SLC36 family, which contains the proton-coupled amino acid transporters (PATs) (2). The human SLC36 family consists of four members: SLC36A1 to SLC36A4 (hPAT1 to hPAT4, respectively). SLC36A1 to SLC36A3 are located on the chromosome 5q33.1, whereas SLC36A4 is present on 11q14.3. These genes encode proteins containing about 500 amino acids, with membrane prediction software predicting that the hPAT proteins contain 9–11 transmembrane domains. In addition to their characterized physiological function as amino acid transporters (Table 1), hPAT1 and hPAT2 are also thought to play a role in pathophysiology and pharmacology.
TABLE 1.
Comparison of transport properties of hPAT1, hPAT2, and hPAT4
ND, no data available.
| PAT1 | hPAT2 | hPAT4 | |
|---|---|---|---|
| Proline affinity | 2.0 ± 0.6 mma | 0.25 ± 0.09 mmb | 3.13 ± 1.13 μm |
| 0.17 ± 0.42 mmc | |||
| Proline transport | Yes | Yes | Yes |
| Alanine affinity | 7.5 ± 0.6 mma | 0.25 ± 0.05 mmb | 1.48 ± 0.79 mm |
| 2.3 ± 0.2 mmd | 0.24 mmc | ||
| Alanine transport | Yes | Yes | Yes |
| Glycine affinity | 7.0 ± 0.7 mma | 0.59 ± 0.04 mmb | >5 mm |
| 2.3 ± 0.2 mmd | |||
| Glycine transport | Yes | Yes | No |
| Tryptophan affinity | No inhibitiond | ND | 1.72 ± 1.02 μm |
| 4.7 ± 0.3 mme | |||
| Tryptophan transport | Noa,e | Noa | Yes |
Both hPAT1 and hPAT2 have been considered as important candidate genes that could be responsible for the hereditary disease iminoglycinuria where the defect observed is in the absorption of proline and glycine. Although it was originally suggested that hPAT1 might be the likely transporter involved in iminoglycinuria (3), more recent genetic studies have implicated hPAT2 (SLC36A2) as the major gene associated with the disease (4).
hPAT1 and hPAT2 are also considered to play a significant role in the absorption of certain drugs. The pharmaceutically active amino acid derivatives that PAT1 can transport include the orally delivered antibiotic and antischizophrenic agent cycloserine (5), the orally active antihyperglycemic drug β-guanidinopropionic acid (6), the experimental anti-insomnia compound gaboxadol (7), the anti-epileptic vigabatrin (8), and the photodynamic therapy agent 5-aminolevulinic acid (9, 10). PAT2-transported drugs include a number of neuroactive compounds, including cycloserine (an antischizophrenic), l-2-azetidine-carboxylate, pipecolic acid, nipecotic acid, and isonipecotic acid (11).
PAT3 and PAT4 are so named purely because they are the third and fourth members of the SLC36 family, respectively, and are considered as “orphan” transporters, i.e. their substrates are unknown. PAT3 was first cloned from mice (12), and mRNA expression was found to be very high in the testis and negligible elsewhere (13). PAT4 was first identified based on sequence similarity to the remaining members of the SLC36 family and seems to have a more ubiquitous expression (14). Here we describe the results of experiments that characterize the function of hPAT4 (SLC36A4) when heterologously expressed in Xenopus laevis oocytes and show for the first time that it is an amino acid transporter with a very high affinity for proline and tryptophan.
EXPERIMENTAL PROCEDURES
All chemicals were purchased from Sigma, Poole, UK, unless otherwise stated. All experiments were performed at room temperature.
Subcloning of the hPAT4 Gene into the Xenopus Oocyte-optimized Vector
The hPAT4 clone was purchased from Geneservice Ltd. (Cambridge, UK, clone ID 5313230) in the pBluescriptR vector. hPAT4 was subcloned into the oocyte expression vector pXT7 (15) via the pGEMT vector system (Promega) with the restriction sites KpnI and EagI added by PCR using the following primers (bold underlined bases indicate the restriction sites): forward, 5′-GCTGGTACCGGCGGCGTCCTGGAGACC-3′, and reverse, 5′-GCCGGCCGGAAGACTCATGATTCTGCTTTTACTATTTCAAACCAG-3′. hPAT1 (clone ID 3307964) and hPAT2 (clone ID 40010274) were also purchased from Geneservice and cloned into the pXT7 vector using a similar strategy as for hPAT4 above and were used as positive controls as indicated in the text.
In Vitro Transcription of cRNA
The template cDNA was linearized using NdeI (New England Biolabs). Linearized template was then used to synthesize cRNA using in vitro transcription (either T7 mMessage mMachine, Ambion, or Ribomax T7 transcription kit, Promega).
Expression of hPATs in X. laevis oocytes
Mature female X. laevis were purchased from the European Xenopus Resource Centre (University of Portsmouth, UK). They were housed and euthanized in accordance with Home Office regulations. The ovaries were harvested and rinsed in OR2 solution (80 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 5 mm HEPES adjusted to pH 7.6 with NaOH). Oocytes were defolliculated by collagenase A (0.1 mg/ml) treatment in calcium-free OR2 solution (80 mm NaCl, 2 mm KCl, 1 mm MgCl2, and 5 mm HEPES adjusted to pH 7.6 with NaOH) for up to 2 h with constant agitation. The oocytes were then rinsed five times with OR2 and then with Barth's solution (88 mm NaCl, 1 mm KCl, 0.82 mm MgSO4, 0.33 mm CaNO3, 2.4 mm NaHCO3, 1 mm CaCl2, and 15 mm HEPES adjusted to pH 7.6 with NaOH, 5 mm sodium pyruvate, and 50 μg/ml gentamycin). Stage V and VI oocytes were selected and kept at 18 °C for at least 2 h prior to injection with 27 nl of cRNA (1 μg/μl). The oocytes were maintained at 18 °C for 3 days with daily replacement of the Barth's solution.
Uptake of Radiolabeled Substrate in hPAT-expressing Xenopus Oocytes
Oocytes (10 per data point) were washed with amino acid-free uptake medium (100 mm choline chloride, 2 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 10 mm MES-Tris at pH 5.5 or 10 mm HEPES-Tris at pH 7.4 or 8.4) for 2 min prior to transfer into the uptake medium (as above but supplemented with 5 μCi/ml 3H-labeled or 20 μCi/ml 14C-labeled amino acid and cold amino acid if required, see “Results” and figure legends for experimental details). The radiolabeled amino acids used were as follows (final concentration in μm): [3H]Pro (0.10), [3H]Trp (0.25), [3H]Ala (0.11), [14C]Cys (50.0), [14C]Ser (6.21), and [14C]Gly (10.1). After incubation for 1 h (except in the case of the time course experiment), the oocytes were washed five times in 1 ml of ice-cold 0.12 m NaCl and individually transferred to wells of a 96-well scintillation plate (PerkinElmer Life Sciences) and lysed with 50 μl of SDS (2% w/v). Scintillation mixture (250 μl of Optiphase, PerkinElmer Life Sciences) was added to each well and mixed thoroughly on a shaking platform. For experiments to investigate sodium dependence, choline chloride was substituted with NaCl (100 mm).
Uptake of Substrate as Assessed by Two-electrode Voltage Clamp
Recordings were performed 3 days after injection. hPAT4-expressing oocytes were placed in a 0.1-ml recording chamber and perfused at ∼15 ml/min with uptake solution (100 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 10 mm HEPES-Tris at pH 7.4). Oocytes were impaled by two agarose-cushioned microelectrodes filled with 3 m KCl (resistance = 0.3–1.0 megaohm) and voltage-clamped at −30 mV using a GeneClamp 500B amplifier (Molecular Devices, Wokingham, Berkshire, UK) with traces recorded on paper. hPAT1-expressing oocytes were used as a positive control, perfused at pH 5.5 as this is the optimal pH for hPAT1 function, with non-injected oocytes providing the negative control.
Indirect Measure of Electrogenicity by Raising Extracellular K+ Concentration
hPAT4-mediated [3H]Pro uptake into oocytes was measured in uptake medium containing either normal potassium (2 mm KCl) or high potassium (50 mm KCl, with the choline chloride reduced to 52 mm to maintain constant osmolarity). hPAT1 (with pH 5.5 uptake medium) was used as a positive control, and non-injected oocytes were the negative control.
Data Presentation and Statistical Analysis of Data
Data are presented as mean ± S.E. of n oocyte preparations (experiments) with at least five oocytes per data point in each individual experiment. In all cases, hPAT4-mediated transport is presented, i.e. the uptake remaining after subtraction of the uptake measured simultaneously in control (non-injected) oocytes. Where data are normalized, the rates are given in the figure legends.
Outlier data were detected using Grubbs' test (QuickCalcs online, GraphPad Software, La Jolla, CA). Significance was established using a two-way unpaired Student's t test unless otherwise stated (SigmaPlot 11, Systat Software Inc., San Jose, CA), with p < 0.05 considered significant. Affinities were calculated using the method of Deves and Boyd (16).
RESULTS
hPAT4 Mediates Uptake of Proline
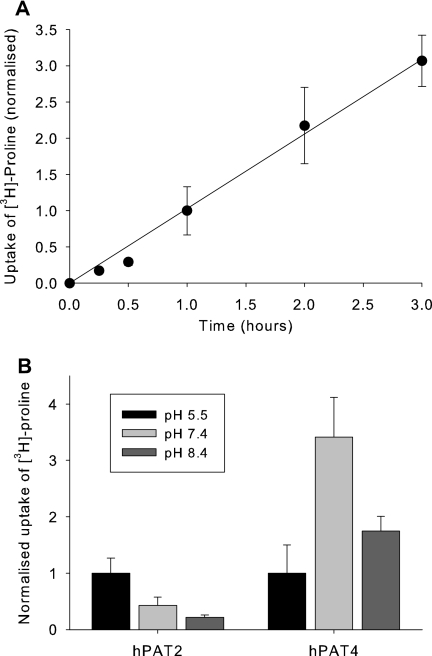
Expression of hPAT4 resulted in an uptake of [3H]proline, linear over the first 3 h (Fig. 1A; for subsequent experiments, 1-h incubations were used) and with maximum transport at pH 7.4 and lowest at pH 5.5 (Fig. 1B). This is in contrast to hPAT2, which showed the expected proton-coupled pH dependence and maximum transport at pH 5.5, which decreases steadily as the pH increases (17).
FIGURE 1.
Time and extracellular pH dependence of hPAT4-mediated proline uptake. A, time dependence of hPAT4-mediated [3H]proline uptake into Xenopus oocytes. Data are normalized to uptake at 1 h, which was 2.96 ± 0.98 fmol/oocyte/h, n = 4 oocyte preparations. B, dependence of hPAT4-mediated [3H]proline uptake on extracellular pH in Xenopus oocytes expressing either hPAT2 or hPAT4. Data are normalized to uptake at extracellular pH 5.5, which was 27.4 ± 7.39 fmol/oocyte/h for hPAT2 and 4.26 ± 2.14 fmol/oocyte/h for hPAT4, n ≥ 3 oocyte preparations.
hPAT4 Is a High Affinity Proline Transporter
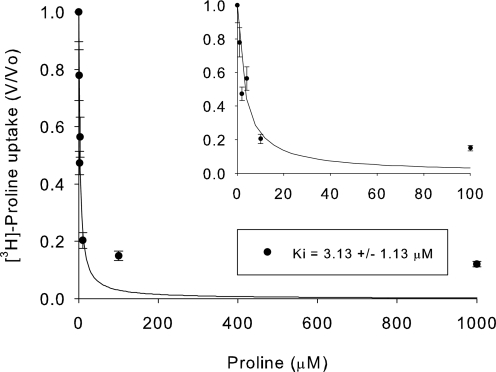
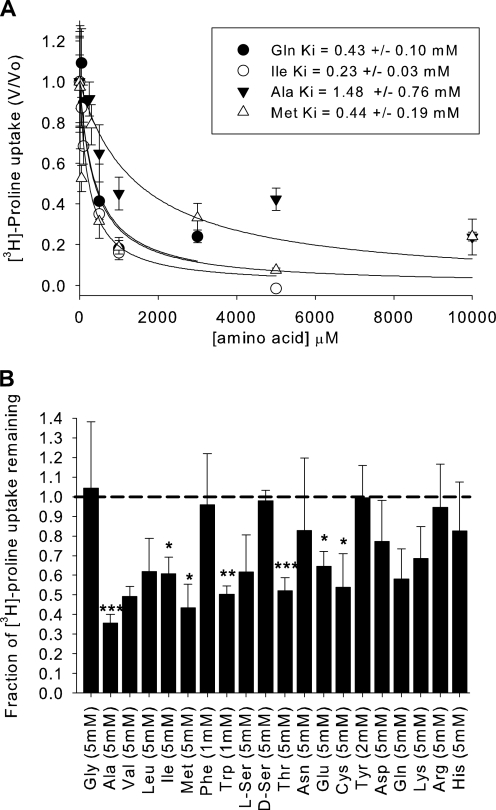
To test the affinity of hPAT4 for proline, a concentration dependence experiment was performed. hPAT4 appears to be a very high affinity transporter with an apparent Km of 3.13 ± 0.13 μm (Fig. 2). As the other characterized SLC36 family members have the prototype substrates of proline, alanine, and glycine, the affinities of the latter two substrates were also tested. The affinity for alanine was much lower than that for proline, with a Ki of 1.48 ± 0.76 mm (Fig. 3A) and >5 mm glycine (data not shown).
FIGURE 2.
Concentration dependence of hPAT4-mediated proline uptake, as determined by inhibition of [3H]proline by increasing concentrations of unlabeled proline. The inset shows 0–100 μm proline at higher magnification for clarity. The apparent Km was 3.13 ± 1.13 μm, n = 3 oocyte preparations.
FIGURE 3.
Concentration dependence and substrate specificity of hPAT4-mediated proline uptake. A, concentration dependence of hPAT4-mediated isoleucine, methionine, glutamine, and alanine inhibition of [3H]proline uptake. The Ki values (in mm) were: Ile, 0.23 ± 0.03; Met, 0.44 ± 0.19; Gln, 0.43 ± 0.10; and Ala, 1.48 ± 0.76, n = 2–4 oocyte preparations. B, inhibition of hPAT4-mediated [3H]proline uptake into Xenopus oocytes by the proteogenic amino acids. Uptake has been normalized to that seen in the absence of inhibitor. *, p < 0.05, **, p < 0.01, ***, p < 0.001, one-sample t test, n ≥ 3 oocyte preparations.
Inhibition of Proline Uptake through hPAT4 by Other Amino Acids
To determine the full substrate range of hPAT4, all of the proteogenic amino acids and d-serine were tested at a single, high concentration (5 mm except for where limited by solubility). The fractional inhibition of hPAT4-mediated l-[3H]proline uptake is shown in Fig. 3B, with the amino acids that statistically inhibited uptake being alanine (p < 0.001, one-sample t test; n = 5 oocyte preparations), isoleucine (p < 0.05, n = 3), methionine (p < 0.05, n = 3), tryptophan (p < 0.01, n = 3), threonine (p < 0.001, n = 6), glutamine (p < 0.05, n = 3), and cysteine (p < 0.05, n = 6).
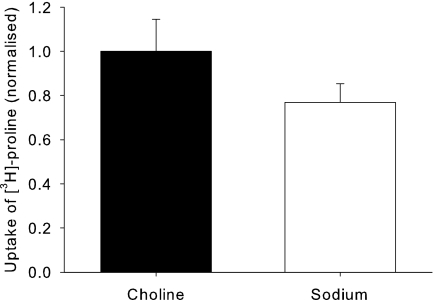
Proline Transport by hPAT4 Is Sodium-independent and Electroneutral
Replacement of choline chloride in the uptake medium by sodium chloride did not significantly change the uptake of [3H]proline (Fig. 4). Time course experiments indicate no evidence of concentrative uptake even after 8 h of uptake (assuming an oocyte volume of ∼1 μl (18), data not shown). In addition, no change in membrane potential above that seen in non-injected controls could be detected by two-electrode voltage clamp in hPAT4-expressing oocytes, unlike for hPAT1 (Table 2). However, due to the low capacity of hPAT4, it is likely that even the current when the transporter is operating at Vmax (i.e. the Imax) will be too low to measure. This was confirmed by the fact that there was no measurable current when an hPAT4-saturating concentration of proline was applied to hPAT1 (20 μm) despite the fact that both isoforms had similar uptakes of tracer (0.1 μm) [3H]Pro under optimal conditions and that hPAT1 is known to be electrogenic.
FIGURE 4.
Uptake of hPAT4-mediated [3H]proline in the absence or presence of sodium, normalized to that seen in the absence of sodium (replaced by choline, actual uptake 27.8 ± 15.2 fmol/oocyte/h). p > 0.05 paired t test, n = 4 oocyte preparations.
TABLE 2.
Characterization of proline transport in hPAT4-expressing Xenopus oocytes by two-electrode voltage clamp electrophysiology, as compared with hPAT1-expressing and control (non-injected) oocytes (n ≥ 3 oocytes per condition)
*, p < 0.05 versus pH5.5 non-injected; #, p < 0.05 versus pH7.4 non-injected; NMS, no measurable signal.
| hPAT1 | hPAT4 | Non-injected | |
|---|---|---|---|
| nA | |||
| pH 5.5, 20 μm Pro | NMS | NMS | NMS |
| pH 5.5, 10 mm Pro | 16.8 ± 2.09* (n = 17) | 4.70 ± 0.60 (n = 10) | 7.60 ± 0.87 (n = 5) |
| pH 7.4, 20 μm Pro | NMS | NMS | NMS |
| pH 7.4, 10 mm Pro | 13.5 ± 1.18# (n = 6) | 9.00 ± 0.93 (n = 12) | 8.67 ± 1.17 (n = 6) |
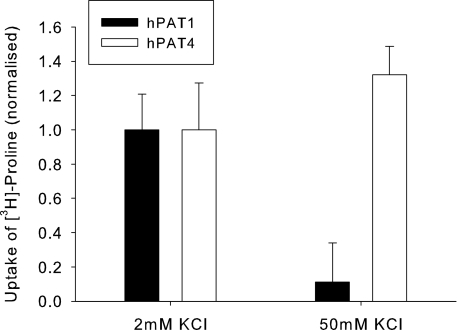
Therefore an indirect approach was taken to try to investigate whether transport by hPAT4 was electrogenic. Uptake was measured under normal (2 mm) and high (50 mm) extracellular potassium concentrations as the latter conditions will act to depolarize the membrane potential. As can be seen in Fig. 5, for the known electrogenic transporter hPAT1, as expected, the mediated transport was greatly reduced by raising the extracellular potassium to 50 mm. However, there was no reduction in hPAT4-mediated proline uptake, suggesting that the transport process is electroneutral.
FIGURE 5.
Uptake of hPAT4-mediated [3H]proline in normal (2 mm) and raised (50 mm) extracellular potassium, normalized to that seen in the normal potassium (mean uptake of 25.3 ± 12.2 and 31.8 ± 24.9 fmol/oocyte/h, respectively, n = 3 oocyte preparations with p < 0.05 for 2 mmversus 50 mm for hPAT1 and p > 0.05 for hPAT4 in each).
Affinity of hPAT4 for the Amino Acids That Inhibit Proline Uptake
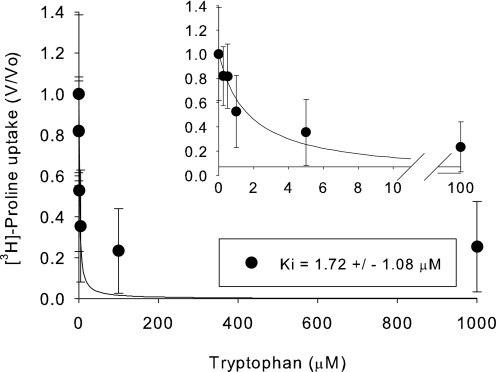
The affinity for all the amino acids that gave significant inhibition of hPAT4-mediated proline uptake was assessed. hPAT4 had a similarly high affinity for tryptophan (1.72 ± 1.09 μm, Fig. 6) as for proline (Fig. 2) and the following lower Ki values (in mm) for Ile (0.23 ± 0.03), Gln (0.43 ± 0.10), Met (0.44 ± 0.19), and Ala (1.48 ± 0.79) (Fig. 3). Gly, Thr, and Cys had very low affinities (Ki > 5 mm, data not shown).
FIGURE 6.
Concentration dependence of hPAT4-mediated tryptophan inhibition of [3H]proline uptake. Ki was 1.72 ± 1.08 μm, n = 2 oocyte preparations.
Transport of Amino Acids Other than Proline by hPAT4
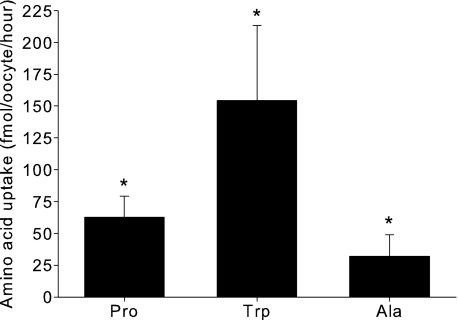
Although proline uptake through hPAT4 is inhibited by a number of amino acids, this does not necessarily mean that those amino acids are transported substrates. To check whether the amino acids Trp, Cys, and Ser were transported, uptake experiments described earlier were performed using radiolabeled substrates. Additionally, Gly and Ala were included in these experiments as they are prototypical PAT substrates. There was significant hPAT4-mediated uptake of Ala and Trp (Fig. 7) but not of Gly, Cys, or Ser (p > 0.05, n ≥ 3 oocyte preparations). The relative rates of transport of Trp and Ala, when normalized to Pro, were 2.7- and 0.5-fold, indicating that experimentally Trp was the best transported substrate for hPAT4 tested here.
FIGURE 7.
hPAT4-mediated uptake of amino acids into Xenopus oocytes (fmol/oocyte/h). *, p < 0.05 as compared with non-injected oocytes, n ≥ 3 oocyte preparations.
Transport of Substrates Other than the 20 Proteogenic Amino Acids
Due to the interest in non-proteogenic amino acid substrates for PAT1 and PAT2, a number of other substrates were tested for inhibition of [3H]proline uptake. γ-Aminobutyric acid (GABA), β-alanine, δ-aminolevulinic acid, and l-Dopa did not significantly inhibit uptake, whereas sarcosine did (p < 0.05, one-sample t test) with an affinity of 1.09 ± 0.35 mm.
DISCUSSION
Proton-coupled Amino Acid Transporter 1 (PAT1)
Evidence for the existence of a transporter with PAT1-like substrate range came initially from Newey and Smyth (19) reporting a glycine-proline carrier, which was subsequently cloned by Sagné et al. (20) as a lysosomal amino acid transporter from rat brain (LYAAT1), the homologue of hPAT1 (SLC36A1) in humans. PAT1 acts as a proton-coupled symporter of small neutral amino acids. Although H+-coupled transport has long been accepted to play an important role in bacteria, plants, and fungi, in recent years, there has been considerable evidence to indicate its significance in mammals (21). PAT1 transport activity is Cl−-independent and partially Na+-dependent due to involvement of the sodium hydrogen exchanger in maintaining the proton electrochemical gradient (22, 23). The substrates for PAT1 have Km values in the mm range (1–10 mm) (23). The main substrates for PAT1 are d- and l-proline, glycine, and l-alanine (3, 17, 21, 24) (Table 1). PAT1 has a higher affinity for proline than for glycine or alanine (25). It is also capable of transporting small unbranched, apolar, zwitterionic (α, β, and γ) amino and imino acids, d- and l-amino acids, heterocyclic amino/imino acids with 4–6-membered rings, and N-methylated amino and imino acids (12, 21, 23).
The PAT1 protein is found in a variety of tissues ranging from the intestinal epithelium to neurons, with varying subcellular localization. In the intestinal epithelia, PAT1 is isolated to the brush border, whereas in the neurons, it is localized mainly in the lysosomal membrane and to a smaller extent in the plasma membrane. In the lysosomes, it exports the products of proteolysis out of the lysosomes (23).
PAT2
The second member of the SLC36 family, PAT2, has also been shown to be a sodium-independent proton-coupled amino acid symporter, with a potential role in the myelination of neurons (3). The main substrates for PAT2 are the small neutral amino acids glycine, alanine, and proline (17), which have a higher affinity than for PAT1 (11, 14) (Table 1). PAT2 has a narrower substrate specificity than PAT1 (26); for example, although both β-amino and γ-amino butyric acids can inhibit PAT1, they have no effect on PAT2 (11).
PAT2 mRNA shows a high abundance in organs such as the kidneys, lungs, and spinal cord and brain (14, 26). The subcellular localization of PAT2 in vivo is still to be determined, but in HeLa cells, it does not localize to lysosomes but seems to be expressed in the plasma membrane (23).
Amino Acid Transport Characteristics of hPAT4
Since its identification at the gene level in 2004 along with SLC36A3 (hPAT3), the ubiquitously expressed fourth member of the SLC36 gene family, SLC36A4 (hPAT4), has been an orphan transporter with no known substrate. Here we show that hPAT4 is a sodium-independent amino acid transporter, like the other two characterized SLC36 family members, hPAT1 and hPAT2. The substrate characteristics of hPAT4 differ subtly from the other studied isoforms in that the affinity for proline is very high at 3.13 μm, whereas those for alanine and glycine are much lower (1.48 and >5 mm, respectively) and in that respect more like hPAT1/hPAT12. In addition, we show that hPAT4 is also significantly inhibited by the amino acids Ile, Met, Thr, Gln, and Cys (unlike hPAT1/hPAT2), and Trp was notable for also having a very low Ki (1.72 μm) as compared with that recorded for hPAT1 (4.7 mm (24)). Unlike for PAT1/PAT2, Trp was transported by hPAT4, whereas Gly was not a transported substrate (see Table 1 for a summary of data).
One of the most noteworthy differences between hPAT4 and hPAT1/hPAT2 is that hPAT4 did not show a maximal rate of transport of proline at acidic pH (pH 5.5) but rather at pH 7.4. Although this could be explained by the hPAT4 protein being inhibited by extracellular protons (for example, the amino acid transporter System A has been shown to be inhibited by a fall in extracellular pH by Bevington et al. (27)), taken together with the facts that transport appeared to be electroneutral and non-concentrative, this suggests that proline transport through hPAT4 is not proton-coupled or electrogenic and is therefore via facilitated diffusion.
Recognition of Other Substrates by hPAT4
Of the other substrates tested, no inhibition of proline uptake by hPAT4 was seen for GABA, δ-aminolevulinic acid, or β-alanine, in comparison with reports for PAT1 (3, 12, 17). The story for PAT2 is more complex as there are differences between mouse and rat PAT2; for example, only the former is inhibited by GABA (11) and with very low affinity (Ki 30.9 mm (17)). Similarly for β-alanine, the inhibitory effect was greater in mouse than rat PAT2, whereas sarcosine was shown to be a good substrate for rat PAT2 (no data for mouse) (11).
For mouse and human PAT1, d-serine is a better substrate than l-serine (3, 12); the same is true for mouse PAT2, but not rat PAT2, for which d- and l-serine invoke equal currents in two electrode voltage-clamped Xenopus oocytes (11). In hPAT4, neither d-serine nor l-serine significantly inhibits proline uptake (Fig. 7). d-Serine is of interest as it is given orally (and has high bioavailability) for treatment of psychiatric disorders such as schizophrenia (28), and hPAT1 may be its route of intestinal uptake, being located in the apical membrane of the enterocyte model cell line Caco-2 (3).
Is Amino Acid Transport the Only Role of hPAT4?
Of the 11 SLC36 homologue genes in Drosophila, two of these genes have been shown to code for functional amino acid transporters when expressed in Xenopus oocytes (29). CG1139 gave a proton-coupled alanine transporter with kinetic properties similar to hPAT1, whereas PATH had a low micromolar substrate affinity and was not proton-coupled, remarkably similar to hPAT4 as described here. In Drosophila, PATH regulates growth through the target of rapamycin (TOR)/S6-kinase pathway, and genetically removing path results in a small fly (29). Although changes in intracellular amino acid concentrations are well known to signal through this pathway (30), the signaling through PATH was despite it being a low capacity transporter that could mediate only a very small predicted change in global intracellular amino acid concentration, suggesting that it may be acting more as a sensor or a “transceptor” (29). Intriguingly, hPAT4 can functionally replace PATH in Drosophila, reversing the reduced growth phenotype (31), suggesting that hPAT4 might also signal through to S6-kinase. As for PATH, it seems likely that under many physiological conditions, hPAT4 will always be saturated, and due to its low capacity, its role as a bulk transporter is open to question. Whether hPAT4 is a transceptor is an interesting and important area of future research due to the implication of the mammalian TOR/S6-kinase signaling pathway in diseases such as cancer and diabetes (32).
Acknowledgments
We thank Professor Isabel Bermudez and Simone Mazzaferro (School of Life Sciences, Oxford Brookes University) for helping with the electrophysiology studies. We are grateful to Drs. Richard Boyd, Clive Wilson, and Deborah Goberdhan, Department of Physiology, Anatomy and Genetics, University of Oxford, for helpful discussion.
This work was supported by a scholarship from the School of Life Sciences, Oxford Brookes University (to S. M. P.).
- SLC
- solute-linked carrier
- PAT
- proton-coupled amino acid transporter
- hPAT
- human PAT
- GABA
- γ-aminobutyric acid.
REFERENCES
- 1. Hediger M. A., Romero M. F., Peng J. B., Rolfs A., Takanaga H., Bruford E. A. (2004) Pflugers. Arch. 447, 465–468 [DOI] [PubMed] [Google Scholar]
- 2. Thwaites D. T., Anderson C. M. (2007) Exp. Physiol. 92, 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen Z., Kennedy D. J., Wake K. A., Zhuang L., Ganapathy V., Thwaites D. T. (2003) Biochem. Biophys. Res. Commun. 304, 747–754 [DOI] [PubMed] [Google Scholar]
- 4. Bröer S., Bailey C. G., Kowalczuk S., Ng C., Vanslambrouck J. M., Rodgers H., Auray-Blais C., Cavanaugh J. A., Bröer A., Rasko J. E. (2008) J. Clin. Invest. 118, 3881–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thwaites D. T., Basterfield L., McCleave P. M., Carter S. M., Simmons N. L. (2000) Br. J. Pharmacol. 129, 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Metzner L., Dorn M., Markwardt F., Brandsch M. (2009) Mol. Pharm. 6, 1006–1011 [DOI] [PubMed] [Google Scholar]
- 7. Larsen M., Holm R., Jensen K. G., Brodin B., Nielsen C. U. (2009) Br. J. Pharmacol. 157, 1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abbot E. L., Grenade D. S., Kennedy D. J., Gatfield K. M., Thwaites D. T. (2006) Br. J. Pharmacol. 147, 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson C. M., Jevons M., Thangaraju M., Edwards N., Conlon N. J., Woods S., Ganapathy V., Thwaites D. T. (2010) J. Pharmacol. Exp. Ther. 332, 220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frølund S., Marquez O. C., Larsen M., Brodin B., Nielsen C. U. (2010) Br. J. Pharmacol. 159, 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kennedy D. J., Gatfield K. M., Winpenny J. P., Ganapathy V., Thwaites D. T. (2005) Br. J. Pharmacol. 144, 28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boll M., Foltz M., Anderson C. M., Oechsler C., Kottra G., Thwaites D. T., Daniel H. (2003) Mol. Membr. Biol. 20, 261–269 [DOI] [PubMed] [Google Scholar]
- 13. Bermingham J. R., Jr., Pennington J. (2004) Mamm. Genome 15, 114–125 [DOI] [PubMed] [Google Scholar]
- 14. Foltz M., Oechsler C., Boll M., Kottra G., Daniel H. (2004) Eur. J. Biochem. 271, 3340–3347 [DOI] [PubMed] [Google Scholar]
- 15. Dominguez I., Itoh K., Sokol S. Y. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8498–8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deves R., Boyd C. A. (1989) Proc. R. Soc. Lond. B Biol. Sci. 237, 85–97 [DOI] [PubMed] [Google Scholar]
- 17. Boll M., Foltz M., Rubio-Aliaga I., Kottra G., Daniel H. (2002) J. Biol. Chem. 277, 22966–22973 [DOI] [PubMed] [Google Scholar]
- 18. Meredith D. (2004) J. Biol. Chem. 279, 15795–15798 [DOI] [PubMed] [Google Scholar]
- 19. Newey H., Smyth D. H. (1964) J. Physiol. 170, 328–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sagné C., Agulhon C., Ravassard P., Darmon M., Hamon M., El Mestikawy S., Gasnier B., Giros B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7206–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Metzner L., Neubert K., Brandsch M. (2006) Amino Acids 31, 111–117 [DOI] [PubMed] [Google Scholar]
- 22. Anderson C. M., Grenade D. S., Boll M., Foltz M., Wake K. A., Kennedy D. J., Munck L. K., Miyauchi S., Taylor P. M., Campbell F. C., Munck B. G., Daniel H., Ganapathy V., Thwaites D. T. (2004) Gastroenterology 127, 1410–1422 [DOI] [PubMed] [Google Scholar]
- 23. Boll M., Daniel H., Gasnier B. (2004) Pflugers. Arch. 447, 776–779 [DOI] [PubMed] [Google Scholar]
- 24. Metzner L., Kottra G., Neubert K., Daniel H., Brandsch M. (2005) FASEB J. 19, 1468–1473 [DOI] [PubMed] [Google Scholar]
- 25. Bröer A., Cavanaugh J. A., Rasko J. E., Bröer S. (2006) Pflugers. Arch. 451, 511–517 [DOI] [PubMed] [Google Scholar]
- 26. Rubio-Aliaga I., Boll M., Vogt Weisenhorn D. M., Foltz M., Kottra G., Daniel H. (2004) J. Biol. Chem. 279, 2754–2760 [DOI] [PubMed] [Google Scholar]
- 27. Bevington A., Brown J., Butler H., Govindji S., M-Khalid K., Sheridan K., Walls J. (2002) Eur. J. Clin. Invest. 32, 590–602 [DOI] [PubMed] [Google Scholar]
- 28. Javitt D. C. (2002) Curr. Opin. Investig. Drugs 3, 1067–1072 [PubMed] [Google Scholar]
- 29. Goberdhan D. C., Meredith D., Boyd C. A., Wilson C. (2005) Development 132, 2365–2375 [DOI] [PubMed] [Google Scholar]
- 30. Christie G. R., Hajduch E., Hundal H. S., Proud C. G., Taylor P. M. (2002) J. Biol. Chem. 277, 9952–9957 [DOI] [PubMed] [Google Scholar]
- 31. Heublein S., Kazi S., Ogmundsdóttir M. H., Attwood E. V., Kala S., Boyd C. A., Wilson C., Goberdhan D. C. (2010) Oncogene 29, 4068–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reynolds B., Laynes R., Ogmundsdóttir M. H., Boyd C. A., Goberdhan D. C. (2007) Biochem. Soc. Trans. 35, 1215–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]