Abstract
Extracellular signal-regulated kinase-1 and -2 (ERK1/2) proteins regulate a variety of cellular functions, including cell proliferation and differentiation, by interacting with and phosphorylating substrate proteins. Two docking sites, common docking (CD/ED) domain and F-site recruitment site (FRS), on ERK proteins have been identified. Specific interactions with the CD/ED domain and the FRS occur with substrates containing a docking site for ERK and JNK, LXL (DEJL) motif (D-domain) and a docking site for ERK, FXF (DEF) motif (F-site), respectively. However, the relative contributions of the ERK docking sites in mediating substrate interactions that allow efficient phosphate transfer are largely unknown. In these studies, we provide a quantitative analysis of ERK2 interactions with substrates using surface plasmon resonance to measure real time protein-protein interactions. ERK2 interacted with ELK-1 (DEF and DEJL motifs), RSK-1 (DEJL motif), and c-Fos (DEF motif) with KD values of 0.25, 0.15, and 0.97 μm, respectively. CD/ED domain mutations inhibited interactions with ELK-1 and RSK-1 by 6-fold but had no effect on interactions with c-Fos. Select mutations in FRS residues differentially inhibited ELK-1 or c-Fos interactions with ERK2 but had little effect on RSK-1 interactions. Mutations in both the ED and FRS docking sites completely inhibited ELK-1 interactions but had no effect on interactions with stathmin, an ERK substrate whose docking site is unknown. The phosphorylation status of ERK2 did not affect interactions with RSK-1 or c-Fos but did inhibit interactions with ELK-1 and stathmin. These studies provide a quantitative evaluation of specific docking domains involved in mediating interactions between ERK2 and protein substrates and define the contributions of these interactions to phosphate transfer.
Keywords: ERK, MAP Kinases (MAPKs), Protein Motifs, Protein Phosphorylation, Protein-Protein Interactions, Signal Transduction, Surface Plasmon Resonance (SPR)
Introduction
The major members of the mitogen-activated protein kinase (MAPK) family consist of the extracellular signal-regulated kinases-1 and -2 (ERK1/2), c-Jun N-terminal kinases (JNKs), p38 mitogen-activated protein kinases, and ERK5 (1). ERK1/2 proteins are serine/threonine kinases involved in signal transduction pathways that mediate cellular proliferation and differentiation in response to growth factors and hormones (2). In vitro studies suggest that ERK1/2 can interact with and phosphorylate over 160 substrates, implicating their involvement in a diverse number of cellular functions (3–6). Therefore, stringent control over ERK interactions with substrate proteins and efficient phosphate transfer are essential for maintaining normal cell physiology. However, unregulated activation of the ERK1/2 pathway is often observed in a variety of cancers, which contributes to the uncontrolled cell proliferation, survival, and resistance to anticancer drugs (7–10). Thus, a better understanding of the mechanisms regulating ERK interactions with substrates may aide in the discovery of ERK-targeted chemotherapeutics that selectively regulate substrates involved in proliferation while preserving other ERK functions in normal cells.
Docking domains located on ERK1/2 and protein substrates have been shown to confer specificity and provide a means to coordinate selective regulation for the mitogen-activated protein kinase signaling cascades. Several ERK1/2 substrate proteins, including the transcription factor ELK-1, p90 ribosomal S6 kinase-1 (RSK-1),2 caspase-9, and the protein-tyrosine phosphatase HePTP, all contain a docking domain known as the D-domain or docking site for ERK and JNK, LXL (DEJL) motif (11–14). The D-domain consists of basic residues followed by a hydrophobic LXL motif and has been shown to interact with ERK1/2 on an acidic region in the C terminus (referred to as the common docking (CD) domain) (15–18). The CD domain residues include Asp-316 and Asp-319 as well as adjacent hydrophobic amino acids that mediate substrate interactions. Adjacent to the aspartates in the crystal structure are residues that form the ED component (Glu-160 and Asp-161 for p38 mitogen-activated protein kinase and Thr-157 and Thr-158 for ERK2) that also coordinate with the CD domain to facilitate substrate-selective interactions with mitogen-activated protein kinases (19). A second docking domain on substrates, known as the docking site for ERK, FXF (DEF) motif or F-site, has been identified on a number of ERK1/2 substrates, including ELK-1, Ksr-1, A-Raf, c-Fos, and nucleoporin proteins (20–24). Substrates containing an F-site have been shown to interact with the F-site recruitment site (FRS) consisting of residues Leu-198, Tyr-231, Leu-232, Leu-235, and Tyr-261 on ERK2 (18, 20–24). ERK2 substrates are likely to function in association with other proteins, which may affect their interactions with regulatory kinases. For example, physiologically relevant heterodimers that form between proteins such c-Fos and c-Jun, which make up the AP-1 complex, or c-Myc and Max may also play a role in determining ERK substrate specificity. Dimerization of c-Fos with c-Jun exposes an N-terminal region lined with basic residues that have been shown to be important for DNA binding and may (as is the case for phosphorylation of c-Jun by JNK) play a role in ERK1/2 protein regulation of the AP-1 complex as well (25). Phosphorylation of c-Fos by ERK1/2 has been shown to enhance c-Fos protein stability in vivo (23, 26, 27). In addition, activation by ERK1/2 enhances the interaction and subsequent phosphorylation of c-Fos by RSK-1 (28). In vitro studies have suggested that c-Fos interacts with the FRS docking domain on ERK1/2 proteins through its DEF motif (29).
Although a large body of evidence supports the interactions of the D-domain and/or F-site of a substrate with the CD/ED domain and FRS of an ERK protein, respectively (17, 21, 29–34), the current knowledge of ERK-substrate interactions has largely been based on studies examining the ability of ERK to interact with or phosphorylate substrate docking domain peptides or pulldown assays that have quantitative limitations (17, 21, 31, 35, 36). To date, the only reported study to quantify the binding affinity of ERK with a substrate protein was the analysis of ERK interactions with the phosphoprotein PEA-15 using fluorescence anisotropy (37). Moreover, little is known of the relative contributions of the known ERK docking sites to binding efficiency of substrate proteins that utilize both CD/ED and FRS domains.
The activation status of ERK1/2 proteins may also regulate the interactions with substrate proteins. Comparison of the structures of ERK2 and diphosphorylated ERK2 as determined by x-ray crystallography has shown extensive refolding of the activation loop, which includes phosphorylated residues Thr-183 and Tyr-185 that confer activity (38). This results in considerable localized conformational changes without affecting the overall structure of the protein. Nonetheless, studies utilizing electron paramagnetic resonance spectroscopy have observed changes in residue side chains located near the ED docking domain (39). In addition, the FRS is situated adjacent to the activation loop and may involve the phosphorylated Tyr-185 in mediating substrate interactions (21). Thus, conformational changes in this domain during activation may regulate ERK1/2 interactions with substrates.
We herein report the use of surface plasmon resonance (SPR) methodologies to determine binding dissociation constants of ERK2 interactions with known ERK2 substrates ELK-1, RSK-1, and c-Fos. These studies reveal that the ERK2 CD/ED domain but not the FRS is involved in RSK-1 interactions, whereas a single residue within the FRS contributes to c-Fos interactions ERK2. ELK-1, however, utilizes both the ED domain and an alternative residue within the FRS site for interaction with ERK2. We also demonstrate quantitative changes in ERK2 interactions with substrates that are dependent on the phosphorylation status of ERK. Finally, we show that binding interactions are not necessarily predictive of whether efficient phosphate transfer will occur.
EXPERIMENTAL PROCEDURES
Bacterial Strains
DNA manipulations were carried out in Escherichia coli strain DH5α (F′, ara D (lac-proAB) rpsL Φ80dlacZ DM15 hasd R17), and E. coli strain BL21 (DE3) (B F−dmc ompT hsdS (rB− mB−) gal λ (DE3)) was utilized for the expression of wild type and mutant ERK2 proteins and substrate proteins ELK-1, RSK-1, c-Fos, and stathmin.
Site-directed Mutagenesis of ERK2
Alterations in the erk2 gene (pETHis6ERK2) were done by site-directed mutagenesis utilizing the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). ERK2 docking domain mutations (T157A, D319N, T157A/D319N, L232A, L198A/L235A, and T157A/L198A/L235A) were verified by DNA sequencing at the Biopolymer Facility, School of Medicine, University of Maryland.
Expression and Purification of ERK2 Wild Type and Mutant Proteins
Transformed E. coli BL21 (DE3) cells with either wild type or mutant erk2 constructs were grown on Luria-Bertani (LB)-ampicillin agar plates, and colonies were used to inoculate LB-ampicillin broth. The cells were grown at 30 °C until an optical density at 600 nm equaled ∼0.8. ERK2 protein expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 30 °C. Cells were lysed using Bugbuster® HT (Novagen) containing 10% (w/v) glycerol, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 10 mm benzamidine, and lysates were clarified by centrifugation at 10,000 rpm. The supernatant was applied to a Talon® Metal Affinity Resin (Clontech) column equilibrated in 50 mm sodium phosphate (pH 7.8) containing 2 mm imidazole, 300 mm NaCl, and 10% (w/v) glycerol and washed with the same buffer containing 20 mm imidazole. ERK2 was eluted in 50 mm sodium phosphate (pH 7.8) containing 250 mm imidazole, 300 mm NaCl, and 10% (w/v) glycerol. The fractions containing ERK2, as determined following SDS-PAGE, were pooled and concentrated with an Amicon stirred ultrafiltration cell (Millipore, Bedford, MA). The concentrated protein was dialyzed against 20 mm Tris-HCl (pH 7.8) containing 10% (w/v) glycerol and further purified over a Q-Sepharose column equilibrated in the same buffer. The column was washed with the same buffer containing 20 mm NaCl, and ERK2 was eluted with a 50–300 mm NaCl gradient. The fractions containing ERK2 were concentrated and dialyzed against 20 mm sodium phosphate containing 150 mm NaCl, 1 mm DTT, and 10% (w/v) glycerol. ERK2 protein concentrations were determined by Bio-Rad RC DC protein assay.
Generation of Active ERK2
Expression of active MKK1 in bacteria and incubation with ERK were as described (40). Briefly, active MKK1 was incubated with wild type ERK2 (20 μm) in kinase buffer (10 mm Hepes (pH 7.4), 1 mm CaCl2, 0.02% (w/v) Triton X-100, 0.2% β-mercaptoethanol, 20 mm MgCl2, and 4 mm ATP) at 30 °C for 1 h. Dually phosphorylated ERK2 was analyzed by SDS-PAGE and estimated to be ∼80% of the total ERK2 protein.
Expression and Purification of Substrate Proteins ELK-1, RSK-1, c-Fos, and Stathmin
The GST-ELK-1 (amino acids 307–428), GST-RSK-1 (amino acids 386–752), His6-stathmin, and His6-c-Fos proteins were expressed and purified as described previously (21, 41–43). The GST tag on RSK-1 was removed following thrombin cleavage (1 unit/μl) in phosphate-buffered saline (PBS) (pH 7.2) for 8 h at room temperature. The reaction was dialyzed against 4 liters of PBS buffer, and RSK-1 was separated from GST using a glutathione-Sepharose column.
Surface Plasmon Resonance Analysis of Protein-Protein Interactions
Protein-protein interactions of ERK2 with substrates were investigated by SPR analysis utilizing a Biacore® 3000 instrument (GE Healthcare). A carboxymethylated dextran chip (CM5, GE Healthcare) was used to immobilize the substrate proteins. A net positive charge is required for direct amine coupling; therefore, the proteins were diluted to a final concentration of 30 μg/ml in 10 mm sodium acetate buffer ranging from pH 4.0 to 5.5 depending on the pKa of the protein. The flow cell was activated by a 1:1 solution of 100 mm N-hydroxysuccinimide and 400 mm N-ethyl-N′-(dimethylaminopropyl)carbodiimide. The protein was injected at 2 μl/min until approximately a 4500 resonance/response unit level of immobilized ligand was reached, and then the flow cell was deactivated with 1 m ethanolamine HCl (pH 8.5). A control flow cell without immobilized ligand was activated and deactivated as indicated above. Degassed 20 mm Hepes (pH 8.0) containing 5 mm MgCl2, 100 mm NaCl, and 0.005% Nonidet P40 was utilized as continuous running buffer for all experiments. ERK2 binding interactions with immobilized substrate were investigated by injecting ERK2 (0.005–10 μm) at a flow rate of 20 μl/min. Each injection was completed in duplicate and in a random order. After injection of ERK2 was completed, running buffer was injected into the flow cell to dissociate ERK2, and the surface of the CM5 chip was regenerated by an injection of 2 mm NaOH. As an additional control, ERK2 was immobilized on the surface of a Ni2+-nitrilotriacetic acid chip utilizing a His6 tag, and substrate proteins were injected at various concentrations in duplicate and in a random order for each experiment. To determine whether the GST tag interfered with steady state binding determinations, untagged RSK-1 was injected (0.005–5.0 μm) in duplicate and in a random order and analyzed for interaction with wild type ERK2. The interaction of myelin basic protein (MBP) with wild type and the FRS ERK2 mutant L198A/L235A was also determined by SPR where MBP was immobilized on a CM5 chip, and ERK2 proteins were the analyte (0.1–5.0 μm). The resulting sensorgrams were analyzed using BIAevaluation 3.1 software (GE Healthcare). The dissociation constant (KD) was calculated using the binding response level at equilibrium (Req) for each analyte concentration as determined by Equation 1 where KA is the association constant, C is analyte concentration, Rmax is the response at saturation, and n is the number of binding sites. To determine the validity of the SPR data, global analysis and fitting of the data were performed utilizing BIAevaluation 3.1 software. Global fitting of the on (ka) and off (kd) rates determined one dissociation constant per concentration series. Analysis of replicate experiments allowed for determination of a one-site binding model fit and calculation of standard error (44). In addition, our SPR experiments were validated by utilizing the self-consistency tests for SPR reported previously (45).
 |
In Vitro Kinase Assays
Active MKK1 (0.5 μg) and ERK2 wild type or mutant proteins (5 μg) were incubated with 0.5 μg of MBP, RSK-1, ELK-1, or c-Fos for 0–60 min at 30 °C in 50 mm Tris-HCl, 10 mm MgCl2, 1 mm EGTA, 2 mm DTT, and 0.01% Brij 35 (pH 7.5) containing 100 μm ATP. In radioactive assays, 20 μm ATP and 2 μCi of [γ-32P]ATP were used. Reactions were stopped with an equal volume of 2× SDS-PAGE sample buffer, and the proteins were resolved by SDS-PAGE and analyzed for substrate phosphorylation by phosphorimaging analysis or autoradiography.
Circular Dichroism Spectra
To determine whether the introduction of mutations at ERK2 docking site residues altered the secondary structure, circular dichroism spectra were generated for wild type and mutant ERK2 proteins in 10 mm sodium phosphate (pH 7.5). The spectra from three scans were averaged for each protein. Spectra were collected at 25 °C on a Jasco J-810 spectropolarimeter from 190 to 260 nm using a scanning speed of 10 nm/min, data pitch of 0.5 nm, response of 2 s, and a 1.0-cm bandwidth. Protein spectra were converted to mean residue ellipticity (degrees cm2 dmol−1) after subtraction of the background spectra generated by the buffer alone.
RESULTS
Quantification of ERK2 Interactions with Substrate Proteins
Our understanding of how ERK1/2 proteins interact with a diverse range of substrates is limited. We have utilized SPR methodologies to provide a quantitative analysis of ERK interactions with substrates and identify features that determine efficient phosphate transfer. Protein binding interactions as determined by SPR are a two-step process. The first event requires the analyte to diffuse from the buffer to the surface of the chip, and the second process is the actual binding event at the surface of the chip of the analyte to the immobilized ligand. The first event, also known as mass transfer, can affect the accurate determination of association and dissociation rates and subsequent calculations of dissociation constants (KD). Because of the inherent difficulty accounting for mass transfer, the determination of accurate binding affinity of protein-protein interactions can be readily accomplished using equilibrium binding to determine the steady state binding characteristics of interacting partners (45). As such, equilibrium binding studies are advantageous due to the fact that binding is unaffected by mass transfer limitations (46).
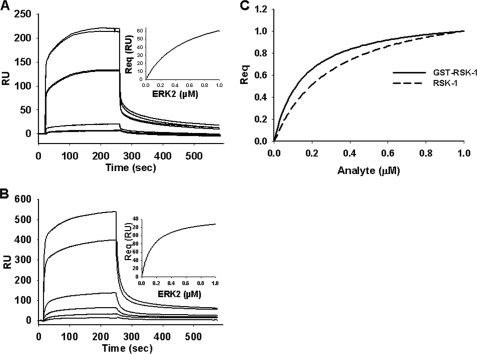
ERK2 interactions were first examined with ELK-1 and RSK-1, two ERK substrates that have been well characterized (18, 30, 31, 33, 47). The equilibrium response level (Req) for each analyte, in this case ERK2, was determined as described under “Experimental Procedures.” Dissociation constants (KD) were determined by plotting steady state binding level as a function of analyte concentration from the sensorgrams (Fig. 1, inset). The resulting KD values for ELK-1 and RSK-1 were 0.25 and 0.15 μm, respectively (Fig. 1 and Table 1). As a control, it was determined whether binding was affected by the generation of a heterogeneous surface due to direct amine coupling to a CM5 chip. In this case, ERK2 was immobilized as the ligand using nickel-nitrilotriacetic acid, and the substrates were the analytes. The calculated KD values were similar when ERK2 was used as the analyte or the ligand, indicating that protein interactions were not affected by adherence to the chip surface (data not shown). Given that the substrates contain a GST tag, additional controls examining interactions with GST alone showed no interactions with ERK2 within the protein concentration range tested for determining ELK-1 and RSK-1 binding (data not shown). In addition, removal of the GST tag from RSK-1 did not have a statistically significant effect on binding interactions with ERK2 with steady state binding affinities for the GST-tagged and untagged RSK-1 of 0.15 ± 0.01 and 0.30 ± 0.1 μm, respectively (Fig. 1C).
FIGURE 1.
ERK2 binding to ELK-1 and RSK-1. Shown is a sensorgram of response units (RU) for ERK2 (0.005–1 μm) interaction with GST-ELK-1 (A) or GST-RSK-1 (B) as a function of time. Insets show steady state binding (Req) as a function of ERK2 concentration. C, normalized steady state binding of GST-tagged RSK-1 and untagged RSK-1 (analyte concentrations from 0 to 1.0 μm).
TABLE 1.
Steady state binding affinity (KD) of protein-protein interactions of substrates with wild type and mutant proteins
Steady state binding affinities (μm) are the mean and S.E. from three to five independent determinations. ERK2 substrate docking sites include residues Asp-319 in the CD domain, Thr-157 in the ED domain, and residues Leu-198, Leu-232, and Leu-235 in the FRS. Docking sites on ERK substrates include the DEJL (D-domain) and DEF (F-site) motifs.
| WT | T157A (ED) | D319N (CD) | T157A/D319N (CD/ED) | L232A (FRS) | L198A/L235A (FRS) | T157A/L198A/L235A (ED + FRS) | |
|---|---|---|---|---|---|---|---|
| ELK-1 (DEJL + DEF) | 0.25 ± 0.2 | 5.0 ± 0.9 | 3.6 ± 0.1 | 1.6 ± 0.01 | 0.37 ± 0.05 | 5.0 ± 0.9 | NBa |
| RSK-1 (DEJL) | 0.15 ± 0.01 | 0.68 ± 0.2 | 1.1 ± 0.2 | 0.98 ± 0.03 | NDb | 0.17 ± 0.03 | 0.85 ± 0.02 |
| c-Fos (DEF) | 0.97 ± 0.2 | 2.7 ± 0.1 | 2.9 ± 0.3 | 1.7 ± 0.3 | NB | 0.34 ± 0.05 | NB |
| Stathmin (unknown) | 0.85 ± 0.2 | ND | ND | ND | ND | ND | 0.76 ± 0.3 |
a NB, no binding interaction.
b ND, not determined.
Contributions of ERK2 CD/ED and FRS Docking Domains for Interactions with Substrate Proteins
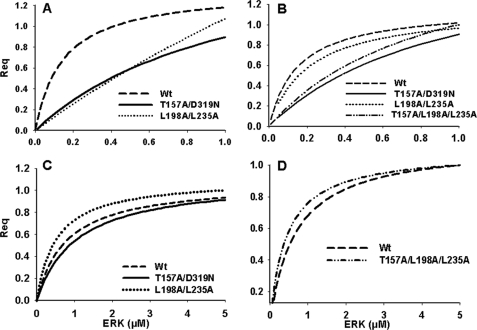
ERK2 docking domains are important for determining substrate specificity in the kinase signaling cascade (18, 30, 48–50). To quantify the importance of the docking domains in these protein-protein interactions, we tested ERK2 that contained mutations in the CD/ED domain (T157A, D319N, and T157A/D319N), the FRS (L232A and L198A/L235A), or both the ED and FRS (T157A/L198A/L235A) for substrate interactions. The D-domain substrate RSK-1 interacts with ERK2 primarily through the CD component of the CD/ED domain; c-Fos through its DEF motif reportedly interacts with the FRS; and ELK-1, which contains both docking motifs, interacts with the CD/ED domain and FRS (23, 31, 51). The ERK-interacting regions of stathmin are currently unknown. Mutations in the CD/ED domain increased the KD for ELK-1 and RSK-1 binding by ∼6-fold as compared with wild type ERK2 (Fig. 2 and Table 1).
FIGURE 2.
Normalized steady state binding (Req) of ELK-1, RSK-1, stathmin, and c-Fos to wild type and mutant ERK2 proteins. A, ELK-1 interactions with wild type or mutants L198A/L235A or T157A/D319N. B, RSK-1 interactions with wild type or mutants L198A/L235A, T157A/D319N, or T157A/L198A/L235A. C, c-Fos interactions with wild type or mutants T157A/D319N or L198A/L235A. D, stathmin interactions with wild type or the T157A/L198A/L235A mutant. Analyte concentrations ranged from 0 to 1.0 μm in A, B, and D and from 0 to 5.0 μm in C.
To determine the individual contributions of the CD and the ED domains, interactions between single ERK2 mutants D319N (CD domain) and T157A (ED domain) with ELK-1 and RSK-1 were investigated. The binding interaction of ELK-1 with ERK2 T157A and D319N mutants resulted in a KD of 5.0 ± 0.9 and 3.6 ± 0.1 μm, respectively, which were statistically different from wild type ERK2 binding affinity and from each other, indicating that the ED domain may contribute more to determining selectivity for the interactions between ELK-1 with ERK2. The ERK2 T157A mutant had a steady state binding affinity of 0.68 ± 0.2 μm with RSK-1 as compared with the D319N mutant, which had a KD of 1.1 ± 0.2 μm (Fig. 2 and Table 1). Although this difference was small, the KD values were statistically different, indicating that the CD domain may contribute more to the interactions between ERK2 and RSK-1.
The binding interaction of RSK-1 with the FRS mutant was identical to that of the wild type ERK2, suggesting RSK-1 does not interact with ERK2 through this motif (Fig. 2 and Table 1). In contrast, the FRS mutations L198A/L235A inhibited ELK-1 binding to ERK2 by ∼20-fold as compared with wild type, whereas the FRS mutation L232A had no effect on binding (Table 1). Mutations in both the ED and FRS (T157A/L198A/L235A) docking domains completely abolished interactions with ELK-1 (Fig. 2 and Table 1), supporting the contribution of both ERK2 docking sites to ELK-1 interactions with Leu-198 and Leu-235 being the important residues within the FRS (21).
In the case of c-Fos, there was little difference in binding affinity between the CD/ED domain mutants and a slight increase in binding to the FRS docking domain mutant L198A/L235A as compared with the wild type protein (Fig. 2 and Table 1). Although this finding is in agreement with previous reports that c-Fos does not utilize the CD/ED domain for interactions with ERK, it does not explain data showing the importance of Leu-198 in ERK-mediated phosphorylation of c-Fos (29). No binding interactions were observed, however, between c-Fos and the ERK2 L232A mutant, which is in agreement with previously reported in vitro kinase data (29). These results indicated that the DEF domain-containing substrate, c-Fos, primarily utilizes Leu-232 for interactions with the FRS on ERK2 and not residues Leu-198 and Leu-235. The binding interactions of c-Fos with the D319N or T157A mutant were 2.9 ± 0.3 and 2.7 ± 0.1 μm, respectively. This is a lower affinity interaction than that of the wild type protein, suggesting that mutation of either residue Thr-157 or Asp-319 causes a conformational change in ERK2 that may contribute to a decrease in substrate binding affinity. Interestingly, mutations in both the ED and FRS docking domains completely abolished ERK2 interactions with c-Fos (Fig. 2 and Table 1). The reason for this is unclear, but it suggests that c-Fos may utilize additional regions on the ERK2 protein that are influenced by these mutations and affect subsequent protein-protein interactions.
Lastly, we tested ERK2 interactions with stathmin, a substrate involved in microtubule formation and whose binding interactions to ERK1/2 proteins are unknown (52). Stathmin protein-protein interactions with ERK2 containing ED and FRS docking domain mutations were similar to those of wild type, indicating that this substrate utilizes alternative docking sites (Fig. 2 and Table 1).
Determination of Wild Type and Mutant ERK2 Catalytic Activity
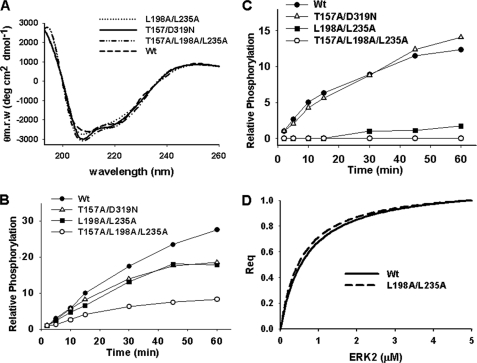
For ERK2 to transmit extracellular signals to mediate cellular responses, ERK1/2 proteins must first properly interact with corresponding substrate proteins in a manner that facilitates efficient catalysis and phosphate transfer. To differentiate between binding events and catalysis, the activity following incubation with constitutively activated MKK1 of each ERK2 mutant was evaluated and compared with the wild type protein. First, to establish that the CD/ED or FRS mutations did not affect the overall ERK2 protein structure, circular dichroism was utilized to determine changes in secondary structure. As shown, all mutant ERK2 proteins were observed to have CD spectra similar to that of the wild type protein, indicating overall structural similarity (Fig. 3A).
FIGURE 3.
Activity of wild type and mutant ERK2 proteins. A, circular dichroism spectral analysis (190–260 nm) of secondary structure of wild type and mutants L198A/L235A, T157A/D319N, and T157A/L198/L235A (1 μm). B, radioactive phosphate incorporation into wild type and mutant ERK2 proteins following incubation with active MKK1 as a function of time. C, phosphorylation of myelin basic protein over time following incubation with wild type or mutant ERK2 proteins plus active MKK1. D, normalized steady state interaction of wild type ERK2 or the L198A/L235A mutant (analyte concentration from 0 to 1.0 μm) with myelin basic protein.
To further determine whether the mutant ERK2 proteins (T157A/D319N, L198A/L235A, and T157A/L198A/L235A) were phosphorylated to the same extent as the wild type protein, MKK1-mediated phosphorylation of ERK2 proteins was monitored over time. As shown in Fig. 3B, phosphorylation of the CD/ED domain mutant or the FRS mutant was decreased compared with the wild type protein. Mutations in both the ED and FRS docking domains further decreased the extent of MEK1-induced phosphorylation (Fig. 3B). These findings indicate that the mutations inhibit MKK1 interactions. The activity of the ERK2 proteins was also compared by monitoring the phosphorylation of MBP as a nonspecific kinase substrate (18). Whereas the CD/ED domain mutant showed MBP kinase activity similar to that of the wild type protein, mutations in the FRS caused a significant decrease in MBP phosphorylation (Fig. 3C). The decreased MBP kinase activity was not due to binding defects as SPR analysis showed that MBP interacted similarly with wild type ERK2 or the FRS mutant (L198A/L235A) with KD values of 0.67 and 0.55 μm, respectively (Fig. 3D). These data suggest that FRS mutations do not affect MBP binding interactions but cause defects in ERK2 catalytic activity.
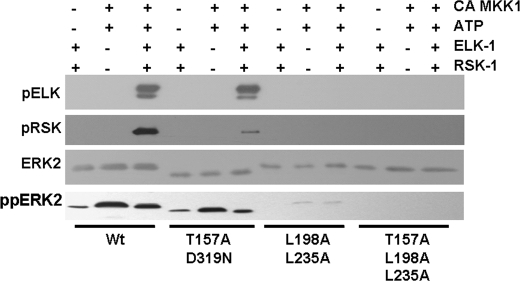
Given the effects of the FRS docking domain mutations on MBP kinase activity, we next tested the ability of the ERK mutants to phosphorylate ELK-1 and RSK-1. As shown in Fig. 4, mutations in the CD/ED domain inhibited RSK-1 phosphorylation at Thr-574 but had little effect on ELK-1 phosphorylation at Ser-383 (Fig. 4). This supports the CD/ED domain as the major determinant of RSK-1 interactions with and phosphorylation by ERK2. In addition, these findings support previous reports that the CD domain on ELK-1 does not play a major role in mediating ERK phosphorylation at Ser-383 (31). ERK2 mutated at the FRS docking domain had no detectable activity toward RSK-1 or ELK-1 (Fig. 4). Although the FRS mutant binds to RSK-1 equally as well as wild type ERK2 (Fig. 2), this interaction is non-productive and does not allow phosphate transfer. Moreover, the lack of MBP kinase activity of the FRS mutants suggests that Leu-198 and Leu-235 are important for general ERK2 catalysis and phosphorylation of substrates.
FIGURE 4.
Analysis of ELK-1 and RSK-1 phosphorylation by wild type or mutant ERK2 proteins. ELK-1 and RSK-1 were incubated with wild type ERK2 or the T157A/D319N, L198A/L235A, or T157A/L198A/L235A mutants in the presence and absence of constitutively active MKK1 (CA MKK1) and 100 μm ATP for 60 min at 30 °C. The reaction was separated by SDS-PAGE and immunoblotted for phosphorylated ELK-1 (pELK) and RSK-1 (pRSK), ERK2, and dually phosphorylated ERK2 (ppERK2).
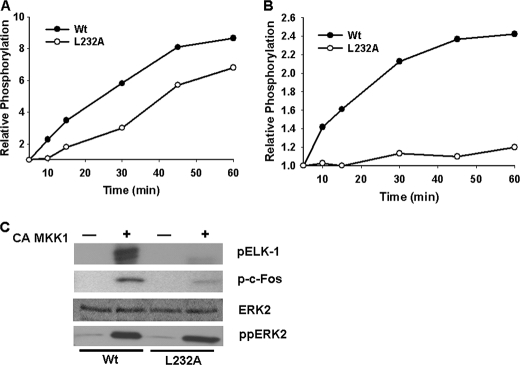
Residue Leu-232 in the FRS was determined to be important for ERK2 interaction with c-Fos but not ELK-1. To evaluate the effect of this mutant on c-Fos phosphorylation, the enzymatic activity of this mutant was compared with that of the wild type protein. First, the rate of MKK1-mediated phosphorylation of the ERK2 L232A mutant was decreased compared with the wild type protein (Fig. 5A). Second, the ability of ERK2 L232A to phosphorylate MBP was greatly reduced as compared with wild type (Fig. 5B). Although SPR analysis revealed that wild type and ERK2 mutant L232A had a similar binding affinity to ELK-1, the ability of the L232A mutant to phosphorylate ELK-1 was inhibited. Likewise, phosphorylation of c-Fos was also inhibited with the ERK2 L232A mutation (Fig. 5C). These findings indicate that residues within the FRS are important not only for substrate interactions but also for enzyme catalysis.
FIGURE 5.
Analysis of ELK-1 and c-Fos phosphorylation by wild type ERK2 or L232A mutant. A, radioactive phosphate incorporation into ERK2 wild type or mutant L232A following incubation with active MKK1 as a function of time. B, phosphorylation of myelin basic protein over time following incubation with wild type or mutant L232A and constitutively active MKK1 (CA MKK1). C, ELK-1 and c-Fos were incubated with wild type ERK2 or the L232A mutant in the presence and absence of constitutively active MKK1 and [γ-32P]ATP for 60 min at 30 °C. The top, middle, and lower panels show phosphate incorporation into ELK-1 (pELK-1) or c-Fos (p-c-Fos) as determined by autoradiography, total ERK2 protein following staining with Coomassie Blue, and immunoblot analysis of phosphorylated ERK2 (ppERK2), respectively.
Binding Interactions of Active ERK2 with Substrates
The next objective was to evaluate whether the activation status of ERK2 affected substrate binding interactions. Previous studies indicate that phosphorylated ERK2 can both promote and inhibit interactions with proteins (21, 53). Moreover, activated ERK binding affinity to cytosolic substrates may decrease to allow translocation to the nucleus (54). Using SPR, we determined whether the activation status of ERK2 plays a role in protein-protein interactions with substrate proteins. The steady state dissociation constants (KD) for substrate interactions with unphosphorylated or phosphorylated ERK2 were determined by plotting steady state binding levels as a function of analyte concentration (Table 2). As shown, RSK-1 and c-Fos interactions were not affected by the activation status of ERK2. However, the KD for the interaction with ELK-1 increased by more than 40-fold with activated ERK2, and no binding was observed with stathmin. Although binding affinities are reduced with ELK-1 and stathmin, activated ERK is still capable of phosphorylating these substrates in vitro (data not shown). These data suggest that each ERK substrate may be unique in its preference for active or inactive ERK2 and that inactive ERK2 may coordinate substrates into signaling complexes prior to activation.
TABLE 2.
Steady state binding affinity (KD) of substrates to inactive unphosphorylated (ERK2) or active phosphorylated ERK2 (ppERK2)
Substrates were immobilized on a CM5 chip with inactive unphosphorylated ERK2 or active phosphorylated ERK2 analyte concentrations ranging from 0 to 1.0 μm. Steady state binding affinities (μm) are the mean and S.E. from three to five independent determinations.
| ERK2 | ppERK2 | |
|---|---|---|
| ELK-1 | 0.25 ± 0.2 | >10 |
| RSK-1 | 0.15 ± 0.01 | 0.12 ± 0.04 |
| c-Fos | 0.97 ± 0.2 | 1.3 ± 0.2 |
| Stathmin | 0.85 ± 0.2 | NBa |
a No binding interaction.
DISCUSSION
In these studies, SPR was utilized to quantify ERK2 interactions with substrates and address the question of how these interactions contribute to phosphate transfer. We demonstrate that ELK-1, RSK-1, c-Fos, and stathmin binding affinity (KD) is in the low to submicromolar range for the interactions of inactive ERK2. This is consistent with low micromolar ERK2 cellular concentrations that have been reported previously (55). We have also identified the relative contributions of the two known ERK docking domains, CD/ED and FRS, to substrate binding. However, our findings demonstrate that binding interactions do not necessarily correlate with effects on substrate phosphorylation. For example, in the case of RSK-1, mutations in the CD/ED domain inhibited both binding affinity to ERK2 and phosphorylation. Based on the large size of RSK-1, it was predicted that other docking sites may form with ERK2. Although this may be true for other phosphorylation sites, mutations in the CD/ED domain appeared to be sufficient to block most of the ERK2-mediated phosphorylation of RSK-1 at Thr-573. In contrast, mutations in the FRS domain (Leu-198 and Leu-235) had no effect on RSK-1 binding interactions, which was to be expected as RSK-1 has not been shown to have a DEF motif. However, these FRS mutants were potent inhibitors of the ability of ERK2 to phosphorylate RSK-1 as well as other substrates tested. These data indicate that FRS residues have global effects on the catalytic functions of ERK and its ability to phosphorylate substrates. FRS residues may also be involved in MKK1/2 protein phosphorylation of ERK2. This was indicated in Fig. 3 where mutations in the CD/ED and FRS domains inhibited the ability of MKK1 to phosphorylate ERK2. Mutations in Leu-198 of the FRS have also been shown to affect ERK2 activation in cell-based assays (29).
The ERK substrate ELK-1 contains both DEJL and DEF motifs, which interact with both the CD/ED and the FRS on ERK proteins. It has been proposed that the FRS mediates ELK-1 phosphorylation of Ser-383 and that the CD/ED domain is involved in other phosphorylation events (31). Although mutations in the CD/ED domain in our studies inhibit ELK-1 interactions in agreement with the previously mentioned studies, the ability for ELK-1 to be phosphorylated on Ser-383 was largely unaffected. Mutations of the FRS caused a larger degree of binding inhibition compared with mutations in the CD/ED domain (∼20- versus 6-fold decrease for FRS versus CD/ED domain, respectively; Table 1), and mutations in both docking domains resulted in complete loss of the protein-protein interaction between ELK-1 and ERK2. This functionally additive or perhaps synergistic effect of the CD/ED domain has been observed previously in kinase assays (31, 56). Thus, although it can also be concluded that both docking domains play a role in ELK-1 interaction, the FRS domain appears to dominate in regard to phosphorylation events at Ser-383.
The results from the SPR analysis of the interactions of c-Fos and ERK2 were unexpected. Although the binding affinity of c-Fos was similar with ERK2 wild type and the individual mutations at the CD/ED domains and mutations at Leu-198 and Leu-235 in the FRS, no interactions were observed with ERK2 containing the L232A mutation. Decreased c-Fos phosphorylation have been reported previously with immunoprecipitated ERK2 containing a mutation at Leu-198, Leu-232, or Tyr-261 in the FRS (29). Our data suggest that c-Fos interactions with ERK2 require the Leu-232 residue in the FRS. Although ERK2 mutations at Leu-198 and Leu-235 did not affect c-Fos binding, the addition of another mutation at Thr-157 completely abolished c-Fos interactions with ERK2. It was initially thought that these three mutations caused a distortion in ERK2 structure that affected substrate interactions. However, circular dichroism data suggested that the overall structures of the mutant proteins were similar to wild type ERK2. Moreover, the lack of effects of the triple ED and FRS domain mutant on ERK2 binding to stathmin also supports a largely intact ERK2 structure. One possibility is that the mutations are creating a localized structural change that is integral to c-Fos binding. These findings also highlight the exquisite selectivity and sensitivity substrates have for ERK2 binding interactions.
The activation status of ERK1/2 proteins has been implicated in regulating the interactions with substrate proteins and other binding partners (21, 53, 54). For example, ELK-1 interactions with active ERK2 were 2-fold higher than with inactive ERK as determined using GST pulldown assays (21). However, the antiapoptotic PEA-15 protein reportedly can bind to inactive ERK2 10-fold more strongly than it does to active ERK2 (53). Although ERK2 activation did not appear to affect interactions with RSK-1 and c-Fos, ELK-1 and stathmin binding were markedly reduced with active ERK2 as compared with binding with inactive ERK2. It has been suggested in previous studies that active ERK binding to cytosolic substrates is reduced to allow nuclear translocation (54). In these studies, cell lysates were incubated with active or inactive GST-tagged ERK2, and interacting proteins were identified by immunoblotting or mass spectrometry. Although immunoblots suggested that ELK-1 and c-Fos interactions did not change with the ERK2 activation status, RSK-1 interactions with active ERK2 were decreased (54). Although our c-Fos data are in agreement with these studies, the discrepancies in ELK-1 and RSK-1 may be due to a lack of direct quantitative comparisons. An advantage of SPR analysis is that it allows for quantitative determination of molecular interactions in an isolated system.
In conclusion, the current studies provide the first quantitative analysis of ERK2 interactions with substrate proteins in a label-free environment. The determination of residues within ERK2 docking domains that facilitate substrate binding provides insight into their role in mediating phosphorylation events. Our findings demonstrate that changes in binding interactions do not always result in changes in substrate phosphorylation and that efficient catalysis also requires proper protein-protein orientation. Nonetheless, opportunities exist to utilize ERK-substrate binding interactions in the development of substrate-selective inhibitors that disrupt clinically relevant protein-protein interactions. For example, RSK-1 is an important mediator of cell proliferation and survival, and agents that target ERK sites involved in regulating RSK-1 could have utility as novel chemotherapeutics.
Acknowledgments
We thank Dr. Natalie Ahn (University of Colorado, Boulder) for the erk2, mek1, rsk-1, and elk-1 plasmids and Dr. Kevin Dalby (University of Texas, Austin) for the stathmin plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant CA120215.
- RSK-1
- p90 ribosomal S6 kinase-1
- DEJL
- docking site for ERK and JNK, LXL
- DEF
- docking site for ERK, FXF
- CD
- common docking
- FRS
- F-site recruitment site
- SPR
- surface plasmon resonance
- MBP
- myelin basic protein.
REFERENCES
- 1. Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. (2001) Endocr. Rev. 22, 153–183 [DOI] [PubMed] [Google Scholar]
- 2. Cobb M. H., Robbins D. J., Boulton T. G. (1991) Curr. Opin. Cell Biol. 3, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 3. Hong S. K., Yoon S., Moelling C., Arthan D., Park J. I. (2009) J. Biol. Chem. 284, 33006–33018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chuderland D., Seger R. (2005) Mol. Biotechnol. 29, 57–74 [DOI] [PubMed] [Google Scholar]
- 5. Kolch W. (2005) Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 6. Yoon S., Seger R. (2006) Growth Factors 24, 21–44 [DOI] [PubMed] [Google Scholar]
- 7. McCubrey J. A., Steelman L. S., Chappell W. H., Abrams S. L., Wong E. W., Chang F., Lehmann B., Terrian D. M., Milella M., Tafuri A., Stivala F., Libra M., Basecke J., Evangelisti C., Martelli A. M., Franklin R. A. (2007) Biochim. Biophys. Acta 1773, 1263–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roberts P. J., Der C. J. (2007) Oncogene 26, 3291–3310 [DOI] [PubMed] [Google Scholar]
- 9. McCubrey J. A., Milella M., Tafuri A., Martelli A. M., Lunghi P., Bonati A., Cervello M., Lee J. T., Steelman L. S. (2008) Curr. Opin. Investig. Drugs 9, 614–630 [PubMed] [Google Scholar]
- 10. Friday B. B., Adjei A. A. (2008) Clin. Cancer Res. 14, 342–346 [DOI] [PubMed] [Google Scholar]
- 11. Akella R., Moon T. M., Goldsmith E. J. (2008) Biochim. Biophys. Acta 1784, 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muñoz J. J., Tárrega C., Blanco-Aparicio C., Pulido R. (2003) Biochem. J. 372, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zúñiga A., Torres J., Ubeda J., Pulido R. (1999) J. Biol. Chem. 274, 21900–21907 [DOI] [PubMed] [Google Scholar]
- 14. Martin M. C., Allan L. A., Mancini E. J., Clarke P. R. (2008) J. Biol. Chem. 283, 3854–3865 [DOI] [PubMed] [Google Scholar]
- 15. Bardwell L. (2006) Biochem. Soc. Trans. 34, 837–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharrocks A. D., Yang S. H., Galanis A. (2000) Trends Biochem. Sci. 25, 448–453 [DOI] [PubMed] [Google Scholar]
- 17. Tanoue T., Adachi M., Moriguchi T., Nishida E. (2000) Nat. Cell Biol. 2, 110–116 [DOI] [PubMed] [Google Scholar]
- 18. Jacobs D., Glossip D., Xing H., Muslin A. J., Kornfeld K. (1999) Genes Dev. 13, 163–175 [PMC free article] [PubMed] [Google Scholar]
- 19. Tanoue T., Maeda R., Adachi M., Nishida E. (2001) EMBO J. 20, 466–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahn N. G. (2009) Nat. Struct. Mol. Biol. 16, 1004–1005 [DOI] [PubMed] [Google Scholar]
- 21. Lee T., Hoofnagle A. N., Kabuyama Y., Stroud J., Min X., Goldsmith E. J., Chen L., Resing K. A., Ahn N. G. (2004) Mol. Cell 14, 43–55 [DOI] [PubMed] [Google Scholar]
- 22. MacKenzie S. J., Baillie G. S., McPhee I., Bolger G. B., Houslay M. D. (2000) J. Biol. Chem. 275, 16609–16617 [DOI] [PubMed] [Google Scholar]
- 23. Murphy L. O., Smith S., Chen R. H., Fingar D. C., Blenis J. (2002) Nat. Cell Biol. 4, 556–564 [DOI] [PubMed] [Google Scholar]
- 24. Polychronopoulos S., Verykokakis M., Yazicioglu M. N., Sakarellos-Daitsiotis M., Cobb M. H., Mavrothalassitis G. (2006) J. Biol. Chem. 281, 25601–25611 [DOI] [PubMed] [Google Scholar]
- 25. Monje P., Marinissen M. J., Gutkind J. S. (2003) Mol. Cell. Biol. 23, 7030–7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen R. H., Juo P. C., Curran T., Blenis J. (1996) Oncogene 12, 1493–1502 [PubMed] [Google Scholar]
- 27. Okazaki K., Sagata N. (1995) EMBO J. 14, 5048–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen R. H., Abate C., Blenis J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10952–10956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dimitri C. A., Dowdle W., MacKeigan J. P., Blenis J., Murphy L. O. (2005) Curr. Biol. 15, 1319–1324 [DOI] [PubMed] [Google Scholar]
- 30. Smith J. A., Poteet-Smith C. E., Malarkey K., Sturgill T. W. (1999) J. Biol. Chem. 274, 2893–2898 [DOI] [PubMed] [Google Scholar]
- 31. Fantz D. A., Jacobs D., Glossip D., Kornfeld K. (2001) J. Biol. Chem. 276, 27256–27265 [DOI] [PubMed] [Google Scholar]
- 32. Biondi R. M., Nebreda A. R. (2003) Biochem. J. 372, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gavin A. C., Nebreda A. R. (1999) Curr. Biol. 9, 281–284 [DOI] [PubMed] [Google Scholar]
- 34. Pawson T., Nash P. (2003) Science 300, 445–452 [DOI] [PubMed] [Google Scholar]
- 35. Fernandes N., Allbritton N. L. (2009) Biochem. Biophys. Res. Commun. 387, 414–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ley R., Hadfield K., Howes E., Cook S. J. (2005) J. Biol. Chem. 280, 17657–17663 [DOI] [PubMed] [Google Scholar]
- 37. Callaway K., Rainey M. A., Dalby K. N. (2005) Biochim. Biophys. Acta 1754, 316–323 [DOI] [PubMed] [Google Scholar]
- 38. Canagarajah B. J., Khokhlatchev A., Cobb M. H., Goldsmith E. J. (1997) Cell 90, 859–869 [DOI] [PubMed] [Google Scholar]
- 39. Hoofnagle A. N., Stoner J. W., Lee T., Eaton S. S., Ahn N. G. (2004) Biophys. J. 86, 395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shapiro P. S., Whalen A. M., Tolwinski N. S., Wilsbacher J., Froelich-Ammon S. J., Garcia M., Osheroff N., Ahn N. G. (1999) Mol. Cell. Biol. 19, 3551–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Richards S. A., Fu J., Romanelli A., Shimamura A., Blenis J. (1999) Curr. Biol. 9, 810–820 [DOI] [PubMed] [Google Scholar]
- 42. Curmi P. A., Maucuer A., Asselin S., Lecourtois M., Chaffotte A., Schmitter J. M., Sobel A. (1994) Biochem. J. 300, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ferguson H. A., Goodrich J. A. (2001) Nucleic Acids Res. 29, E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cornish-Bowden A. (2001) Methods 24, 181–190 [DOI] [PubMed] [Google Scholar]
- 45. Schuck P., Minton A. P. (1996) Trends Biochem. Sci. 21, 458–460 [DOI] [PubMed] [Google Scholar]
- 46. Phillips S. K., Cheng Q. J. (2008) in Molecular Biomethods Handbook (Walker J. M., Rapley R. eds) 2nd Ed., pp. 809–820, Humana Press, Totowa, NJ [Google Scholar]
- 47. Barsyte-Lovejoy D., Galanis A., Sharrocks A. D. (2002) J. Biol. Chem. 277, 9896–9903 [DOI] [PubMed] [Google Scholar]
- 48. Sheridan D. L., Kong Y., Parker S. A., Dalby K. N., Turk B. E. (2008) J. Biol. Chem. 283, 19511–19520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou T., Sun L., Humphreys J., Goldsmith E. J. (2006) Structure 14, 1011–1019 [DOI] [PubMed] [Google Scholar]
- 50. Zhou B., Wu L., Shen K., Zhang J., Lawrence D. S., Zhang Z. Y. (2001) J. Biol. Chem. 276, 6506–6515 [DOI] [PubMed] [Google Scholar]
- 51. Yang S. H., Yates P. R., Whitmarsh A. J., Davis R. J., Sharrocks A. D. (1998) Mol. Cell. Biol. 18, 710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lovrić J., Dammeier S., Kieser A., Mischak H., Kolch W. (1998) J. Biol. Chem. 273, 22848–22855 [PubMed] [Google Scholar]
- 53. Callaway K., Abramczyk O., Martin L., Dalby K. N. (2007) Biochemistry 46, 9187–9198 [DOI] [PubMed] [Google Scholar]
- 54. Chuderland D., Marmor G., Shainskaya A., Seger R. (2008) J. Biol. Chem. 283, 11176–11188 [DOI] [PubMed] [Google Scholar]
- 55. Fujioka A., Terai K., Itoh R. E., Aoki K., Nakamura T., Kuroda S., Nishida E., Matsuda M. (2006) J. Biol. Chem. 281, 8917–8926 [DOI] [PubMed] [Google Scholar]
- 56. Galanis A., Yang S. H., Sharrocks A. D. (2001) J. Biol. Chem. 276, 965–973 [DOI] [PubMed] [Google Scholar]