Abstract
Human group IIA-secreted phospholipase A2 (sPLA2-IIA) is an important regulator of cytokine-mediated inflammatory responses in both in vitro and in vivo models of rheumatoid arthritis (RA). However, treatment of RA patients with sPLA2-IIA inhibitors shows only transient benefit. Using an activity-impaired sPLA2-IIA mutant protein (H48Q), we show that up-regulation of TNF-dependent PGE2 production and cyclooxygenase-2 (COX-2) induction by exogenous sPLA2-IIA in RA fibroblast-like synoviocytes (FLSs) is independent of its enzyme function. Selective cytosolic phospholipase A2-α (cPLA2-α) inhibitors abrogate TNF/sPLA2-IIA-mediated PGE2 production without affecting COX-2 levels, indicating arachidonic acid (AA) flux to COX-2 occurs exclusively through TNF-mediated activation of cPLA2-α. Nonetheless, exogenous sPLA2-IIA, but not H48Q, stimulates both AA mobilization from FLSs and microparticle-derived AA release that is not used for COX-2-dependent PGE2 production. sPLA2-IIA-mediated AA production is inhibited by pharmacological blockade of sPLA2-IIA but not cPLA2-α. Exogenous H48Q alone, like sPLA2-IIA, increases COX-2 protein levels without inducing PGE2 production. Unlike TNF, sPLA2-IIA alone does not rapidly mobilize NF-κB or activate phosphorylation of p38 MAPK, two key regulators of COX-2 protein expression, but does activate the ERK1/2 pathway. Thus, sPLA2-IIA regulates AA flux through the cPLA2-α/COX-2 pathway in RA FLSs by up-regulating steady state levels of these biosynthetic enzymes through an indirect mechanism, rather than direct provision of substrate to the pathway. Inhibitors that have been optimized for their potency in enzyme activity inhibition alone may not adequately block the activity-independent function of sPLA2-IIA.
Keywords: Arachidonic Acid, Cyclooxygenase (COX) Pathway, Inflammation, Phospholipase A, Prostaglandins, Enzyme Function, Human Fibroblast-like Synoviocytes, Microparticles, Rheumatoid Arthritis
Introduction
Phospholipase A2 (PLA2)2 enzymes regulate the provision of arachidonic acid (AA) to the cyclooxygenase (COX) and lipoxygenase biosynthetic pathways, the products of which, in turn, are critical autocrine and paracrine regulators of diverse physiological processes in mammals. Of the 23 currently known mammalian PLA2 enzymes, in vivo gene deletion studies in mice have established the widely expressed intracellular enzyme cytosolic PLA2-α (cPLA2-α, Group IVA PLA2) as an important enzyme in providing AA substrate to COX and lipoxygenase because deletion of this gene product abrogates eicosanoid production in cells stimulated ex vivo (1, 2). Significantly, cPLA2-α gene deletion markedly reduces disease severity in the collagen-induced arthritis model of rheumatoid arthritis (RA), suggesting cPLA2-α has a key role in the pathogenesis of RA (2).
The contribution of the remaining 18 PLA2 enzymes to AA metabolism and to immune-mediated, inflammatory pathology is less clear. Macrophages from Group V secreted PLA2 (sPLA2)-deficient mice show impaired production of both COX- and lipoxygenase-derived eicosanoid products in response to the inflammatory stimulus zymosan (3), whereas deletion of Group X sPLA2 results in impaired eicosanoid release into the lungs following ovalbumin challenge (4). sPLA2-IIA, the best studied of the 10 mammalian sPLA2 enzymes, is not expressed in certain mouse strains with restricted expression in others compared with either rats or humans (5, 6), making classical genetic deletion experiments impractical for this enzyme. Despite this, a proinflammatory role for sPLA2-IIA in arthritis has been confirmed by recent genetic studies showing that arthritis is attenuated in sPLA2-IIA knock-out mice, relative to congenic wild-type mice, in a K/BxN serum transfer model of arthritis (7). Surprisingly, these studies also showed that Group V sPLA2 has an anti-inflammatory role in this model of arthritis (7). Transgenic expression of human sPLA2-IIA in mice results in spontaneous atherosclerosis (8) that is transferable to non-transgenic mice by transplantation of transgenic bone marrow (9). Thus, aberrant expression of the human enzyme, in vivo, induces inflammatory pathology. These animals do not develop spontaneous arthritis (10), however, transgenic expression of human sPLA2-IIA leads to earlier onset and more severe arthritis in a TNF transgenic, spontaneous arthritis model (11) implicating aberrant expression of sPLA2-IIA as a positive regulator of cytokine-mediated joint inflammation. Furthermore, transgenic expression of human sPLA2-IIA in mice results in increased severity in the K/BxN serum transfer arthritis model (7).
sPLA2-IIA is markedly induced in the serum of patients with immune-mediated conditions, including RA and in tissues of patients with certain cancers (12–14). Serum enzyme activity and concentration correlate with disease severity in RA (15), synovial tissue expression of sPLA2-IIA correlates with histological markers of inflammation (16), and several other sPLA2 enzymes are also expressed in RA synovial tissue (17) and synovial fluid (7). Exogenous addition of sPLA2-IIA to cultured RA synovial cells, at concentrations found in the synovial fluids of RA patients, enhances both TNF-stimulated PGE2 production and up-regulation of the inducible cyclooxygenase, COX-2 by an unknown mechanism (18). However, blockade of enzyme activity with a potent inhibitor of sPLA2-IIA, Group V and Group X sPLA2 (in a randomized, double-blinded, placebo-controlled study) shows only transient benefit in patients with active RA (19)). Thus, despite compelling preclinical and early phase clinical data (19), the utility of sPLA2-IIA blockade in the treatment of arthritis is not well supported by the most recent clinical evidence.
Here we show for the first time in cells that mediate inflammatory synovitis in RA that although exogenous sPLA2-IIA contributes to AA flux in these cells in culture, exogenous sPLA2-IIA-amplified cytokine-mediated PGE2 production is sPLA2-IIA enzyme activity-independent and is thus mediated by a signaling function of the enzyme that indirectly up-regulates levels of the cPLA2-α/COX-2 pathway enzymes.
EXPERIMENTAL PROCEDURES
Materials
sPLA2-IIA protein was expressed, purified, and quantified as described (18). cPLA2-α inhibitors pyrrolidine-1 (20, 21) and pyrrophenone (22, 23) were synthesized as described. Pyrrolidine-1 inhibited purified, recombinant cPLA2-α in a vesicle assay with an IC50 of 70 nm, and AA release in ionomycin-stimulated Madin-Darby canine kidney cells with an IC50 of 800 nm. It showed no detectable inhibition of purified, recombinant human sPLA2-IIA, Group V or Group X sPLA2 at 10 μm concentration, and no physiologically relevant inhibition of recombinant cytosolic PLA2-γ or the calcium-independent PLA2, iPLA2β (20). Pyrrophenone inhibited recombinant cPLA2-α in a mixed-micelle assay with an IC50 of 80 nm (24) without significant inhibition of all five remaining human cPLA2 isoforms in this assay.3 It inhibited AA release and PGE2 production in ionophore-stimulated THP-1 cells with an IC50 of 25 nm (25) and had no appreciable inhibition of murine cytosolic PLA2-β (24) or purified, recombinant human Group IB sPLA2 or sPLA2-IIA at 200 μm (25). The sPLA2 inhibitor c(2NapA)LS(2NapA)R was synthesized as previously described (Auspep, Melbourne, Australia) (26). LY311727 was a kind gift from Eli Lilly and Co. (Indianapolis, IN). The iPLA2-β inhibitor, bromoenol lactone, was obtained from Sigma.
Construction of sPLA2-IIA Catalytic Site Mutant H48Q
The sPLA2-IIA cDNA (a kind gift from J. Seilhamer) (27) was subcloned into pBlueScribe(+) and histidine 48 was substituted for glutamine by oligonucleotide-directed mutagenesis of the His codon (non-coding strand oligonucleotide sequence 5′-AGCAACAGTCCTGAGTGACAC-3′). Mutagenesis was carried out with an in vitro mutagenesis kit (Amersham Biosciences) based on the method of Eckstein and co-workers (28). The nucleotide sequence of the mutagenized construct was confirmed and the cDNA was cloned into the zinc-inducible mammalian expression vector pMTSV40polyABam (pLEN) (29). The resultant plasmid (pMIK-1) was co-transfected with pRSV2-neo, carrying a G418 resistance gene, into Chinese hamster ovary (CHO) cells by calcium phosphate precipitation. Following several rounds of G418 selection, the resultant cell culture pool was used to express H48Q, and the protein was purified from conditioned medium by affinity chromatography (AKTA Explorer purification system, GE Healthcare) as described for sPLA2-IIA (18), and quantified by ELISA (12).
sPLA2 Enzyme Activity Assay
sPLA2 enzyme activity was measured with a colorimetric microtiter plate, mixed micelle assay (Cayman Chemical, Ann Arbor, MI) using diheptanoylthiophosphatidylcholine as substrate (30) with the following modifications. Briefly, enzyme (10 μl, 2.5 μg/ml, sPLA2-IIA or H48Q) diluted in assay buffer (10 mm CaCl2, 100 mm KCl, 0.3 mm Triton X-100, 1 mg/ml of BSA, 25 mm Tris-HCl, pH 7.5) was added to each well containing the free-thiol detection reagent 5,5′-dithio-bis-(2-nitrobenzoic acid) (10 μl, 10 mm 5,5′-dithio-bis-(2-nitrobenzoic acid) in 0.4 m Tris-HCl, pH 8.0) and 5 μl of assay buffer. Phospholipid substrate was reconstituted in assay buffer to a final concentration of 1.66 mm with vortexing until the solution was clear, then preheated to 40 °C. Assays were performed at 40 °C, started by addition of substrate (200 μl/well) and A405 was measured every 3 min over a 60-min time course (Spectramax 250 microtiter plate reader, Molecular Devices, Sunnyvale CA). Assays were performed in triplicate relative to blank wells containing assay buffer and data were analyzed using SoftMax Pro version 1.1 software in kinetic mode.
Fibroblast-like Synoviocytes
Synovial tissue was obtained from patients undergoing joint surgery and who were diagnosed with RA according to American Rheumatism Association criteria (31) using procedures approved by the St. Vincent's Hospital Ethics Committee. Fibroblast-like synoviocyte (FLS) cultures were established as described (18) and used between passages 3 and 10. Cells, CD14-negative and 4-prolylhydroxylase-positive by immunohistochemistry and CD21-negative by RT-PCR, were grown in Ham's/DMEM containing 10% FBS and used at 80–90% confluence.
PGE2 Assay
Cells, grown in 96-well plates were stimulated, medium was harvested and stored at −80 °C prior to PGE2 assay. Cells were lysed in wells by resuspension in ice-cold lysis buffer (40 μl) containing 1% (v/v) Nonidet P-40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 1 mm EGTA, 50 μg/ml of aprotinin, 200 μm leupeptin, 1 mm PMSF in PBS. Lysates from triplicate experiments were combined and stored at −80 °C prior to protein determination (Bio-Rad DC Protein Assay, Bio-Rad). PGE2 in medium was determined by enzyme immunoassay (Cayman Chemical) as previously described (18) and expressed as picograms of PGE2/mg of total cellular protein.
Nuclear Protein Extracts
FLSs, grown in 150-cm2 flasks, were stimulated and nuclear extracts were prepared as described (32) with minor modification as follows. Cells were harvested with trypsin/EDTA, centrifuged (4000 × g, 1 min), supernatants were discarded, cell pellets were washed with PBS (1 ml), recentrifuged, and placed on ice. Cells were resuspended in ice-cold Buffer A (175 μl, 10 mm KCl, 1.5 mm MgCl2, 100 mm EDTA, 1 mm DTT, 1 μm PMSF, 100 μg/ml of aprotinin, 100 mg/ml of leupeptin, 10 mm HEPES, pH 7.9) and incubated on ice for 5 min. Nonidet P-40 (9 μl, 10% v/v) was added, samples were vortexed for 10 s, centrifuged (20 s, 13,790 × g), and supernatants were discarded. Pellets were washed gently with ice-cold buffer A (150 μl), centrifuged, and supernatants were discarded. Buffer C (40 μl, 420 mm KCl, 1.5 mm MgCl2, 100 mm EDTA, 1 mm DTT, 1 mm PMSF, 100 μg/ml of aprotinin, 100 μg/ml of leupeptin, 10 mm HEPES, pH 7.9) was added, samples were vortexed (10 s) and incubated with orbital shaking for 30 min on ice. Samples were centrifuged (13,790 × g, 15 min, 4 °C), supernatants were divided into aliquots and stored at −80 °C prior to use. Protein concentration was determined by the Bradford protein assay (Bio-Rad).
Western Blot Analysis
Cells were grown in 24-well plates, stimulated, and medium was harvested and stored at −80 °C. Cells were lysed, triplicate wells were combined and protein determinations made as described above. For phosphoprotein determination experiments, cells were grown in 75-cm2 flasks, treated, and washed once with ice-cold PBS (10 ml/flask) containing 10 mm orthovanadate. Cells were scraped into ice-cold PBS (1 ml) containing 5.3 mm EDTA, 10 mm sodium orthovanadate, 50 mm disodium β-glycerophosphate, and 10 mm NaF. Following centrifugation the cells were lysed in 1% (v/v) Nonidet P-40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 1 mm EGTA, 50 μg/ml of aprotinin, 200 μm leupeptin, 1 mm PMSF, 10 mm sodium orthovanadate, 50 mm disodium β-glycerophosphate, and 10 mm NaF in PBS. Protein (15–20 μg/well) was electrophoresed on polyacrylamide gels (4–20% BisTris (2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol) gels, Novex) according to the manufacturer's instructions and transferred to nitrocellulose as described (18). Primary Abs for cPLA2-α, COX-1, COX-2, and β-actin and secondary Abs were used as described previously (18). Murine monoclonal anti-human c-Jun Ab (Santa Cruz Biotechnology, Santa Cruz, CA; number sc-822) was used at 0.2 μg/ml and detected with sheep anti-mouse IgG-horseradish peroxidase (HRP) (GE Healthcare, 1/3000). Rabbit polyclonal Abs were used at the following concentrations: anti-human IκB-α (Santa Cruz, number sc-371), 10 ng/ml; anti-mouse IκB-β (Santa Cruz, number sc-945), 400 ng/ml; anti-human NF-κB p50 (Santa Cruz, number sc-114), 4 μg/ml; anti-human NF-κB p65 (Santa Cruz, number sc-109), 1 μg/ml; anti-human phospho-p38 MAP kinase Ab (phospho-Thr180/phospho-Tyr182, New England Biolabs, number 9211), 1/1000 dilution; anti-human p38α MAP kinase (Santa Cruz, number sc-535), 33 ng/ml, anti-human phospho-ERK (phospho-Thr202/phosphoTyr204) (Cell Signaling Technologies, number 9101), 1/10,000 dilution; and anti-rat ERK1 (Santa Cruz, number sc-94, ERK-2 cross-reactive), 10 ng/ml. These Abs were detected with donkey anti-rabbit IgG horseradish peroxidase conjugate (GE Healthcare, 1/3000 dilution). Bands were visualized using chemiluminescence (Renaissance chemiluminescent reagent Plus, New England Nuclear) with detection on x-ray film (Hyperfilm ECL, GE Healthcare). Bands were scanned (PDI densitometer, Molecular Dynamics) and density was quantified with IPLabgelH software (Macintosh version 1.5g).
NF-κB Gel Shift Assays
NF-κB binding to a double-stranded consensus NF-κB binding site oligonucleotide with top strand sequence (5′-AGTTGAGGGGACTTTCCCAGGC-3′), 5′-end-labeled with [γ-32P]ATP (GE Healthcare), and T4 polynucleotide kinase (Promega) was measured by electromobility shift assay (EMSA) as described (33). Briefly, nuclear extracts (∼5 μg of total protein), prepared as described above, were added to a binding reaction (20 μl) containing (polydeoxyinosine (dI)-deoxycytidine (dC))· (polydI-dC) (GE Healthcare) at 0.25 μg/μg of total protein, 1–2 ng of 32P-labeled double-stranded oligonucleotide probe and DNA binding buffer (20 mm HEPES, pH 7.9, 1 mm EDTA, pH 8.0, 60 mm KCl, 1 mm DTT, glycerol (12% v/v)). The binding reaction was incubated at room temperature for 30 min prior to electrophoresis on non-denaturing polyacrylamide gels (5% polyacrylamide) in 0.25 × Tris borate EDTA (TBE) buffer, pH 8.3, at 150 V for 2–3 h. Gels were dried and bands were imaged with x-ray film (X-Omat AR, Kodak, Sydney, Australia). NF-κB EMSA bands were confirmed by NF-κB cold-competitor studies and supershift of NF-κB EMSA bands, using anti-p65 and anti-p50 Abs, were performed on nuclear extracts from TNF-α-stimulated FLSs.
AA Mobilization Assays
Cells were grown in 96-well plates and labeled with [5,6,8,9,11,12,14,15-3H]AA (PerkinElmer Life Sciences; NET-298Z) in Ham's/DMEM containing 10% FBS for 16 h at 37 °C. Cells were washed 3 times in PBS containing 0.1% (w/v) BSA (fatty acid free, Sigma) and stimulated for the indicated times in Ham's/DMEM containing 0.1% (w/v) BSA. Treatments were added simultaneously with stimulants. Medium was harvested, cells were detached with trypsin/EDTA, and tritium was determined in medium and cells by scintillation counting (LS6000 TA, Beckman, Sydney, Australia). Data are expressed as % total radioactivity (cells plus medium) released into medium.
Thin Layer Chromatography
Samples (50 μl), to which unlabeled AA (1 μg) had been added, were spotted onto silica gel plates (Merck, Darmstadt, Germany), air dried, and eluted in chloroform:methanol:acetic acid:water (90:8:1:0.8) (34). Plates were air-dried and developed in iodine vapor to identify the AA spot. Eluted samples were cut into seven equal segments in order of increasing Rf value and tritium was determined by scintillation counting (LS6000 TA, Beckman, Sydney, Australia). Data for each segment were expressed as % total radioactivity recovered from each sample for each segment.
Statistical Analysis
Data were analyzed and plotted using Prism Graphpad version 4.0. Statistical significance was determined using the Student's paired t test unless otherwise stated.
RESULTS
Exogenous sPLA2-IIA Up-regulates TNF-mediated PGE2 Production and COX-2 Protein by an Activity-independent Mechanism
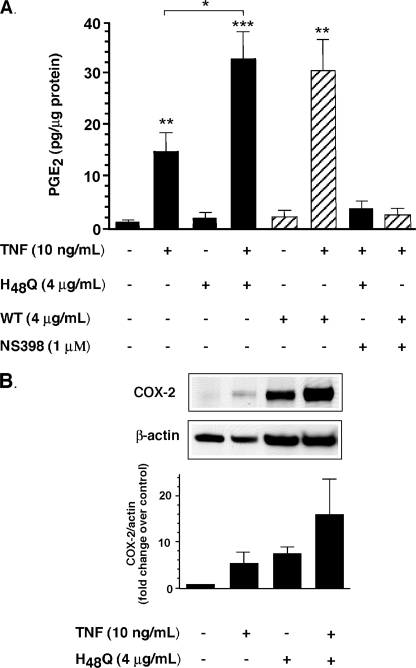
To determine whether sPLA2-IIA enzyme activity was necessary for up-regulation of TNF-dependent PGE2 production and COX-2, we constructed an “activity-impaired” mutant of sPLA2-IIA (H48Q) by site-directed mutagenesis as described under “Experimental Procedures.” Quantification of H48Q was determined relative to a sPLA2-IIA standard by ELISA (35). The ELISA was validated for H48Q by quantitative amino acid analysis of 2 independent samples of sPLA2-IIA and H48Q, followed by quantitation of each sample in the same ELISA. Equivalent concentrations were obtained for each sample by both methods (data not shown). Importantly, the mol % amino acid composition for each amino acid obtained for both sPLA2-IIA and H48Q in the amino acid analysis was not significantly different from the theoretical composition calculated from their known protein sequences (p = 1.0000, p = 0.9926 for sPLA2-IIA and H48Q respectively, Student's paired t test), confirming that both proteins were >99% pure. This mutation has been reported by others to have 2–4% residual enzyme activity (36). In our hands purified H48Q had 1% residual sPLA2-IIA activity, with a specific activity of 0.28 ± 0.12 μmol of diheptanoylthiophosphatidylcholine/min/mg of protein (data are mean ± S.D. of three experiments performed in triplicate) relative to 27.8 ± 2.3 μmol of diheptanoylthiophosphatidylcholine/min/mg of protein for sPLA2-IIA. Our purified H48Q protein lacks the proliferative capacity of purified sPLA2-IIA in the LNCaP prostate cancer cell line (37), confirming that the 1% residual enzyme activity we measure in our H48Q preparations is insufficient to recapitulate the effects of fully active sPLA2-IIA in these cells. In FLSs, H48Q alone like sPLA2-IIA, had no effect on PGE2 production (Fig. 1A) at enzyme concentrations that are found in synovial fluid (18). TNF alone resulted in a 15-fold stimulation of PGE2. H48Q increased this stimulation to ∼30-fold, as did sPLA2-IIA. In both cases PGE2 production was completely abrogated by the COX-2-selective inhibitor NS-398. In a side by side experiment with sPLA2-IIA, H48Q-mediated PGE2 production was dose-dependent with both mutant and native enzyme having no effect at concentrations below 100 ng/ml (7 nm) (data not shown). In agreement with our earlier work (18), TNF alone up-regulated steady state COX-2 protein levels (Fig. 1B). H48Q alone (Fig. 1B), as with sPLA2-IIA (18), also increased COX-2 protein, despite having no effect on PGE2 production (Fig. 1A). As we have also shown for sPLA2-IIA, H48Q, in combination with TNF, synergistically up-regulated COX-2 protein (Fig. 1B) without any effect on COX-1 (data not shown).
FIGURE 1.
Up-regulation of cytokine-dependent PG production and COX-2 does not require sPLA2-IIA enzyme activity. A, FLS cells, grown to 80–90% confluence, were stimulated with TNF (10 ng/ml), alone or in combination with the activity-impaired mutant of sPLA2-IIA (H48Q) (4 μg/ml), or with sPLA2-IIA (WT) (4 μg/ml) in the presence or absence of the COX-2-selective inhibitor NS-398 (1 μm) for 16 h in DMEM/Ham's F-12 containing 0.1% BSA. PGE2 concentration was measured in cell culture supernatants and total cellular protein was determined as described under “Experimental Procedures.” Data are combined mean ± S.E. of triplicate determinations from cell cultures derived from each of 4 patients. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Student's unpaired t test) relative to unstimulated cells unless indicated. B, cells were treated for 16 h as indicated, lysed, and protein extracts were subjected to electrophoresis and Western blot analysis as described under “Experimental Procedures.” A representative Western blot from one cell culture (RA79) is shown. Bands were quantified by densitometry as described. COX-2 density for each sample was normalized relative to β-actin density and the COX-2/β-actin ratio for each treatment was then normalized relative to control for each cell culture. Data are mean ± S.E. of three independent cell cultures.
cPLA2-α Mediates sPLA2-IIA-dependent PGE2 Production but Not COX-2 Up-regulation
To determine whether PGE2 production in response to TNF and sPLA2-IIA was dependent on cPLA2-α, we used a pharmacological approach with well characterized pyrrolidine inhibitors that selectively block human cPLA2-α activity over other human cPLA2 isoforms, iPLA2-β or sPLA2 activities (20–25). Pyrrophenone (5 μm) completely blocked the PGE2 response to TNF alone, substantially suppressed the response to TNF/sPLA2-IIA and showed a small but significant inhibition of the sPLA2-IIA/TNF response at 1 μm (Fig. 2A). Pyrrolidine-1 abrogated PGE2 production in response to sPLA2-IIA/TNF stimulation at all concentrations tested (Fig. 2B), without any effect on basal PGE2 production. However, pyrrophenone, at concentrations that abrogate PGE2 production, did not significantly block sPLA2-IIA-mediated COX-2 up-regulation (Fig. 2C). Thus provision of AA to COX-2 for both TNF-dependent and sPLA2-IIA up-regulated PGE2 production appears to be mediated by cPLA2-α.
FIGURE 2.
PGE2 production, but not COX-2 up-regulation is dependent on cPLA2-α enzyme activity. 80–90% confluent FLSs were stimulated with TNF (10 ng/ml), sPLA2-IIA (4 μg/ml) either alone or in combination in the presence or absence of the cPLA2-α-selective inhibitor (A) pyrrophenone or (B) pyrrolidine-1 at the concentrations shown for 16 h in Ham's/DMEM medium containing 0.1% BSA. Medium and cells were harvested, PGE2 in medium was determined and the protein concentration in cell lysates determined as described under “Experimental Procedures.” Data are mean ± S.E. of triplicate determinations from cell cultures derived from each of four patients. **, p < 0.01; ***, p < 0.001 (Student's unpaired t test) relative to unstimulated cells unless indicated. C, cells were grown in 24-well plates and stimulated as described above. Cells were harvested, lysates were electrophoresed, transferred to nitrocellulose, probed with Abs, and labeled proteins were visualized on x-ray film by enhanced chemiluminescence, blots were scanned and densitometry performed and analyzed as described under “Experimental Procedures.” Representative blots from one cell culture are shown. Densitometry data are mean ± S.E. normalized relative to unstimulated cells from experiments performed on cell cultures derived from three separate patients.
Effect of sPLA2-IIA on AA Mobilization
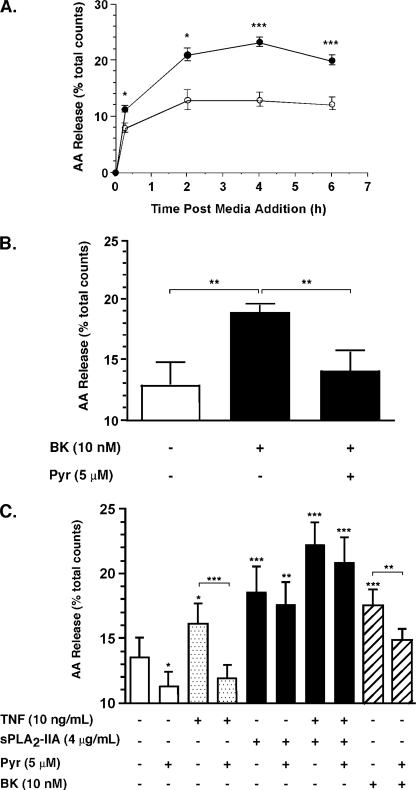
In light of these data and reports that exogenous sPLA2-IIA does not efficiently mobilize AA in attachment dependent cells in culture (38), the effect of TNF and exogenous sPLA2-IIA on AA mobilization was examined by [3H]AA release assays. First, the assay was validated by examining the response of FLSs to the known AA-mobilizing agonist bradykinin (BK) (Fig. 3A). BK stimulation (10 nm, 15 min) resulted in increased [3H]AA release from 8.0 ± 0.8% (mean ± S.E. of duplicate experiments from 4 independent cell cultures) in untreated cells to 11.3 ± 0.6% total counts incorporated (p < 0.05), consistent with a previous report (39). Release peaked by 2 h post-stimulation (21.0 ± 1.2% total counts, p < 0.05 relative to unstimulated cells). The basal level of AA release also increased rapidly with time peaking at 2 h (12.9 ± 1.8% total counts) with similar kinetics to stimulated cells. Subsequent studies were terminated 2 h post-stimulation. Under these conditions, pyrrophenone (5 μm) blocked BK-dependent AA release (Fig. 3B).
FIGURE 3.
Effect of sPLA2-IIA on AA mobilization in RA FLSs. Human synovial FLSs were labeled with [3H]AA and stimulated (A) with (closed circles) or without (open circles) BK (10 nm) or (B, C) as indicated in the presence or absence of pyrrophenone (Pyr) for 2 h prior to harvesting medium and cells for AA release determination as described under “Experimental Procedures.” Data are mean ± S.E. of two independent experiments combined, each comprising duplicate determinations from four independent cultures. *, p < 0.05; **, p < 0.05; ***, p < 0.001 (Student's paired t test) relative to control unless indicated. Total radioactivity incorporated into cells ranged from 8,185–38,367 dpm.
We next examined the effect of sPLA2-IIA, alone or in combination with TNF in the presence or absence of pyrrophenone on AA release. Pyrrophenone alone showed a small but significant reduction in basal AA release (Fig. 3C). Basal AA release was unaffected by the calcium-independent Group VIA PLA2 (iPLA2-β) inhibitor, bromoenol lactone (40) (10 μm) (data not shown). TNF stimulation resulted in a 1.2-fold increase in AA release that was abrogated by pyrrophenone. Surprisingly, at concentrations that fail to stimulate PGE2 production, sPLA2-IIA alone increased AA release by 1.5-fold and the increase was not inhibited by pyrrophenone. sPLA2-IIA in combination with TNF resulted in a 1.7-fold increase over untreated cells that was not significantly affected by pyrrophenone. As with previous experiments, BK stimulated AA release by 1.3-fold and this was inhibited by pyrrophenone.
The dose responsiveness of sPLA2-IIA-mediated AA release and the effect of selective sPLA2 inhibition on the response were then determined (Fig. 4A). sPLA2-IIA dose dependently induced AA release at concentrations above 1 μg/ml (71 nm) but was ineffective at 100 ng/ml (7 nm) concentration. Co-incubation with the selective sPLA2 inhibitor c(2NapA)LS(2NapA)R (26) (1 μm) resulted in significant inhibition of the response at a molar ratio of inhibitor to enzyme approaching 1:1 and complete inhibition of the response at a molar ratio of 3.5:1. Because this inhibitor also suppresses the activity-independent functions of exogenous sPLA2-IIA, viz. sPLA2-IIA-mediated up-regulation of cytokine-dependent PGE2 production (26), the effect of H48Q on AA release was determined in a side by side experiment with sPLA2-IIA. H48Q was ineffective at concentrations where sPLA2-IIA stimulates AA mobilization (Fig. 4B), confirming that sPLA2-IIA enzyme activity mediates the response.
FIGURE 4.
sPLA2-IIA-dependent AA mobilization requires sPLA2-IIA enzyme activity. FLSs were labeled with [3H]AA, incubated for 2 h: A, in the presence or absence of the sPLA2-IIA inhibitor c(2Nap)LS(2Nap)R (C2), at 1 μm concentration (26) and in the presence or absence of sPLA2-IIA as indicated; or B, in the presence or absence of sPLA2-IIA or the activity-impaired mutant H48Q. AA release was measured as described under “Experimental Procedures.” Data are mean ± S.E. of duplicate determinations from three to four cell cultures derived from separate patients and are representative of two independent experiments. **, p < 0.01; ***, p < 0.001 (Student's unpaired t test) relative to unstimulated cells unless indicated. Total radioactivity incorporated into cells ranged from: A, 18,733–40,749 dpm; B, 12,523–35,086 dpm.
FLSs are known to spontaneously release microparticles into culture medium (41), and purified microparticles derived from other cell types, particularly platelets, are known to amplify inflammation in arthritis models (42). It is possible that the AA mobilization measured here could reflect microparticle release from FLSs in addition to free AA. To evaluate this possibility, we examined the distribution of tritium in phospholipid (PL), arachidonic acid, and other lipid mediator fractions of conditioned medium derived from labeled cells following 2 h stimulation, by thin layer chromatography (TLC). More than half (63%) of the tritium released into supernatants of resting labeled cells remained associated with phospholipids on TLC (Table 1), whereas 20% coeluted with AA. The remaining 17% was evenly distributed between these fractions. TNF stimulation showed no change in the distribution of tritium in phospholipids and a trend to increased tritium in AA. In the presence of sPLA2-IIA, the distribution of AA in phospholipids was significantly reduced relative to unstimulated cells with a trend to increased tritium distributed evenly between AA and other lipid mediators. This distribution pattern was largely recapitulated in cells stimulated with TNF + sPLA2-IIA (Table 1).
TABLE 1.
Distribution of 3H in supernatants of FLSs as measured by thin layer chromatography
| Sample |
3H in TLC fractions (% total 3H) |
||
|---|---|---|---|
| PL (Rf = < 0.07) | AA (Rf > 0.81) | Other (0.07 ≤ Rf ≤ 0.81) | |
| [3H]AA controla | 4.6 ± 0.2b | 86 ± 1 | 9.6 ± 0.8 |
| No stimulus control | 63 ± 5 | 20 ± 9 | 17 ± 8 |
| TNF | 52 ± 5 | 39 ± 6 | 8 ± 2 |
| sPLA2-IIA | 16 ± 1c | 43 ± 17 | 41 ± 16 |
| TNF + sPLA2-IIA | 15 ± 2c | 60 ± 18 | 25 ± 16 |
a Purified [3H]AA standard. Total 3H in sample ∼6800 dpm for each experiment.
b Data are mean ± S.E. of data from three separate experiments. In the case of FLSs, data represent 3 independent FLS cultures. Total 3H in FLS samples ranged from 1169 to 2781 dpm.
c p < 0.001 relative to “no stimulus” control (two-way analysis of variance using data from cell culture experiments, Bonferonni's multiple comparison test).
Effect of sPLA2-IIA on NF-κB Mobilization, p38 MAPK, and ERK MAPK Activation
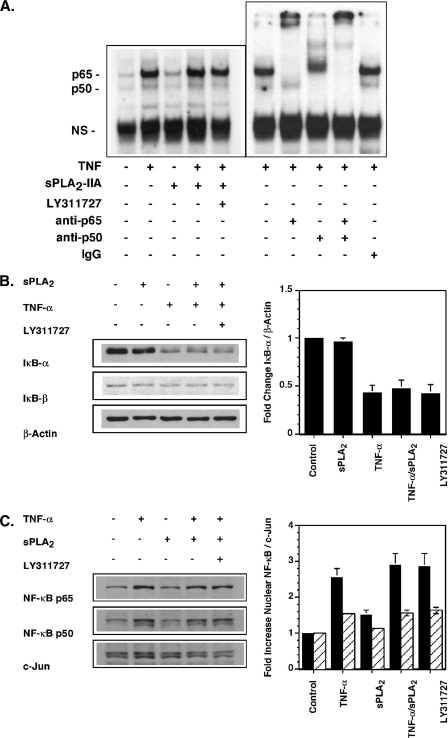
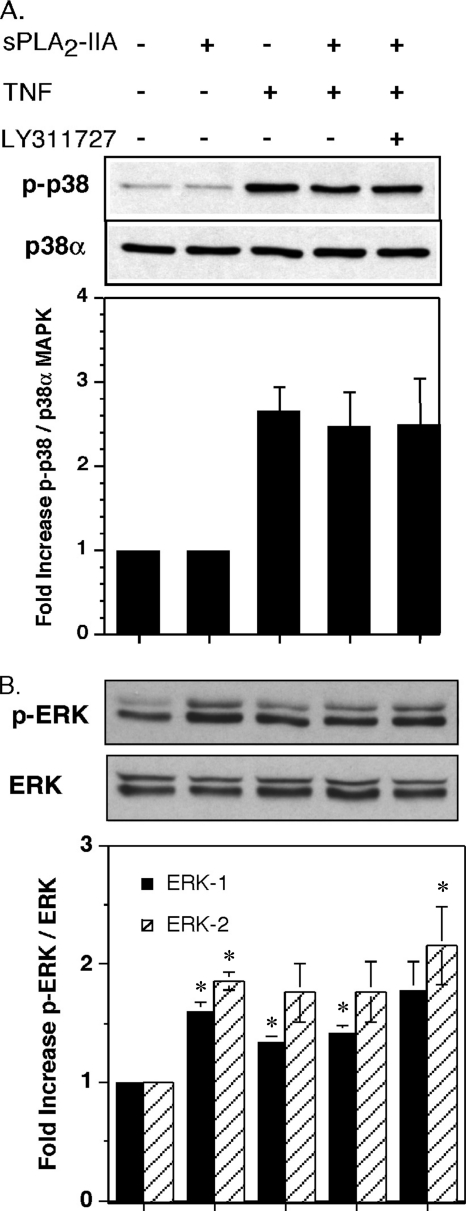
Enhanced TNF-dependent PGE2 production in the presence of sPLA2-IIA appears to result from induction of COX-2 rather than sPLA2-IIA-mediated increased AA flux through the COX-2 pathway. COX-2 protein induction in FLSs is regulated in response to certain agonists at the level of transcription via NF-κB activation (43), ERK MAPK activation (44), and/or post-transcriptionally through regulation of mRNA stability that requires phosphorylation of the MAPK p38 (45). To determine whether sPLA2-IIA was activating these pathways in FLSs, the effect of exogenous enzyme on rapid activation of the NF-κB pathway, p38, and ERK phosphorylation was determined. An EMSA was established to measure direct binding of nuclear proteins to a consensus NF-κB DNA binding sequence. Supershift assays with anti-p65 and anti-p50 Abs (Fig. 5A) and competition experiments with cold binding sequence (data not shown) demonstrated the specificity of this assay for NF-κB subunits. Unlike TNF, sPLA2-IIA alone had no effect on NF-κB binding to DNA (Fig. 5A), the mobilization of NF-κB subunits in the cytoplasm as measured by IκB-α degradation (Fig. 5B), or NF-κB subunit accumulation in the nucleus (Fig. 5C). Furthermore, sPLA2-IIA in combination with TNF had no additional effect over stimulation with TNF alone in these assays. The potent and selective sPLA2 enzyme activity inhibitor LY311727, at a concentration (10 μm) that blocks enzyme activity and also blocks COX-2 up-regulation in FLSs (17), did not modulate NF-κB mobilization by TNF/sPLA2-IIA (Fig. 5). In addition, sPLA2-IIA alone, again unlike TNF, did not induce p38 phosphorylation (Fig. 6A), nor did it modulate TNF-dependent phosphorylation. As with NF-κB mobilization, blockade of sPLA2-IIA with LY311727 had no effect on p38 phosphorylation in the presence of TNF/sPLA2-IIA (Fig. 6A). Unstimulated FLSs show significant basal ERK activation that was further stimulated by treatment with sPLA2-IIA or TNF alone (Fig. 6B). However, sPLA2-IIA did not augment TNF-dependent ERK phosphorylation, despite TNF being at a low (submaximal) concentration (50 pg/ml). In contrast to its effect of abrogating COX-2 up-regulation (18), LY311727 had no effect on TNF/sPLA2-IIA-mediated ERK activation (Fig. 6B).
FIGURE 5.
sPLA2-IIA does not activate or enhance TNF activation of the NF-κB pathway in RSF. A, EMSA. Single flasks of 90% confluent FLSs were stimulated for 1 h in DMEM/Ham's F-12 containing 0.1% BSA with TNF (10 pg/ml), sPLA2-IIA (5 μg/ml), TNF/sPLA2-IIA or TNF/sPLA2-IIA with LY311727 (10 μm). Nuclear protein extracts were prepared and binding to a radiolabeled NF-κB consensus binding sequence was determined by EMSA as described under “Experimental Procedures.” Binding specificity to detected bands was confirmed by supershift assays with anti-p65 or anti-p50 Abs relative to an isotype-matched control Ab in extracts of TNF-stimulated cells as indicated. B, IκBα degradation. Single flasks of 90% confluent FLSs (n = 3) in DMEM/Ham's F-12 containing 0.1% BSA were stimulated for 15 min with sPLA2-IIA (5 μg/ml), TNF (50 pg/ml), TNF/sPLA2-IIA or TNF/sPLA2-IIA with LY311727 (10 μm). Total cell lysates were prepared. IκBα, IκBβ, and β-actin protein were detected by Western blot analysis. The ratio of IκBα to β-actin protein was quantified by densitometry and is normalized to the ratio measured in unstimulated cells. Data are mean ± S.E. (n = 3). C, nuclear NF-κB p65 and p50 protein in FLSs. Single flasks of 90% confluent RSF (n = 3) in DMEM/Ham's F-12 containing 0.1% BSA were stimulated for 1 h with TNF (10 pg/ml), sPLA2-IIA (5 μg/ml), TNF/sPLA2-IIA or TNF/sPLA2-IIA with LY311727 (10 μm). Nuclear protein extracts were prepared. Nuclear c-Jun, NF-κB p65 and p50 protein were detected by Western blot analysis. The ratio of nuclear NF-κB p65 and p50 to c-Jun protein was quantified by densitometry and normalized to the ratio measured in unstimulated cells. Data are mean ± S.E. (n = 3 independent FLS cultures).
FIGURE 6.
sPLA2-IIA does not activate p38 MAP kinase in FLSs but activates ERK. Single flasks of 90% confluent FLSs (n = 3) in DMEM/Ham's F-12 containing 0.1% BSA were stimulated for 15 min with sPLA2-IIA (5 μg/ml), TNF (50 pg/ml), TNF/sPLA2-IIA or TNF/sPLA2-IIA with LY311727 (10 μm). Total cell lysates were prepared, and A, phosphorylated p38 and total p38α MAP kinase protein, or B, p-ERK and total ERK were detected by Western blot analysis as described under “Experimental Procedures.” Representative Western blots and the ratio of phospho- to total MAP kinase protein normalized relative to unstimulated cells is shown. Data are mean ± S.E. (n = 3 independent FLS cultures). Control phospho-MAPK/total MAPK ratios varied between cultures from 0.281 to 0.455 for p38, 0.432 to 0.671 for ERK-1, and 0.454 to 0.831 for ERK-2 (*, p < 0.05 relative to control, Student's paired t test).
DISCUSSION
These data establish for the first time in cultured cells relevant to the pathogenesis of RA that the regulation of TNF-dependent PG production by exogenous sPLA2-IIA does not depend on its enzyme function. sPLA2-IIA mutant, H48Q, which retains only 1% of sPLA2-IIA enzyme activity, is as effective as the fully functional enzyme in up-regulating PGE2 production and in superinducing TNF-mediated COX-2 production (Fig. 1). H48Q alone up-regulates the production of COX-2 without increasing PGE2 production (Fig. 1B), as does sPLA2-IIA (18). It is very unlikely that the sPLA2-IIA and H48Q effects are mediated by low-level contaminants because the effects we measure on PGE2 production and COX-2 up-regulation by sPLA2-IIA are completely abrogated by LY311727 (10 μm) (18), indicating that they are intrinsic to sPLA2-IIA. Importantly, our earlier study also showed that the augmentation of TNF-induced PGE2 production by wild-type sPLA2-IIA depends on the amount of sPLA2-IIA added. For example, addition of 1 μg/ml of sPLA2-IIA together with TNF led to only 50% as much PGE2 production as did addition of 10 μg/ml of sPLA2-IIA together with TNF (18). Our observations that 4 μg/ml of H48Q gives the same level of PGE2 production as 4 μg/ml of wild-type sPLA2-IIA (Fig. 1A) and that the H48Q response is dose-dependent, shows that the augmentation of PGE2 production is not due to the residual 1% enzymatic activity of H48Q. We have recently shown4 that complete abrogation of sPLA2-IIA enzyme activity with the covalent active site modifier bromophenacylbromide also did not affect PGE2 production. Although these findings may appear to conflict with our observation that LY311727 (a potent inhibitor of sPLA2-IIA catalytic activity) also blocks PGE2 production and COX-2 up-regulation (18), they argue that LY311727 acts as a dual-function sPLA2-IIA inhibitor. Our finding is consistent with other observations that LY311727 can inhibit other catalytic activity-independent functions of sPLA2-IIA such as M-type receptor binding (46). Interestingly, the cyclic peptide inhibitor c2, demonstrated to block AA mobilization here, also blocks PGE2 production in FLS (26), indicating that it is a dual-function sPLA2-IIA inhibitor also.
Our studies provide important and unexpected insights into the regulation of the AA metabolism in RA FLSs. First (Fig. 7), AA mobilization in resting FLSs appears high (10–15% over 2 h) in comparison to other resting cell lines (1–2%) (47). Basal AA mobilization is partially suppressible by inhibitors of cPLA2-α, (Fig. 3C), but not by inhibitors of sPLA2-IIA (Fig. 4A) or iPLA2-β (data not shown). Analysis of the distribution of tritium in conditioned medium from these cells (Table 1) suggests that the majority of mobilized AA remains esterified in phospholipids. The apparently high basal AA mobilization is thus likely due to microparticle release. The 20% of tritium coeluting with AA correlates well with the proportion of AA mobilization that is suppressible by cPLA2-α inhibition (Fig. 3C) suggesting that basal AA release is likely cPLA2-α-dependent. Under resting conditions, a small amount of PGE2 production is detectable, which is not suppressible by COX-2 selective inhibitors (18), suggesting PGE2 production likely couples to COX-1 in these circumstances.
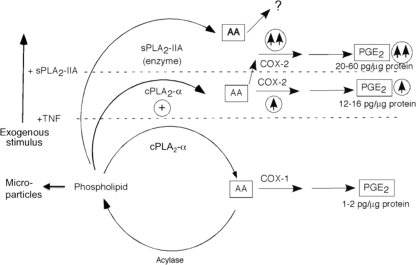
FIGURE 7.
Model of sPLA2-IIA function in FLS AA metabolism. Unstimulated cells, AA is mobilized in microparticles esterified to phospholipids. Low level AA release is mediated by cPLA2-α and is reincorporated into phospholipid pools by acylase(s). AA flux to PGE2 is very low and is via COX-1. TNF-stimulated cells, TNF. cPLA2-α is activated (⊕) and COX-2 expression is induced (↑ in circle), stimulating AA flux through the COX-2 pathway to stimulate PGE2 production. TNF + sPLA2-IIA, sPLA2-IIA enzyme activity increases AA mobilization that is not coupled to PGE2 production. sPLA2-IIA-mediated signaling superinduces COX-2 (↑↑ in circle) resulting in increased cPLA2-α-dependent AA flux through COX-2 to PGE2. sPLA2-IIA alone increases AA release and induces COX-2 without increasing PGE2 production. The figure is modeled after Fitzpatrick and Soberman (51).
Second (Fig. 7), AA mobilization following stimulation with TNF or BK is dependent on enhanced cPLA2-α activity because pyrrophenone blocks agonist-induced AA mobilization (Fig. 3C). Thus, TNF alone, although inefficient at rapidly mobilizing calcium in most cells, activates cPLA2-α, probably via enhancing cPLA2-α phosphorylation, as established in other model cell lines (2). cPLA2-α activation results in an ∼50% increase over basal in AA mobilization (Fig. 3C) with this increase likely distributed to AA, because no change is seen in the distribution of tritium in phospholipid relative to unstimulated cells (Table 1). Increased steady-state levels of COX-2 protein (Figs. 1B and 2C) also contribute to the 8–10-fold increase in COX-2-dependent PGE2 production (Figs. 1A and 2B) seen on TNF stimulation.
Third (Fig. 7), increased AA mobilization by sPLA2-IIA (Figs. 3C and 4) is mediated directly by its enzyme function not by cPLA2-α. H48Q fails to mobilize AA (Fig. 4B) and pyrrophenone does not block the effect (Fig. 3C). Importantly the distribution of mobilized tritium esterified in phospholipids is significantly lower in sPLA2-IIA-stimulated cells than that seen in unstimulated cells, indicating that exogenous sPLA2-IIA mobilizes AA from microparticle phospholipid pools. The trend to increased tritium distribution into other lipid mediators suggests that sPLA2-IIA-derived AA may be metabolized into eicosanoids. Because sPLA2-IIA alone does not increase PGE2 production (Fig. 1A) and the majority of the tritium eluted at Rf values between 0.67 and 0.81 (data not shown), these data are consistent with the metabolites being hydroxyeicosatetraenoic acids, although further work is necessary to confirm this. Importantly, increased AA mobilization in FLSs requires concentrations of exogenous sPLA2-IIA above 70 nm (1 μg/ml), 7–70-fold higher than the concentrations required for activity-dependent enhanced proliferation in prostate cancer cells (37). Exogenous sPLA2-IIA is not as potent in AA mobilization from resting cells as some other sPLA2 forms present in RA synovial tissue, notably Group X sPLA2 (38). However, effective sPLA2-IIA concentrations are within the range of concentrations measured in RA synovial fluids suggesting that sPLA2-IIA may contribute to AA mobilization in pathological conditions such as RA, where enzyme concentrations are high.
FLSs appear more sensitive to AA mobilization by exogenous sPLA2-IIA than other attachment-dependent cells. No detectable AA release was observed in CHO cells (38) or HEK 293 cells (47) at sPLA2-IIA concentrations up to 1 or 10 μg/ml, respectively, even following 6 h stimulation. Thus, FLSs have a stable metabolic “phenotype” not seen in other attachment-dependent cells that allows exogenous sPLA2-IIA-mediated AA mobilization. Although it is known that microparticle membranes express phosphatidylserine on their surface, the structural characteristics of FLS membranes have not yet been studied in detail, so whether membrane lipid asymmetry has been stably altered in FLSs remains to be determined.
AA mobilized by sPLA2-IIA, either alone or in combination with TNF, is not utilized for PGE2 production, despite the induction of COX-2. In the case of sPLA2-IIA stimulation alone, no increased PGE2 production is observed (Fig. 1A) and in the case of costimulation with TNF, abrogation of sPLA2-IIA enzyme activity by mutagenesis does not affect PGE2 production (Fig. 1A) and all of the observed increase in PGE2 production is suppressible by cPLA2-α inhibitors (Fig. 2, A and B). Thus under all conditions examined, only cPLA2-α activation can account for AA flux to PGE2.
The functional coupling of cPLA2-α and COX-2, also seen in other cell lines (48), is particularly striking in FLSs: despite effectively blocking TNF or bradykinin-mediated AA mobilization, inhibition of cPLA2-α in the presence of both TNF and sPLA2-IIA has no significant effect on AA mobilization (Fig. 3C), although effectively blocking all PGE2 production (Fig. 2). Under these conditions, over 20% of incorporated AA is mobilized from cells, yet only a very small proportion of released AA contributes to PGE2 production, all of it generated by cPLA2-α activity. Exogenous sPLA2-IIA is thus not functionally coupled to the COX pathway as has been commonly proposed (49–51), but rather indirectly regulates PG production pathways in these cells (Fig. 7). Our data also argue against regulation of cPLA2-α activity by sPLA2-IIA as has been found with other cell types (52), because sPLA2-IIA alone is unable to induce PGE2 production, despite up-regulating COX-2. In the presence of sPLA2-IIA alone, provision of AA to COX-2 by cPLA2-α is the rate-limiting step in PGE2 production. In contrast, in the presence of TNF, PGE2 production is limited by the amount of COX-2. Thus the rate-limiting step in the pathway may be either cPLA2-α or COX-2 depending on the cellular context.
Fourth (Fig. 7), the contribution of sPLA2-IIA to the 10–60-fold increased PGE2 production over basal levels seen on costimulation with TNF can be fully explained by enzyme activity-independent superinduction of the steady state levels of COX-2. The H48Q mutation does not affect the ability of sPLA2-IIA to induce COX-2 (Fig. 1B). As with NS-398 (18), concentrations of pyrrophenone that completely suppress PGE2 production do not affect the induction of COX-2 by sPLA2-IIA (Fig. 2C). It follows then, that cPLA2-α or sPLA2-IIA-derived AA or its metabolites do not regulate COX-2 protein levels in sPLA2-IIA-stimulated RA FLSs. This is in contrast to COX-2 up-regulation by IL-15 (53) or by certain agonists in other cell types (54, 55) whereby stimulus-induced PGE2 further up-regulates COX-2.
Although exogenous sPLA2-IIA up-regulates COX-2 protein in some model cell lines, the effect is cell-type specific (50), and apart from one case, nerve growth factor-stimulated rat serosal mast cells, where up-regulation also appears independent of enzyme activity (56), the mechanism is unknown. We have ruled out rapid activation of NF-κB or p38 MAPK by sPLA2-IIA, two pathways known to mediate agonist-dependent COX-2 up-regulation in FLSs (43, 57); sPLA2-IIA alone activates the ERK MAPK pathway. Although modest, ERK activation demonstrates that sPLA2-IIA regulates intracellular signaling and suggests broader effects on RA FLS function than the regulation of AA metabolism alone. In addition, this finding together with our observation that sPLA2-IIA, in the absence of cytokine stimulation, does not induce PGE2 production (Fig. 1A) (18), indicates that sPLA2-IIA-mediated ERK activation alone is insufficient to stimulate prostaglandin production in these cells, despite induction of COX-2. In our hands, MEK inhibitors PD98059 and UO126, whereas completely suppressing TNF/sPLA2-IIA-mediated PGE2 production did not suppress COX-2 induction (data not shown), suggesting that blockade of the ERK pathway alone is insufficient to affect COX-2 induction under these conditions. In contrast, we have previously shown that blockade of sPLA2-IIA function with LY311727 is sufficient to suppress both PGE2 production and COX-2 induction in the presence of TNF (18), yet LY311727 is unable to suppress ERK phosphorylation under these conditions (Fig. 6B). It is thus likely that TNF is sufficient to stimulate ERK and that sPLA2-IIA effects on PGE2 production and COX-2 expression occur “downstream” of ERK activation. However, the importance of sPLA2-IIA-mediated ERK activation in COX-2 up-regulation remains to be determined.
Our data predict that sPLA2-IIA induces COX-2 expression via an indirect signaling mechanism mediated through direct interaction with a cellular component(s). The identity of this component(s) in RA FLSs is unknown at present, however, in our hands, immunofluorescence studies demonstrate that exogenous sPLA2-IIA binds to the RA FLS cell surface and is very rapidly (within seconds) internalized demonstrating that the enzyme does bind to FLS cellular components.4 Receptor-mediated sPLA2 function has been best established for Group IB sPLA2 in mice using both biochemical and genetic approaches (58, 59). Murine Group IB sPLA2 and sPLA2-IIA both bind the murine 180-kDa M-type sPLA2 receptor with high affinity (1–10 nm) (59). However, human sPLA2-IIA is reported to have a binding affinity for the human M-type receptor that is too weak for sPLA2-IIA to be a physiological ligand in human cells (60). An alternative model is that sPLA2-IIA is internalized via binding to heparan sulfate proteoglycans, particularly glypican-1 in caveolae, followed by subsequent AA release and/or up-regulation of COX-2 (50, 61). It has been reported that sPLA2-IIA localizes to caveolin-containing vesicles as well as the Golgi apparatus in one “normal” synovial cell line following adenoviral transfection with the sPLA2-IIA cDNA, suggesting that this internalization pathway may be operative in these cells (17). However, there is no evidence that perturbation of this pathway has any effect on PGE2 production. Infection of FLSs with adenoviral vectors alone induces both COX-2 and PGE2 production (44) further complicating the interpretation of viral overexpression approaches.
In summary, our data show that human sPLA2-IIA, when added with TNF to RA FLSs, results in enhanced PGE2 production that does not require the enzyme activity of sPLA2-IIA. This finding, coupled with recent findings that sPLA2-IIA can participate in intracellular AA release when stably expressed at lower concentrations than those required exogenously (23, 47) and that some indole inhibitors are cell impermeable (47) and therefore incapable of blocking intracellular effects, suggest that clinical studies with inhibitors that are known to be both cell permeable and to potently block sPLA2-IIA-dependent signaling, may show greater benefit in the treatment of RA.
Acknowledgments
We are grateful to Chitra De Silva for excellent assistance with cell culture and Dr. Siiri E. Iismaa for helpful discussions. Quantitative amino acid analysis was performed by the Australian Proteome Analysis Facility (Macquarie University node), Sydney.
This work was supported, in whole or in part, by National Institutes of Health Grants HL50040 and HL3625 (to M. H. G.), National Health and Medical Research Council Grants 980263 (to K. F. S. and P. M. Brooks) and 222870 (to K. F. S., G. G. G., and H. P. McN), Cancer Council NSW Grant RG07-17 (to K. F. S., G. G. G., Q. Dong, and P. J. Russell), and a grant from the Rebecca Cooper Foundation (to K. F. S.), Australia.
M. H. Gelb, unpublished data.
L. Lee, P.-W. Lei, K. J. Bryant, E. P. Huang, S. Harrop, P. M. Curmi, A. P. Duff, W. B. Church, and K. F. Scott, manuscript in preparation.
- PLA2
- phospholipase A2
- sPLA2
- secreted phospholipase A2
- cPLA2
- cytosolic phospholipase A2
- sPLA2-IIA
- group IIA-secreted phospholipase A2
- RA
- rheumatoid arthritis
- FLS
- fibroblast-like synoviocyte
- cPLA2-α
- cytosolic phospholipase A2-α
- AA
- arachidonic acid
- iPLA2β
- group VIB calcium-independent phospholipase A2
- Ab
- antibody
- BK
- bradykinin
- S.E.
- standard error of the mean
- EMSA
- electromobility shift assay.
REFERENCES
- 1. Bonventre J. V., Huang Z., Taheri M. R., O'Leary E., Li E., Moskowitz M. A., Sapirstein A. (1997) Nature 390, 622–625 [DOI] [PubMed] [Google Scholar]
- 2. Hirabayashi T., Murayama T., Shimizu T. (2004) Biol. Pharm. Bull. 27, 1168–1173 [DOI] [PubMed] [Google Scholar]
- 3. Satake Y., Diaz B. L., Balestrieri B., Lam B. K., Kanaoka Y., Grusby M. J., Arm J. P. (2004) J. Biol. Chem. 279, 16488–16494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henderson W. R., Jr., Chi E. Y., Bollinger J. G., Tien Y. T., Ye X., Castelli L., Rubtsov Y. P., Singer A. G., Chiang G. K., Nevalainen T., Rudensky A. Y., Gelb M. H. (2007) J. Exp. Med. 204, 865–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kennedy B. P., Payette P., Mudgett J., Vadas P., Pruzanski W., Kwan M., Tang C., Rancourt D. E., Cromlish W. A. (1995) J. Biol. Chem. 270, 22378–22385 [DOI] [PubMed] [Google Scholar]
- 6. Sawada H., Murakami M., Enomoto A., Shimbara S., Kudo I. (1999) Eur. J. Biochem. 263, 826–835 [DOI] [PubMed] [Google Scholar]
- 7. Boilard E., Lai Y., Larabee K., Balestrieri B., Ghomashchi F., Fujioka D., Gobezie R., Coblyn J. S., Weinblatt M. E., Massarotti E. M., Thornhill T. S., Divangahi M., Remold H., Lambeau G., Gelb M. H., Arm J. P., Lee D. M. (2010) EMBO Mol. Med. 2, 172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ivandic B., Castellani L. W., Wang X. P., Qiao J. H., Mehrabian M., Navab M., Fogelman A. M., Grass D. S., Swanson M. E., de Beer M. C., de Beer F., Lusis A. J. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 1284–1290 [DOI] [PubMed] [Google Scholar]
- 9. Tietge U. J., Pratico D., Ding T., Funk C. D., Hildebrand R. B., Van Berkel T. J., Van Eck M. (2005) J. Lipid Res. 46, 1604–1614 [DOI] [PubMed] [Google Scholar]
- 10. Grass D. S., Felkner R. H., Chiang M. Y., Wallace R. E., Nevalainen T. J., Bennett C. F., Swanson M. E. (1996) J. Clin. Invest. 97, 2233–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chapdelaine J. M., Ciofalo V. B., Grass D. S., Felkner R., Wallace R. E., Swanson M. E. (1995) Arthritis Rheum. 38, S293 [Google Scholar]
- 12. Smith G. M., Ward R. L., McGuigan L., Rajkovic I. A., Scott K. F. (1992) Br. J. Rheumatol. 31, 175–178 [DOI] [PubMed] [Google Scholar]
- 13. Nevalainen T. J., Haapamäki M. M., Grönroos J. M. (2000) Biochim. Biophys. Acta 1488, 83–90 [DOI] [PubMed] [Google Scholar]
- 14. Scott K. F., Sajinovic M., Hein J., Nixdorf S., Galettis P., Liauw W., de Souza P., Dong Q., Graham G. G., Russell P. J. (2010) Biochimie 92, 601–610 [DOI] [PubMed] [Google Scholar]
- 15. Lin M. K., Farewell V., Vadas P., Bookman A. A., Keystone E. C., Pruzanski W. (1996) J. Rheumatol. 23, 1162–1166 [PubMed] [Google Scholar]
- 16. Jamal O. S., Conaghan P. G., Cunningham A. M., Brooks P. M., Munro V. F., Scott K. F. (1998) Ann. Rheum. Dis. 57, 550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Masuda S., Murakami M., Komiyama K., Ishihara M., Ishikawa Y., Ishii T., Kudo I. (2005) FEBS J. 272, 655–672 [DOI] [PubMed] [Google Scholar]
- 18. Bidgood M. J., Jamal O. S., Cunningham A. M., Brooks P. M., Scott K. F. (2000) J. Immunol. 165, 2790–2797 [DOI] [PubMed] [Google Scholar]
- 19. Bradley J. D., Dmitrienko A. A., Kivitz A. J., Gluck O. S., Weaver A. L., Wiesenhutter C., Myers S. L., Sides G. D. (2005) J. Rheumatol. 32, 417–423 [PubMed] [Google Scholar]
- 20. Ghomashchi F., Stewart A., Hefner Y., Ramanadham S., Turk J., Leslie C. C., Gelb M. H. (2001) Biochim. Biophys. Acta 1513, 160–166 [DOI] [PubMed] [Google Scholar]
- 21. Seno K., Okuno T., Nishi K., Murakami Y., Watanabe F., Matsuura T., Wada M., Fujii Y., Yamada M., Ogada T., Okada T., Hashizume H., Kii M., Hara S., Hagashita S., Nakamoto S., Yamada K., Chikazawa Y., Ueno M., Teshirogi I., Ono T., Ohtani O. (2000) J. Med. Chem. 43, 1041–1044 [DOI] [PubMed] [Google Scholar]
- 22. Seno K., Okuno T., Nishi K., Murakami Y., Yamada K., Nakamoto S., Ono T. (2001) Bioorg. Med. Chem. Lett. 11, 587–590 [DOI] [PubMed] [Google Scholar]
- 23. Ni Z., Okeley N. M., Smart B. P., Gelb M. H. (2006) J. Biol. Chem. 281, 16245–16255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghosh M., Loper R., Ghomashchi F., Tucker D. E., Bonventre J. V., Gelb M. H., Leslie C. C. (2007) J. Biol. Chem. 282, 11676–11686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ono T., Yamada K., Chikazawa Y., Ueno M., Nakamoto S., Okuno T., Seno K. (2002) Biochem. J. 363, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Church W. B., Inglis A. S., Tseng A., Duell R., Lei P. W., Bryant K. J., Scott K. F. (2001) J. Biol. Chem. 276, 33156–33164 [DOI] [PubMed] [Google Scholar]
- 27. Seilhamer J. J., Pruzanski W., Vadas P., Plant S., Miller J. A., Kloss J., Johnson L. K. (1989) J. Biol. Chem. 264, 5335–5338 [PubMed] [Google Scholar]
- 28. Nakamaye K. L., Eckstein F. (1986) Nucleic Acids Res. 14, 9679–9698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kushner P. J., Hort E., Shine J., Baxter J. D., Greene G. L. (1990) Mol. Endocrinol. 4, 1465–1473 [DOI] [PubMed] [Google Scholar]
- 30. Reynolds L. J., Hughes L. L., Dennis E. A. (1992) Anal. Biochem. 204, 190–197 [DOI] [PubMed] [Google Scholar]
- 31. Arnett F. C., Edworthy S. M., Bloch D. A., McShane D. J., Fries J. F., Cooper N. S., Healey L. A., Kaplan S. R., Liang M. H., Luthra H. S., Medsger T. A., Jr., Mitchell D. M., Neustadt D. H., Pinals R. S., Schaller J. G., Sharp J. T., Wilder R. L., Hunder G. G. (1988) Arthritis Rheum. 31, 315–324 [DOI] [PubMed] [Google Scholar]
- 32. Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lehmann T., Murphy C., Zahra D. G., Handel M. L. (2002) J. Rheumatol. 29, 787–795 [PubMed] [Google Scholar]
- 34. Nugteren D. H., Hazelhof E. (1973) Biochim. Biophys. Acta 326, 448–461 [DOI] [PubMed] [Google Scholar]
- 35. Green J. A., Smith G. M., Buchta R., Lee R., Ho K. Y., Rajkovic I. A., Scott K. F. (1991) Inflammation 15, 355–367 [DOI] [PubMed] [Google Scholar]
- 36. Edwards S. H., Thompson D., Baker S. F., Wood S. P., Wilton D. C. (2002) Biochemistry 41, 15468–15476 [DOI] [PubMed] [Google Scholar]
- 37. Sved P., Scott K. F., McLeod D., King N. J., Singh J., Tsatralis T., Nikolov B., Boulas J., Nallan L., Gelb M. H., Sajinovic M., Graham G. G., Russell P. J., Dong Q. (2004) Cancer Res. 64, 6934–6940 [DOI] [PubMed] [Google Scholar]
- 38. Bezzine S., Koduri R. S., Valentin E., Murakami M., Kudo I., Ghomashchi F., Sadilek M., Lambeau G., Gelb M. H. (2000) J. Biol. Chem. 275, 3179–3191 [DOI] [PubMed] [Google Scholar]
- 39. Cisar L. A., Mochan E., Schimmel R. (1993) Cell. Signal. 5, 463–472 [DOI] [PubMed] [Google Scholar]
- 40. Ackermann E. J., Conde-Frieboes K., Dennis E. A. (1995) J. Biol. Chem. 270, 445–450 [DOI] [PubMed] [Google Scholar]
- 41. Pásztói M., Nagy G., Géher P., Lakatos T., Tóth K., Wellinger K., Pócza P., György B., Holub M. C., Kittel A., Pálóczy K., Mazán M., Nyirkos P., Falus A., Buzas E. I. (2009) Arthritis Res. Ther. 11, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boilard E., Nigrovic P. A., Larabee K., Watts G. F., Coblyn J. S., Weinblatt M. E., Massarotti E. M., Remold-O'Donnell E., Farndale R. W., Ware J., Lee D. M. (2010) Science 327, 580–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crofford L. J., Tan B., McCarthy C. J., Hla T. (1997) Arthritis Rheum. 40, 226–236 [DOI] [PubMed] [Google Scholar]
- 44. Crofford L. J., McDonagh K. T., Guo S., Mehta H., Bian H., Petruzelli L. M., Roessler B. J. (2005) J. Gene Med. 7, 288–296 [DOI] [PubMed] [Google Scholar]
- 45. Santos L. L., Lacey D., Yang Y., Leech M., Morand E. F. (2004) J. Rheumatol. 31, 1038–1043 [PubMed] [Google Scholar]
- 46. Boilard E., Rouault M., Surrel F., Le Calvez C., Bezzine S., Singer A., Gelb M. H., Lambeau G. (2006) Biochemistry 45, 13203–13218 [DOI] [PubMed] [Google Scholar]
- 47. Mounier C. M., Ghomashchi F., Lindsay M. R., James S., Singer A. G., Parton R. G., Gelb M. H. (2004) J. Biol. Chem. 279, 25024–25038 [DOI] [PubMed] [Google Scholar]
- 48. Murakami M., Kambe T., Shimbara S., Kudo I. (1999) J. Biol. Chem. 274, 3103–3115 [DOI] [PubMed] [Google Scholar]
- 49. Murakami M., Kambe T., Shimbara S., Yamamoto S., Kuwata H., Kudo I. (1999) J. Biol. Chem. 274, 29927–29936 [DOI] [PubMed] [Google Scholar]
- 50. Murakami M., Kudo I. (2002) J. Biochem. 131, 285–292 [DOI] [PubMed] [Google Scholar]
- 51. Fitzpatrick F. A., Soberman R. (2001) J. Clin. Invest. 107, 1347–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Han W. K., Sapirstein A., Hung C. C., Alessandrini A., Bonventre J. V. (2003) J. Biol. Chem. 278, 24153–24163 [DOI] [PubMed] [Google Scholar]
- 53. Min S. Y., Hwang S. Y., Jung Y. O., Jeong J., Park S. H., Cho C. S., Kim H. Y., Kim W. U. (2004) J. Rheumatol. 31, 875–883 [PubMed] [Google Scholar]
- 54. Hughes-Fulford M., Tjandrawinata R. R., Li C. F., Sayyah S. (2005) Carcinogenesis 26, 1520–1526 [DOI] [PubMed] [Google Scholar]
- 55. Bradbury D. A., Newton R., Zhu Y. M., El-Haroun H., Corbett L., Knox A. J. (2003) J. Biol. Chem. 278, 49954–49964 [DOI] [PubMed] [Google Scholar]
- 56. Tada K., Murakami M., Kambe T., Kudo I. (1998) J. Immunol. 161, 5008–5015 [PubMed] [Google Scholar]
- 57. Faour W. H., Mancini A., He Q. W., Di Battista J. A. (2003) J. Biol. Chem. 278, 26897–26907 [DOI] [PubMed] [Google Scholar]
- 58. Cupillard L., Mulherkar R., Gomez N., Kadam S., Valentin E., Lazdunski M., Lambeau G. (1999) J. Biol. Chem. 274, 7043–7051 [DOI] [PubMed] [Google Scholar]
- 59. Hanasaki K. (2004) Biol. Pharm. Bull. 27, 1165–1177 [DOI] [PubMed] [Google Scholar]
- 60. Ancian P., Lambeau G., Mattéi M. G., Lazdunski M. (1995) J. Biol. Chem. 270, 8963–8970 [DOI] [PubMed] [Google Scholar]
- 61. Murakami M., Kudo I. (2004) Biol. Pharm. Bull. 27, 1158–1164 [DOI] [PubMed] [Google Scholar]