Abstract
Gain- and loss-of-function experiments have illustrated that the family of myogenic regulatory factors is necessary and sufficient for the formation of skeletal muscle. Furthermore, MyoD required cellular aggregation to induce myogenesis in P19 embryonal carcinoma stem cells. To determine the mechanism by which stem cells can be directed into skeletal muscle, a time course of P19 cell differentiation was examined in the presence and absence of exogenous MyoD. By quantitative PCR, the first MyoD up-regulated transcripts were the premyogenic mesoderm factors Meox1, Pax7, Six1, and Eya2 on day 4 of differentiation. Subsequently, the myoblast markers myogenin, MEF2C, and Myf5 were up-regulated, leading to skeletal myogenesis. These results were corroborated by Western blot analysis, showing up-regulation of Pax3, Six1, and MEF2C proteins, prior to myogenin protein expression. To determine at what stage a dominant-negative MyoD/EnR mutant could inhibit myogenesis, stable cell lines were created and examined. Interestingly, the premyogenic mesoderm factors, Meox1, Pax3/7, Six1, Eya2, and Foxc1, were down-regulated, and as expected, skeletal myogenesis was abolished. Finally, to identify direct targets of MyoD in this system, chromatin immunoprecipitation experiments were performed. MyoD was observed associated with regulatory regions of Meox1, Pax3/7, Six1, Eya2, and myogenin genes. Taken together, MyoD directs stem cells into the skeletal muscle lineage by binding and activating the expression of premyogenic mesoderm genes, prior to activating myoblast genes.
Keywords: Cell Differentiation, Gene Expression, Helix-Loop-Helix Transcription Factors, Skeletal Muscle, Stem Cell
Introduction
Myogenesis has become a paradigm for studying how one factor can reproducibly regulate several genetic subprograms required to drive cell differentiation, exemplified by the myogenic regulatory factors (MRFs).5 This highly related gene family consists of four members termed MyoD, Myf-5, MRF-4, and myogenin. MRF family members are sufficient to trans-differentiate a variety of mesodermal and ectodermal cell types into skeletal muscle (1–7). The MRFs contain a highly conserved basic helix-loop-helix domain that is important for DNA binding and dimerization, respectively (8, 9). The MRFs form heterodimers with members of the ubiquitous E-protein basic helix-loop-helix subfamily, which on their own cannot turn on the myogenic program, and bind to E-box motifs (DNA sequence CANNTG). These E-boxes are found in the regulatory regions of many muscle-specific genes (2, 9–14).
The transcriptional activation domain of MyoD (1–54 amino acids) (15) is important for maximal interaction with the histone acetyltransferases P300/CBP and PCAF in vitro (16), and this interaction is required for the activation of muscle-specific gene expression (17). Furthermore, studies illustrate that two amino acids in the basic domain of MyoD, an alanine and threonine, are essential to activate the transcription of target genes (18, 19). Finally, it was shown that the histidine/cysteine-rich and the amphipathic α-helix (helix III) domains of MyoD can regulate a subset of genes, distinct from the genes regulated by the classical N-terminal activation domain, through an interaction with the Pbx and Meis homeodomain proteins located adjacent to the myogenin promoter (20, 21).
The MyoD and E protein heterodimer was shown to favor a VCASCTGT consensus site (where V is anything but T, and S represents G or C) (22). Another group has identified CASKTG as the MyoD E-box consensus sequence, where K represents T or G (23). Recently, Cao et al. (24) have confirmed that there is a preference for E-boxes with internal CC or GC sequences. Furthermore, MyoD binding does not always correlate with transcriptional activation (9, 15). MyoD, with the help of other transcription factors and/or cofactors, can bind to E-box sites prior to transcriptional activation (25, 26). Recently, it was shown that MyoD binds to many of the skeletal muscle-specific genes during the course of differentiation (24). However, MyoD can also bind to thousands of additional sites genome-wide and induce regional histone acetylation (24).
There are several other transcription factor gene families that are expressed prior to MRFs in the premyogenic mesoderm and regulate myogenesis, including Pax, Gli, Six, Eya, Meox, and Foxc families. They are expressed in the somite and dermomyotome (27–31) and found to play indirect and/or direct roles in the activation of the MRFs (32–38). Interpreting the function of Gli, Pax, Six, Meox, Eya, and Fox proteins in skeletal myogenesis utilizing knock-out methods has been complicated because at least two isoforms for each gene family are present in developing somites. Therefore, to alleviate this obstacle, several groups have published double knock-outs of Gli2/3, Pax3/7, Six1/2, Meox1/2, Eya1/2, and Foxc1/2. All six studies revealed a phenotype of more pronounced muscle deficiencies than the single knock-out phenotypes alone (32, 38–43).
Knock-out experiments have elucidated many genetic targets of the MRFs, albeit mostly structural proteins, involved in muscle fiber contraction. Moreover, they have shown that the MRFs have overlapping functional roles in myogenesis. For instance, homozygous deletion of either Myf-5 or MyoD in mice does not result in a muscle-deficient animal (44, 45) but MyoD−/−;Myf-5−/−;MRF4−/− neonatal mice are born with no detectable skeletal muscle and no myoblasts (46, 47). On the other hand, a homozygous deletion mutant of MRF-4 and/or myogenin does not affect myoblast formation but does affect the formation of muscle fibers (48, 49). Altogether, this indicates early and late functionally redundant roles for the MRFs in regulating skeletal myogenesis.
Several studies have isolated additional genetic targets of the MRFs using C2C12 cells or fibroblast myogenic conversion assays on null MRF backgrounds (50–52). These studies have revealed novel information on the complex hierarchy of MRF transcriptional networks in myogenesis. The in vitro cell culture systems used in the former studies are models of satellite cell proliferation and differentiation and thus cannot examine the initial specification and commitment of skeletal muscle that is required prior to the formation of myoblasts.
P19 cells (53) are used as a model to study in vitro embryonic development, including cardiac and skeletal myogenesis as well as neurogenesis. P19 cells are embryonal carcinoma stem cells derived from the inner cell mass of a mouse embryo. In the presence of DMSO, aggregated P19 cells develop into beating cardiomyocytes and bipolar skeletal myocytes, which fuse into myotubes, that are physiologically and biochemically analogous to their embryonic counterparts (54). P19-derived cardiac and skeletal muscle show similar cell morphologies to embryonic muscle being mono- and multinucleated, respectively, and express embryo-specific isoforms of several genes. Previously, using dominant-negative and overexpression assays in P19 cells, we have uncovered a regulatory loop between Pax3, Gli2, and Meox1 in activating skeletal myogenesis (36, 37), which is initiated by Wnt signaling via β-catenin (55).
Although ectopic expression of MyoD is sufficient to force nonmuscle cells to complete the myogenic program (7), the pathway by which MyoD converts stem cells into skeletal muscle is unknown. Here, using the P19 cell culture system, we have elucidated novel targets of MyoD. We show that MyoD directly regulates the expression of premyogenic mesoderm factors and that a dominant-negative MyoD is sufficient to prevent their up-regulation in differentiating stem cells.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
The MyoD/EnR construct was created by removal of the transcriptional activation domain (1–54 amino acids) of MyoD (15), by PCR of murine MyoD cDNA utilizing the oligonucleotides 5′-GGATCCATGGCCCTCCTGAAACCGGAG and 3′-CTCGAGGTCGATCTCTCAAAGCACC. The 5′- and 3′-oligonucleotides contained BamHI and XhoI sites (boldface), respectively, to facilitate cloning. The Δ(1–54 amino acids) MyoD cDNA was then cloned into the PGK-1 vector that contains the phosphoglycerate kinase-1 promoter (56). Subsequently, the 198-amino acid N-terminal repression domain (EnR) of the mouse En-2 protein was isolated from En-2 cDNA by PCR utilizing the oligonucleotides 5′-AAGGATCCATGGAGGAGAAGGATTCCAAG and 3′-AAGGATCCCCCAGAGTGGCGCTGGCT and subcloned into the pGEM-T EasyTM (Promega) vector. The 5′- and 3′-oligonucleotides contained BamHI sites (boldface) to facilitate subcloning. The EnR was excised with BamHI and blunt end ligated to the BamHI site of PGK-Δ(1–54 amino acids)MyoD. PGK-MyoD, PGK-Puro, and PGK-LacZ constructs were created as described previously (6, 57).
Cell Culture and Transfections
P19 cells were cultured as described previously (58). Stable PGK-MyoD cell lines, termed P19(MyoD) cells, were described previously, and the PGK-MyoD/EnR cells were prepared using a similar protocol (6). Briefly, a mixture of 2.04 μg of PGK-MyoD/EnR or PGK vector alone, 0.09 μg of PGK-puro, 0.17 μg of PGK-LacZ, and 0.77 μg of B17 was transfected into P19 cells by adding a mixture of DNA and FuGENETM 6 to 2.5 × 106 cells in 35-mm tissue culture plates.
P19(Control) and P19(MyoD/EnR) cells were differentiated in the presence of 0.8% DMSO or 1 μm retinoic acid as described previously (36, 59, 60). P19(Control) and P19(MyoD) cells were differentiated in the absence of DMSO. P19(MyoD), P19(MyoD/EnR), and P19(Control) cells were aggregated in Petri dishes for 4 days and then plated onto tissue culture dishes. Drugs were added only during cellular aggregation.
Immunofluorescence
Myosin heavy chain (MyHC), neurofilament 68 (61), and nuclei were detected as described previously (36, 60) utilizing monoclonal MF20 antibody supernatant, monoclonal anti-neurofilament 68 clone NR4 antibodies (Sigma), and Hoechst dye, respectively. Immunofluorescence was visualized with an Olympus BX50 microscope. Images were captured on a Roger Scientific Cool Snap camera and processed utilizing Image Pro-Plus 5.1 (Media Cybernetics) and Canvas 9 and 11 software.
Reverse Transcription and Q-PCR
Total RNA was isolated using the RNeasy kit (Qiagen, Missisauga, Ontario, Canada) following the protocol described by the manufacturer. 1 μg of purified RNA was used as a template for the first strand DNA synthesis reaction using the QuantiTect reverse transcription kit (Qiagen, Missisauga, Ontario, Canada) following the protocol described by the manufacturers. Real time quantitative PCRs (Q-PCR) were performed as described previously (62), using the FastStart SYBR Green master mix (Roche Applied Sciences). Primer pairs used for quantitative detection of gene expression were selected from Primer Bank (accessed September 2008; Primer 3 input, version 0.4.0, available on line) and are listed in supplemental Table S1. Primer pairs for the ChIP experiments were designed based on ChIP sequencing results provided by the laboratory of Dr. Stephen Tapscott (Seattle) (24). Primer pairs used for quantitative detection of binding were selected from Primer Bank (accessed September 2008; Primer 3, available on line) and are listed in supplemental Table S2. All primers were verified for optimal efficiency. All reactions and data analysis were performed on the ABI 7300 system (Applied Biosystems) using SDS software. All reactions were performed in duplicate, unless otherwise stated, and the results shown are the means ± S.E. of three independent experiments, unless otherwise stated. mRNA levels for each treatment was normalized to β-actin levels for the corresponding day and treatment.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) assays were performed as described previously (62) with minor modifications. P19(Control) and P19(MyoD) cell lines were aggregated for 4 days in the absence of DMSO, and cells were harvested for ChIP analysis on days 0, 4, or 6. The cells were cross-linked with 1% formaldehyde for 1 h. Relative enrichment of binding sites was compared with the IgG negative control. Immunoprecipitation was analyzed from 30 to 60 μg of chromatin using Q-PCR, as described above, with 5 μg of MyoD antibody (catalog no. sc760X, Santa Cruz Biotechnology) or 5 μg of rabbit IgG antibody (catalog no. PP64, Millipore).
Northern Blot, Southern Blot, and Reverse Transcription-PCR
Protocols and DNA probes utilized, with the exception of Pax3, have been described previously (36, 37, 58, 65–67). Densitometry was carried out using ImageJ software (National Institutes of Health). Histograms were plotted for each lane of the respective Northern blot, and base lines were manually inserted, and the area under the curve was taken. All values were normalized to 18 S. N values indicate how many differentiations were performed in total. Standard error was calculated using Microsoft Excel software.
Oligonucleotides to amplify Pax3 were 5′-CTGCACTCAAGGGACTCCTC and 3′-GTGAAGGCGAGACGAAAAAG at an annealing temperature of 60 °C. First strand reactions were tested for linearity with each set of oligonucleotides, and negative controls included RT experiments in the absence of RNA or reverse transcriptase enzyme and PCR in the absence of a first strand reaction.
Western Blot Analysis
P19(MyoD/EnR), P19(MyoD), or P19(Control) cells were grown in monolayers or differentiated for 4–6 days, as indicated. For P19(MyoD/EnR), Western blots were performed as described previously (68). For P19(MyoD) cells, total protein was harvested with modified RIPA buffer (50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 1 mm PMSF, and 1× final concentration of protein inhibitor mixture, Roche Applied Science). Protein (25 or 50 μg, for Pax3 detection) was separated using the NuPAGE 4–12% BisTris gel (Invitrogen) in a 1× NuPAGE MOPS Running Buffer prepared from a 20× stock (50 mm MOPS, 50 mm Tris, 0.1% SDS, 1 mm EDTA (pH 7.7)). Proteins were transferred to an immunoblot PVDF membrane (Bio-Rad). MyoD/EnR and β-actin proteins were detected with anti-MyoD (1:300 dilution; Santa Cruz Biotechnology) and anti-β-actin (1:12,000 dilution; Sigma) antibodies and visualized with HRP-conjugated secondary antibodies (Chemicon). Pax3, Six1, MEF2C, myogenin, MyoD, and α-tubulin proteins were detected with anti-Pax3 (1:300 dilution; catalog no. MAB2457, R&D Systems), anti-Six1 (1:125 dilution) (69), anti-MEF2C (1:500 dilution; catalog no. sc-13266X, Santa Cruz Biotechnology), anti-myogenin (1:500 dilution; F5D hybridoma), anti-MyoD (1:300 dilution; catalog no. 554130, Pharmingen), and anti-α-tubulin (1:1000 dilution; catalog no. T6199, Sigma) antibodies and visualized with HRP-conjugated secondary antibodies. Note that the signal for Pax3-specific antibodies was enhanced using Western blot signal enhancer (catalog no. 21050, Qentix) before blocking with 5% milk in TBST), as per the manufacturer's instructions.
Statistical Analysis
Statistical differences between means were calculated using the Student's t test. P values of p < 0.05 were considered significant.
RESULTS
MyoD Was Sufficient to Induce the Expression of Premyogenic Mesoderm Factors
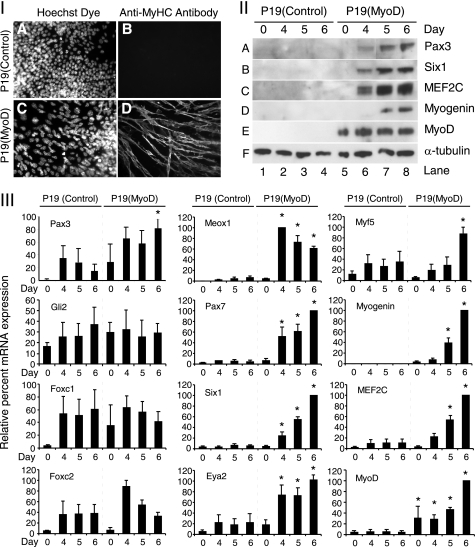
To identify the putative targets of MyoD in a stem cell system, we examined P19 cells overexpressing MyoD, termed P19(MyoD), and P19(Control) cells. P19(MyoD) cells have been characterized in the past and shown to undergo myogenesis and up-regulate Pax3 transcript levels (60, 67). P19(Control) and P19(MyoD) cells were aggregated in the absence of DMSO and fixed on day 6 for staining with MF20 antibodies specific for MyHC. In agreement with previous results (6, 60), P19(Control) cells did not differentiate into skeletal muscle (Fig. 1I, panel B), although P19(MyoD) cells showed robust differentiation (Fig. 1I, panel D).
FIGURE 1.
Overexpression of MyoD in aggregated P19 cells induced the expression of the premyogenic mesoderm genes and MRFs. P19(Control) and P19(MyoD) cells were differentiated in the absence of DMSO. Cultures were either fixed on day 6 for immunofluorescence or harvested for RNA or protein on days 0 and 4–6. I, cells were stained with Hoechst dye to visualize the nuclei (panels A and C) and with anti-MyHC antibody to visualize muscle (panels B and D). Magnification ×400. II, Western blots with 25 μg (50 μg for Pax3) of total protein were probed with antibodies of factors indicated on the right. III, gene expression was analyzed using Q-PCR for the genes indicated. The data were normalized to the expression of β-actin and are expressed relative to day 0 P19(Control) cells, with the maximum expression normalized to 100%. Error bars represent the average ± S.E. (n = 3; *, p < 0.05).
To determine which premyogenic mesoderm transcripts were up-regulated by MyoD, Q-PCR was conducted with total RNA collected on days 0 and 4–6. As expected, MyoD transcripts were overexpressed in P19(MyoD) cells and not in P19(Control) cells (Fig. 1III, bottom right). Interestingly, transcripts initially expressed in the paraxial mesoderm and developing somite of the embryo, such as Meox1, Pax7, Six1, and Eya2, termed premyogenic mesoderm factors, displayed enhanced levels in P19(MyoD) cells compared with P19(Control) cells on day 4 (Fig. 1III). Subsequently, in agreement with previous results, the myoblast markers, myogenin, MEF2C, and Myf-5, were found to be up-regulated by days 5 or 6. Pax3 was up-regulated by MyoD at all time points but not significantly until day 6. Gli2 and Foxc1/2 were not appreciably up-regulated over control cells. Thus, in aggregated P19 cells, MyoD up-regulated premyogenic mesoderm factors prior to up-regulating myoblast marker gene expression.
To verify whether the protein expression levels mirrored the changes seen at the RNA level, a Western blot analysis was performed with anti-Pax3, -Six1, -MEF2C, -myogenin, and -MyoD antibodies on total protein collected on days 0 and 4–6 of differentiation. As expected, MyoD protein was expressed throughout the time course in P19(MyoD) cells and not in P19(Control) cells (Fig. 1II, panel E, lanes 5–8). Pax3, Six1, and MEF2C protein levels were detected by day 4 (Fig. 1II, A–C, lanes 6–8), followed by the detection of the myogenin protein by day 5 (Fig. 1II, D, lanes 7 and 8). Thus, there was no evidence of post-transcriptional control of protein expression in this system. Taken together, MyoD up-regulated the expression of the premyogenic mesoderm factors prior to the up-regulation of myogenin during the induction of myogenesis in aggregated P19 stem cells.
Dominant-negative MyoD Inhibited Skeletal Myogenesis and Expression of Premyogenic Mesoderm Factors
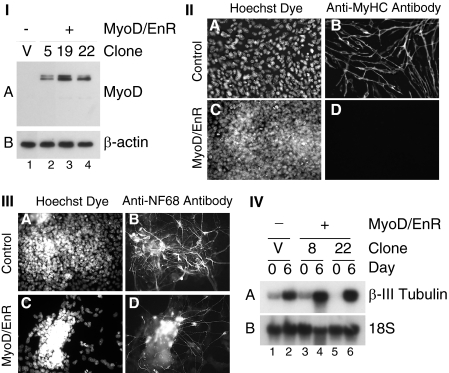
To identify putative targets of the MRF family of transcription factors, we created a dominant-negative MyoD mutant by replacing the transcriptional activation domain of MyoD (15) with the mouse engrailed-2 (En-2) repression domain. The En-2 repression domain works through histone acetyltransferase-dependent and -independent mechanisms to actively repress transcription, preventing rescue from other MRF family members (70). This construct was then transfected and stably integrated into the P19 cell genome, and 12 clones were isolated, termed P19(MyoD/EnR) cells. Western blot analysis was performed with anti-MyoD antibodies on cells grown in monolayer to ensure that the MyoD/EnR construct was expressed appropriately (Fig. 2I, lanes 2–4). The MyoD/EnR protein appeared as a doublet indicating possible alternative post-translational modification, such as acetylation or phosphorylation (71, 72).
FIGURE 2.
Overexpression of MyoD/EnR inhibited the formation of skeletal myocytes but not neurons in aggregated P19 cells. I, P19(Control) (lane 1) and P19(MyoD/EnR) cells (lanes 2–4) were grown in monolayers, and total protein was harvested. Western blots with 30 μg of total protein were probed with antibodies indicated on the right. II, P19(Control) cells (panels A and B) and P19(MyoD/EnR) cells (panels C and D) were differentiated in the presence of 0.8% DMSO. On day 9 of the differentiation, cells were fixed and stained with Hoechst dye to detect nuclei (panels A and C) and anti-MyHC antibody (panels B and D). III and IV, P19(Control) and P19(MyoD/EnR) cells were differentiated in the presence of 1 μm retinoic acid. III, on day 6 of the differentiation, cells were fixed and stained with Hoechst dye to detect nuclei (panels A and C) and anti-NF68 antibody (panels B and D). IV, Northern blots with 4 μg of RNA were probed with the factors indicated on the right. Lanes are indicated at the bottom of I and IV. Magnification ×400 for II and III.
P19(MyoD/EnR) and P19(Control) cells were differentiated in the presence of DMSO, an inducer of myogenesis, and immunofluorescence was performed on day 9 with anti-MyHC antibodies. As expected, all 12 P19(MyoD/EnR) clones did not differentiate into skeletal myocytes compared with the P19(Control) cells, which differentiated into skeletal myocytes (Fig. 2II, panel D compared with panel B, one representative clone). Moreover, 6 of 12 (50%) P19(MyoD/EnR) clones did not differentiate into cardiomyocytes (data not shown). Therefore, the overexpression of MyoD/EnR in P19 cells inhibits skeletal myogenesis, and to a variable degree can repress cardiomyogenesis.
To ensure that the overexpression of MyoD/EnR in P19 cells was not inhibitory for all differentiation processes, we tested whether MyoD/EnR affected the formation of neurons. We aggregated P19(MyoD/EnR) and P19(Control) cells in the presence of 1 μm retinoic acid, an inducer of neurogenesis. On day 6, cells were fixed and visualized for immunofluorescence utilizing anti-NF68 antibodies, which are specific for the neuronal intermediate filament protein, neurofilament 68. Abundant NF68 staining was observed in the presence and absence of MyoD/EnR (Fig. 2III, panels B and D). Furthermore, by Northern blot analysis, neuronal β-III tubulin expression was up-regulated during neurogenesis to similar levels in P19(Control) and P19(MyoD/EnR) cells (Fig. 2IV, panel A). Therefore, MyoD/EnR did not appreciably inhibit neurogenesis.
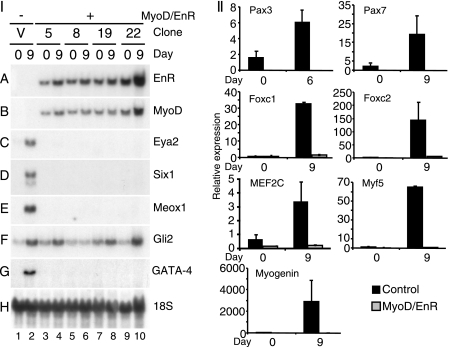
To identify genes inhibited by MyoD/EnR, cells were differentiated in the presence of DMSO, and total RNA was harvested on days 0, 6, and 9, as indicated. By Northern blot analysis, there were high levels of MyoD/EnR in P19(MyoD/EnR) cell lines and undetectable levels in P19(Control) cells, shown both by EnR- and MyoD-specific probes (Fig. 3I, panels A and B). MyoD expression was not seen in control cells on day 9 due to the short exposure time (Fig. 3I, panel B). However, endogenous MyoD expression was ablated in all P19(MyoD/EnR) cells when longer exposures were examined (data not shown). In P19(Control) cells, there was strong expression on day 9 of the premyogenic mesoderm transcription factors Eya2, Six1, and Meox1 that was not detectable in P19(MyoD/EnR) clones (Fig. 3I, panels C–E, respectively). The mRNA transcript levels of Gli2 were moderately affected by the presence of the MyoD/EnR protein. On day 9, Gli2 was down-regulated in the majority of P19(MyoD/EnR) clones compared with P19(Control) cells (Fig. 3I, panel F) by 46 ± 12% (S.E.; n = 15). The extent of down-regulation of expression across several clones was analyzed by densitometry and is summarized in Table 1.
FIGURE 3.
Overexpression of MyoD/EnR in P19 cells inhibited the expression of premyogenic mesoderm factors. P19(Control) and P19(MyoD/EnR) cells were differentiated in the presence of 0.8% DMSO. Total RNA was harvested on day 0 and 6 or 9. I, Northern blots with 12 μg of RNA were probed with the factors indicated on the right. II, RNA was analyzed using Q-PCR. The data have been normalized to the expression of β-actin and are expressed relative to day 0 P19(Control) cells. Error bars represent the average ± S.E. of two independent differentiations of two independent clones.
TABLE 1.
The extent of down-regulation of genes (%) in P19(MyoD/EnR) compared with P19(Control) cultures was quantified from Northern blot analysis by densitometry
| Gene | Down regulation | ±S.E. | n value |
|---|---|---|---|
| % | % | ||
| Eya2 | 99 | 1 | 15 |
| Six1 | 97 | 3 | 16 |
| Meox1 | 92 | 7 | 13 |
| Gata-4 | 64 | 22 | 10 |
| Pax3 | 63 | 14 | 16 |
| Gli2 | 46 | 12 | 15 |
Utilizing Q-PCR analysis, we examined the mRNA expression of Pax3/7, Foxc1/2, MEF2C, myogenin, and Myf-5. Interestingly, the expression of all of these factors was severely down-regulated (Fig. 3II). Altogether, in differentiating cell lines, the presence of MyoD/EnR protein ablated the expression of premyogenic mesoderm factors as well as that of myoblast markers.
Because cardiomyogenesis was down-regulated in half of P19(MyoD/EnR) clones (data not shown) by immunofluorescence analysis, the mRNA transcript levels of GATA-4 was examined by Northern blot analysis. In P19(MyoD/EnR) clones, with no detectable cardiomyocyte formation via immunohistochemistry, mRNA transcripts of GATA-4 were not detectable on day 9 of differentiation when compared with P19(Control) cells (Fig. 3I, panel G). Clones capable of cardiomyogenesis exhibited normal levels of GATA-4 (data not shown). Densitometry of GATA-4 transcript levels on day 9 of differentiation for all of the clones revealed that GATA-4 was down-regulated 64 ± 22% (S.E.; n = 10) in P19(MyoD/EnR) cells relative to P19(Control) cells. Therefore, GATA-4 was down-regulated in approximately half of the clones resulting in the loss of cardiomyogenesis.
To determine at what time point MyoD/EnR disrupts the expression of the transcription factors assayed above, Northern blots and RT-PCR were performed with RNA from a time course of differentiation. The presence of MyoD/EnR inhibited the premyogenic mesoderm factors, Eya2, meox1, Pax3, and Six1 throughout the time course (supplemental Fig. S1). In summary, the expression of a dominant-negative MyoD mutant disrupted the proper expression of premyogenic mesoderm factors and subsequent skeletal myogenesis.
MyoD Binds Directly to the Regulatory Regions of the Premyogenic Mesoderm Genes
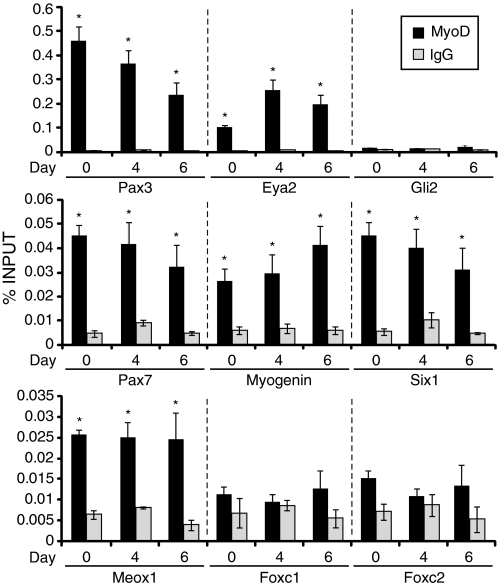
To determine whether the observed changes in gene expression were due to direct or indirect effects of MyoD, ChIP experiments were performed. Primers were designed based on previous ChIP sequencing results (24) showing regions of genes bound by MyoD in C2C12 myoblasts. Using P19(MyoD) cells, ChIP experiments illustrated that MyoD bound significantly to the regulatory regions of Pax3, Eya2, Pax7, Six1, myogenin, and Meox1 when compared with the IgG control, at all time points examined (Fig. 4). The genes with the highest % input bound by MyoD were Pax3 and Eya2, followed by Pax7, myogenin, and finally Meox1. In contrast, MyoD binding to the regulatory regions of Foxc1/2 and Gli2 was not significant, in agreement with their lack of up-regulation by MyoD (Fig. 1). Therefore, MyoD binds directly to the regulatory regions of the majority of premyogenic mesoderm genes.
FIGURE 4.
MyoD binds directly to premyogenic mesoderm transcription factors. P19(Control) and P19(MyoD) stable cell lines were differentiated in the absence of DMSO, and ChIP was performed using an anti-MyoD antibody to identify enriched MyoD targets in P19(MyoD) cells on day 0, 4, and 6 of differentiation. Q-PCR was used to analyze the chromatin isolated for the genes indicated. Error bars represent the average ± S.E. (n = 3; *, p < 0.05). Statistical analysis was performed using the Student's t test versus IgG.
DISCUSSION
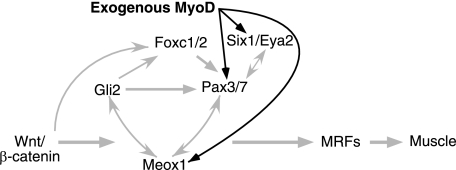
In this study, we have discovered a novel activity for MyoD in regulating the expression of premyogenic mesoderm factors, including Meox1, Six1, Eya2, and Pax3/7. Furthermore, by ChIP assay, MyoD was observed associated with regulatory regions of Meox1, Six1, Eya2, Pax3/7, as well as myogenin (summarized in Table 2). Altogether, from this study we can formulate a model in which exogenous MyoD can transform multipotent stem cells into skeletal muscle, by first binding and activating the expression of a subset of premyogenic mesoderm genes (Fig. 5).
TABLE 2.
Summary of premyogenic mesoderm genes regulated directly or indirectly by MyoD
| Gene | Up-regulated by MyoDa | Down-regulated by MyoD/EnRb | Bound directly by MyoD in ChIPc |
|---|---|---|---|
| Foxc1 | − | + | − |
| Foxc2 | − | + | − |
| Gli2 | − | +/− | − |
| Pax3 | + | + | + |
| Pax7 | + | + | + |
| Eya2 | + | + | + |
| Meox1 | + | + | + |
| Six1 | + | + | + |
a + indicates up-regulated; − indicates not up-regulated.
b + indicates down-regulated; +/− indicates partially down-regulated.
c + indicates bound; − indicates not bound.
FIGURE 5.
Simplified model showing how exogenous MyoD can enter into the regulatory network controlling P19 cell myogenesis, directly up-regulating a subset of premyogenic mesoderm factors. Gray arrows indicate regulatory pathways identified in previous studies (32–37, 55, 73, 76, 77). The black arrows represent the direct regulation of Pax3/7, Six1, Eya2, and Meox1 by MyoD identified in this study. The MRFs once expressed may also feedback onto the premyogenic mesoderm genes. MyoD/EnR would presumably function similar to MyoD but would also have further reaching dominant-negative effects.
Previous results have shown that Wnt signaling via β-catenin up-regulates premyogenic mesoderm factors Gli2, Pax3, Six1, Meox1, Eya2, and Foxc1/2 (73–77). Eya2 acts genetically upstream of Pax3 in the formation of the hypaxial lip of the dermomyotome, and it is a cofactor of Six for the activation of Pax3 expression and migration (42). Pax3 and Meox1 regulate their own expression, although Pax3 can also regulate the activity of Meox1 and Six1 but not Gli2 (36, 37). On the other hand, Meox1 can regulate the expression of Pax3, Six1, and Gli2 (32, 37). Meox1, Pax3, Six1, and Gli2 have all been shown via direct or indirect mechanisms to positively regulate the MRFs (32–35). In addition, we have recently demonstrated that Foxc1/2 transcripts are detected early, and in an overlapping pattern with Wnt3a, prior to Pax3 expression during P19 cell differentiation into skeletal muscle. The expression of Foxc1 is regulated by Shh and Wnt signaling in this system (76).
The new data added to this model from this study (Fig. 5, black arrows) illustrate that MyoD enters this regulatory network to direct stem cells into the skeletal muscle lineage by first binding and activating the expression of premyogenic mesoderm genes, leading to myogenin and Myf-5 up-regulation and subsequent myogenesis. No evidence for post-transcriptional control was found when comparing changes in mRNA and protein expression levels. Although this paper focused on MyoD, it is possible that the other MRFs could also redirect stem cells into skeletal muscle through a similar mechanism.
The transcription factors Six1, Meox1, Eya2, Gli2, Foxc1/2, and Pax3/7 are expressed in the early somite and in the dermomyotome (27–31, 39, 78) prior to the epithelial mesenchymal transition and migration of cells into the myotome. Initially, several factors found to be expressed in the somite were thought be restricted to the dermomyotome, but studies have shown that they are also found in the myotome. Six1 is localized to the myotome in developing mice, chicken, and humans (40, 79–81). Eya2 is expressed in the myotome in human and chick (80, 81), and Meox1 is expressed in mouse myotome (31). In quail and mouse, Gli2 becomes restricted to the myotome and ventral dermomyotome (39, 82). Pax3/7 is found in mouse and human myotome (41, 80). In mouse, Foxc1/2 are expressed only in the paraxial mesoderm and somites (83, 84). Thus, it is possible that a feedback loop between MyoD and the premyogenic mesoderm factors is important for the amplification and maintenance of their expression. It is possible that other MRFs may possess a similar activity.
A role for MRFs in the maintenance of premyogenic mesoderm expression is supported by the activation of Six1 by forced expression of MyoD or Myf-5 into MyoD−/−;Myf-5−/− fibroblasts (51) and by the loss of MRF expression in Six1/Six4 null mice (40). Furthermore, in proliferating C212 cells Eya1, which is also expressed in somites (30), was shown to be a direct target of MyoD (52). Finally, our finding that myogenin is not up-regulated by MyoD until premyogenic mesoderm genes, including Six1, are expressed is consistent with the recent finding that Six1 can regulate MyoD function in C2C12 cells (69).
It was originally thought that satellite cells were derived from cells that mark the fetal myoblasts in the chick and mouse (85, 86). But more recent studies have implicated a primitive satellite cell progenitor population that expresses Pax3 and Pax7 (41, 87, 88). Furthermore, it was shown that essentially all satellite cells in an adult originate from MyoD-positive progenitors (89). This suggests a potential role for MyoD in maintaining premyogenic mesoderm expression. Finally, Zammit et al. (90) illustrated that activated satellite cells initially coexpress Pax7 and MyoD. A subset of cells lose MyoD and can return to a quiescent Pax7-positive state. Thus, a role for MyoD in maintaining the expression of premyogenic mesoderm genes may potentially be important at various stages of embryogenesis and/or satellite cell regeneration.
The loss of skeletal myogenesis in P19(MyoD/EnR) cells is likely due to the dominant-negative function of MyoD/EnR, capable of inhibiting the other MRFs. We would predict that knockdown of MyoD alone would not result in a loss of skeletal myogenesis, in agreement with results in the embryo (47).
MEF2C mRNA expression was found to be down-regulated in several P19(MyoD/EnR) cell lines (Fig. 2II, panel E, and data not shown). This is in agreement with the literature demonstrating the following: 1) MEF2C is expressed in the myotome (91); 2) MEF2C is a genetic target for the MRFs (6, 52, 92); and 3) the promoter of MEF2C contains an essential E-box that, if mutated, severely down-regulates its expression in somites (92). As MEF2C also plays a critical role in cardiomyogenesis (93) and in the positive regulation of Nkx2.5 and GATA-4 (66, 94, 95), the absence of MEF2C (Fig. 3II) may explain the inhibition of cardiomyogenesis and the down-regulation of GATA-4 and Nkx2.5 observed (Fig. 3 and data not shown). Furthermore, Meox1 and Gli2 were sufficient to induce cardiomyogenesis in P19 cells (65), and regulators of Gli activation could modulate Nkx2.5 expression (96, 97). Therefore, the loss of a combination of these factors could negatively affect cardiac muscle development in our system.
The finding that neurogenesis was unaffected in P19(MyoD/EnR) cells indicates a specificity of MyoD/EnR for E-boxes associated with myogenesis and not neurogenesis. For example, NeuroD and Mash1/2 are basic helix-loop-helix factors that regulate neurogenesis (59, 63, 63). Because neurogenesis still occurred in P19(MyoD/EnR) cells, it is unlikely that MyoD/EnR bound to the neuron-specific E boxes.
In summary, MyoD directs stem cells into the myogenic lineage via an embryonic pathway, by first directly up-regulating premyogenic mesoderm factor expression. This regulatory network may be important for the maintenance of premyogenic mesoderm factor expression during myogenesis.
Supplementary Material
Acknowledgments
We thank Sophie Boisvenue for excellent technical assistance and support, Andrew Richetti for statistical analysis assistance, and John Mardinly for imaging software assistance. We thank Diba Ebadi for critically reading the manuscript.
This work was supported in part by Grant MOP-84458 from Canadian Institutes of Health Research (to I. S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- MRF
- myogenic regulatory factor
- MyoD/EnR
- replacement of the MyoD activation domain with the engrailed repressor domain
- P19(MyoD/EnR)
- P19 cells overexpressing MyoD/EnR
- P19(MyoD)
- P19 cells overexpressing MyoD
- MyHC
- myosin heavy chain
- MRF
- myogenic regulatory factor
- EnR
- N-terminal En-2 repression domain
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Q-PCR
- quantitative PCR.
REFERENCES
- 1. Davis R. L., Weintraub H., Lassar A. B. (1987) Cell 51, 987–1000 [DOI] [PubMed] [Google Scholar]
- 2. Olson E. N. (1990) Genes Dev. 4, 1454–1461 [DOI] [PubMed] [Google Scholar]
- 3. Weintraub H., Tapscott S. J., Davis R. L., Thayer M. J., Adam M. A., Lassar A. B., Miller A. D. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5434–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braun T., Buschhausen-Denker G., Bober E., Tannich E., Arnold H. H. (1989) EMBO J. 8, 701–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miner J. H., Wold B. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ridgeway A. G., Wilton S., Skerjanc I. S. (2000) J. Biol. Chem. 275, 41–46 [DOI] [PubMed] [Google Scholar]
- 7. Tapscott S. J., Davis R. L., Thayer M. J., Cheng P. F., Weintraub H., Lassar A. B. (1988) Science 242, 405–411 [DOI] [PubMed] [Google Scholar]
- 8. Brennan T. J., Edmondson D. G., Olson E. N. (1990) J. Cell Biol. 110, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. (1990) Cell 60, 733–746 [DOI] [PubMed] [Google Scholar]
- 10. Brennan T. J., Olson E. N. (1990) Genes Dev. 4, 582–595 [DOI] [PubMed] [Google Scholar]
- 11. Lassar A. B., Davis R. L., Wright W. E., Kadesch T., Murre C., Voronova A., Baltimore D., Weintraub H. (1991) Cell 66, 305–315 [DOI] [PubMed] [Google Scholar]
- 12. Murre C., McCaw P. S., Baltimore D. (1989) Cell 56, 777–783 [DOI] [PubMed] [Google Scholar]
- 13. Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S., et al. (1991) Science 251, 761–766 [DOI] [PubMed] [Google Scholar]
- 14. Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. (1989) Cell 58, 823–831 [DOI] [PubMed] [Google Scholar]
- 15. Weintraub H., Dwarki V. J., Verma I., Davis R., Hollenberg S., Snider L., Lassar A., Tapscott S. J. (1991) Genes Dev. 5, 1377–1386 [DOI] [PubMed] [Google Scholar]
- 16. Sartorelli V., Huang J., Hamamori Y., Kedes L. (1997) Mol. Cell. Biol. 17, 1010–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Puri P. L., Sartorelli V., Yang X. J., Hamamori Y., Ogryzko V. V., Howard B. H., Kedes L., Wang J. Y., Graessmann A., Nakatani Y., Levrero M. (1997) Mol. Cell 1, 35–45 [DOI] [PubMed] [Google Scholar]
- 18. Brennan T. J., Chakraborty T., Olson E. N. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5675–5679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weintraub H., Genetta T., Kadesch T. (1994) Genes Dev. 8, 2203–2211 [DOI] [PubMed] [Google Scholar]
- 20. Berkes C. A., Bergstrom D. A., Penn B. H., Seaver K. J., Knoepfler P. S., Tapscott S. J. (2004) Mol. Cell 14, 465–477 [DOI] [PubMed] [Google Scholar]
- 21. Heidt A. B., Rojas A., Harris I. S., Black B. L. (2007) Mol. Cell. Biol. 27, 5910–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blackwell T. K., Weintraub H. (1990) Science 250, 1104–1110 [DOI] [PubMed] [Google Scholar]
- 23. Huang J., Blackwell T. K., Kedes L., Weintraub H. (1996) Mol. Cell. Biol. 16, 3893–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G. J., Parker M. H., MacQuarrie K. L., Davison J., Morgan M. T., Ruzzo W. L., Gentleman R. C., Tapscott S. J. (2010) Dev. Cell 18, 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bergstrom D. A., Penn B. H., Strand A., Perry R. L., Rudnicki M. A., Tapscott S. J. (2002) Mol. Cell 9, 587–600 [DOI] [PubMed] [Google Scholar]
- 26. Rampalli S., Li L., Mak E., Ge K., Brand M., Tapscott S. J., Dilworth F. J. (2007) Nat. Struct. Mol. Biol. 14, 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goulding M. D., Chalepakis G., Deutsch U., Erselius J. R., Gruss P. (1991) EMBO J. 10, 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Williams B. A., Ordahl C. P. (1994) Development 120, 785–796 [DOI] [PubMed] [Google Scholar]
- 29. Oliver G., Wehr R., Jenkins N. A., Copeland N. G., Cheyette B. N., Hartenstein V., Zipursky S. L., Gruss P. (1995) Development 121, 693–705 [DOI] [PubMed] [Google Scholar]
- 30. Xu P. X., Woo I., Her H., Beier D. R., Maas R. L. (1997) Development 124, 219–231 [DOI] [PubMed] [Google Scholar]
- 31. Candia A. F., Hu J., Crosby J., Lalley P. A., Noden D., Nadeau J. H., Wright C. V. (1992) Development 116, 1123–1136 [DOI] [PubMed] [Google Scholar]
- 32. Mankoo B. S., Skuntz S., Harrigan I., Grigorieva E., Candia A., Wright C. V., Arnheiter H., Pachnis V. (2003) Development 130, 4655–4664 [DOI] [PubMed] [Google Scholar]
- 33. Spitz F., Demignon J., Porteu A., Kahn A., Concordet J. P., Daegelen D., Maire P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14220–14225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tajbakhsh S., Rocancourt D., Cossu G., Buckingham M. (1997) Cell 89, 127–138 [DOI] [PubMed] [Google Scholar]
- 35. Gustafsson M. K., Pan H., Pinney D. F., Liu Y., Lewandowski A., Epstein D. J., Emerson C. P., Jr. (2002) Genes Dev. 16, 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ridgeway A. G., Skerjanc I. S. (2001) J. Biol. Chem. 276, 19033–19039 [DOI] [PubMed] [Google Scholar]
- 37. Petropoulos H., Gianakopoulos P. J., Ridgeway A. G., Skerjanc I. S. (2004) J. Biol. Chem. 279, 23874–23881 [DOI] [PubMed] [Google Scholar]
- 38. Kume T., Deng K., Hogan B. L. (2000) Development 127, 1387–1395 [DOI] [PubMed] [Google Scholar]
- 39. McDermott A., Gustafsson M., Elsam T., Hui C. C., Emerson C. P., Jr., Borycki A. G. (2005) Development 132, 345–357 [DOI] [PubMed] [Google Scholar]
- 40. Grifone R., Demignon J., Houbron C., Souil E., Niro C., Seller M. J., Hamard G., Maire P. (2005) Development 132, 2235–2249 [DOI] [PubMed] [Google Scholar]
- 41. Relaix F., Rocancourt D., Mansouri A., Buckingham M. (2005) Nature 435, 948–953 [DOI] [PubMed] [Google Scholar]
- 42. Grifone R., Demignon J., Giordani J., Niro C., Souil E., Bertin F., Laclef C., Xu P. X., Maire P. (2007) Dev. Biol. 302, 602–616 [DOI] [PubMed] [Google Scholar]
- 43. Winnier G. E., Kume T., Deng K., Rogers R., Bundy J., Raines C., Walter M. A., Hogan B. L., Conway S. J. (1999) Dev. Biol. 213, 418–431 [DOI] [PubMed] [Google Scholar]
- 44. Rudnicki M. A., Braun T., Hinuma S., Jaenisch R. (1992) Cell 71, 383–390 [DOI] [PubMed] [Google Scholar]
- 45. Braun T., Rudnicki M. A., Arnold H. H., Jaenisch R. (1992) Cell 71, 369–382 [DOI] [PubMed] [Google Scholar]
- 46. Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. (1993) Cell 75, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 47. Kassar-Duchossoy L., Gayraud-Morel B., Gomès D., Rocancourt D., Buckingham M., Shinin V., Tajbakhsh S. (2004) Nature 431, 466–471 [DOI] [PubMed] [Google Scholar]
- 48. Hasty P., Bradley A., Morris J. H., Edmondson D. G., Venuti J. M., Olson E. N., Klein W. H. (1993) Nature 364, 501–506 [DOI] [PubMed] [Google Scholar]
- 49. Patapoutian A., Yoon J. K., Miner J. H., Wang S., Stark K., Wold B. (1995) Development 121, 3347–3358 [DOI] [PubMed] [Google Scholar]
- 50. Wyzykowski J. C., Winata T. I., Mitin N., Taparowsky E. J., Konieczny S. F. (2002) Mol. Cell. Biol. 22, 6199–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ishibashi J., Perry R. L., Asakura A., Rudnicki M. A. (2005) J. Cell Biol. 171, 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B. D. (2005) Genes Dev. 19, 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McBurney M. W., Jones-Villeneuve E. M., Edwards M. K., Anderson P. J. (1982) Nature 299, 165–167 [DOI] [PubMed] [Google Scholar]
- 54. Skerjanc I. S. (1999) Trends Cardiovasc. Med. 9, 139–143 [DOI] [PubMed] [Google Scholar]
- 55. Petropoulos H., Skerjanc I. S. (2002) J. Biol. Chem. 277, 15393–15399 [DOI] [PubMed] [Google Scholar]
- 56. Adra C. N., Boer P. H., McBurney M. W. (1987) Gene 60, 65–74 [DOI] [PubMed] [Google Scholar]
- 57. Rogerson P. J., Jamali M., Skerjanc I. S. (2002) FEBS Lett. 524, 134–138 [DOI] [PubMed] [Google Scholar]
- 58. Wilton S., Skerjanc I. S. (1999) In Vitro Cell. Dev. Biol. Anim. 35, 175–177 [DOI] [PubMed] [Google Scholar]
- 59. Skerjanc I. S., Wilton S. (2000) FEBS Lett. 472, 53–56 [DOI] [PubMed] [Google Scholar]
- 60. Skerjanc I. S., Slack R. S., McBurney M. W. (1994) Mol. Cell. Biol. 14, 8451–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Debus E., Weber K., Osborn M. (1983) Differentiation 25, 193–203 [DOI] [PubMed] [Google Scholar]
- 62. Savage J., Conley A. J., Blais A., Skerjanc I. S. (2009) Stem Cells 27, 1231–1243 [DOI] [PubMed] [Google Scholar]
- 63. Horton S., Meredith A., Richardson J. A., Johnson J. E. (1999) Mol. Cell. Neurosci. 14, 355–369 [DOI] [PubMed] [Google Scholar]
- 64. Lee J. E., Hollenberg S. M., Snider L., Turner D. L., Lipnick N., Weintraub H. (1995) Science 268, 836–844 [DOI] [PubMed] [Google Scholar]
- 65. Gianakopoulos P. J., Skerjanc I. S. (2005) J. Biol. Chem. 280, 21022–21028 [DOI] [PubMed] [Google Scholar]
- 66. Skerjanc I. S., Petropoulos H., Ridgeway A. G., Wilton S. (1998) J. Biol. Chem. 273, 34904–34910 [DOI] [PubMed] [Google Scholar]
- 67. Ridgeway A. G., Petropoulos H., Wilton S., Skerjanc I. S. (2000) J. Biol. Chem. 275, 32398–32405 [DOI] [PubMed] [Google Scholar]
- 68. Gianakopoulos P. J., Skerjanc I. S. (2009) In Vitro Cell. Dev. Biol. Anim. 45, 566–572 [DOI] [PubMed] [Google Scholar]
- 69. Liu Y., Chu A., Chakroun I., Islam U., Blais A. (2010) Nucleic Acids Res. 38, 6857–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tolkunova E. N., Fujioka M., Kobayashi M., Deka D., Jaynes J. B. (1998) Mol. Cell. Biol. 18, 2804–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Puri P. L., Sartorelli V. (2000) J. Cell. Physiol. 185, 155–173 [DOI] [PubMed] [Google Scholar]
- 72. Polesskaya A., Duquet A., Naguibneva I., Weise C., Vervisch A., Bengal E., Hucho F., Robin P., Harel-Bellan A. (2000) J. Biol. Chem. 275, 34359–34364 [DOI] [PubMed] [Google Scholar]
- 73. Capdevila J., Tabin C., Johnson R. L. (1998) Dev. Biol. 193, 182–194 [DOI] [PubMed] [Google Scholar]
- 74. Fan C. M., Lee C. S., Tessier-Lavigne M. (1997) Dev. Biol. 191, 160–165 [DOI] [PubMed] [Google Scholar]
- 75. Petropoulos H. (2002) Commitment and Differentiation during Skeletal Myogenesis, Ph.D. thesis, University of Western Ontario, London, Ontario, Canada [Google Scholar]
- 76. Savage J., Voronova A., Mehta V., Sendi-Mukasa F., Skerjanc I. S. (2010) Differentiation 79, 31–40 [DOI] [PubMed] [Google Scholar]
- 77. Wagner J., Schmidt C., Nikowits W., Jr., Christ B. (2000) Dev. Biol. 228, 86–94 [DOI] [PubMed] [Google Scholar]
- 78. Borycki A., Brown A. M., Emerson C. P., Jr. (2000) Development 127, 2075–2087 [DOI] [PubMed] [Google Scholar]
- 79. Laclef C., Hamard G., Demignon J., Souil E., Houbron C., Maire P. (2003) Development 130, 2239–2252 [DOI] [PubMed] [Google Scholar]
- 80. Fougerousse F., Durand M., Lopez S., Suel L., Demignon J., Thornton C., Ozaki H., Kawakami K., Barbet P., Beckmann J. S., Maire P. (2002) J. Muscle Res. Cell Motil. 23, 255–264 [DOI] [PubMed] [Google Scholar]
- 81. Heanue T. A., Reshef R., Davis R. J., Mardon G., Oliver G., Tomarev S., Lassar A. B., Tabin C. J. (1999) Genes. Dev. 13, 3231–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Borycki A. G., Mendham L., Emerson C. P., Jr. (1998) Development 125, 777–790 [DOI] [PubMed] [Google Scholar]
- 83. Swiderski R. E., Reiter R. S., Nishimura D. Y., Alward W. L., Kalenak J. W., Searby C. S., Stone E. M., Sheffield V. C., Lin J. J. (1999) Dev. Dyn. 216, 16–27 [DOI] [PubMed] [Google Scholar]
- 84. Winnier G. E., Hargett L., Hogan B. L. (1997) Genes Dev. 11, 926–940 [DOI] [PubMed] [Google Scholar]
- 85. Cossu G., Molinaro M., Pacifici M. (1983) Dev. Biol. 98, 520–524 [DOI] [PubMed] [Google Scholar]
- 86. Hartley R. S., Bandman E., Yablonka-Reuveni Z. (1992) Dev. Biol. 153, 206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kassar-Duchossoy L., Giacone E., Gayraud-Morel B., Jory A., Gomès D., Tajbakhsh S. (2005) Genes Dev. 19, 1426–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gros J., Manceau M., Thomé V., Marcelle C. (2005) Nature 435, 954–958 [DOI] [PubMed] [Google Scholar]
- 89. Kanisicak O., Mendez J. J., Yamamoto S., Yamamoto M., Goldhamer D. J. (2009) Dev. Biol. 332, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zammit P. S., Relaix F., Nagata Y., Ruiz A. P., Collins C. A., Partridge T. A., Beauchamp J. R. (2006) J. Cell Sci. 119, 1824–1832 [DOI] [PubMed] [Google Scholar]
- 91. Edmondson D. G., Lyons G. E., Martin J. F., Olson E. N. (1994) Development 120, 1251–1263 [DOI] [PubMed] [Google Scholar]
- 92. Dodou E., Xu S. M., Black B. L. (2003) Mech. Dev. 120, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 93. Lin Q., Schwarz J., Bucana C., Olson E. N. (1997) Science 276, 1404–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Reecy J. M., Li X., Yamada M., DeMayo F. J., Newman C. S., Harvey R. P., Schwartz R. J. (1999) Development 126, 839–849 [DOI] [PubMed] [Google Scholar]
- 95. Searcy R. D., Vincent E. B., Liberatore C. M., Yutzey K. E. (1998) Development 125, 4461–4470 [DOI] [PubMed] [Google Scholar]
- 96. Zhang X. M., Ramalho-Santos M., McMahon A. P. (2001) Cell 106, 781–792 [PubMed] [Google Scholar]
- 97. Goodrich L. V., Scott M. P. (1998) Neuron 21, 1243–1257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.