Abstract
Mechanical loading of muscles by intrinsic muscle activity or passive stretch leads to an increase in the production of reactive oxygen species (1, 2). The NAD-dependent protein deacetylase SIRT1 is involved in the protection against oxidative stress by enhancing FOXO-driven Sod2 transcription (3–5). In this report, we unravel a mechanism triggered by mechanical stretch of skeletal muscle cells that leads to an EGR1-dependent transcriptional activation of the Sirt1 gene. The resulting transient increase in SIRT1 expression generates an antioxidative response that contributes to reactive oxygen species scavenging.
Keywords: Gene Expression, Gene Regulation, Oxidative Stress, Reactive Oxygen Species (ROS), SIRT, Skeletal Muscle
Introduction
Skeletal muscles show a rapid response to changes in mechanical environment. Mechanical stimuli are translated to biochemical signals that promote adaptations in muscle mass, structure, and function. This response is mediated by mechanosensitive signaling pathways that provoke changes in gene expression that promote cell growth and survival.
Muscle contractility provokes an increase in ROS2 production due to increased mitochondrial activity and through activation of NADPH oxidase at the membrane (6, 7). Passive mechanical stretch of the diaphragmatic muscle is also associated with increased ROS production (8). As a result, activation of the ROS-responsive MAPK signaling pathway and downstream effectors as the proinflammatory transcriptional factors AP-1 and NFκB (9–11) occurs.
High levels of ROS lead to cell damage by lipid and protein oxidation and contributes to muscle fatigue and consequently loss of contractile force production (1, 8). The mechanisms by which muscles performing constant activity, i.e. the heart and diaphragm, are protected against excessive ROS generation remain elusive.
SIRT1, the mammalian orthologue of Sir2, is an NAD-dependent protein deacetylase, which has been shown to increase lifespan in model organisms as yeast, worms, and flies (12–14). In mammals, SIRT1 is involved in a plethora of physiological processes such as metabolism (15–17), neurogenesis (18), and gametogenesis (19) and has a beneficial effect on age-associated pathologies (20). Particularly in the heart, it has been reported that SIRT1 participates in protection against age-associated oxidative stress by up-regulation of Mn-SOD (21).
At the cellular level, SIRT1 has a prosurvival effect by deacetylating p53 and the FOXO (Forkhead box type O transcription factor) transcription factors. Deacetylation by SIRT1 inhibits p53 apoptotic response and switches FOXO transcription activity toward genes involved in cell cycle arrest, DNA repair and resistance to oxidative stress and away from proapoptotic genes (3, 4, 22, 23).
Mechanisms controlling Sirt1 expression at the transcriptional level have been reported to occur in response to nutrient availability and DNA damage. In actively growing cells, the cell cycle regulator E2F1 is responsible for cell cycle-dependent SIRT1 fluctuations, DNA damage stabilizes E2F1 and induces Sirt1 transcription; in turn, deacetylation of E2F1 by SIRT1 inhibits E2F1 transcriptional and apoptotic activities (24). In end-differentiated cells, Sirt1 expression is repressed by p53 binding to positions 168 and 178 of the Sirt1 promoter; by nutrient deprivation, FOXO3a translocates to the nucleus, interacts with p53 on the Sirt1 promoter, and abolishes the repression on Sirt1 transcription imposed by p53 (25).
In this report, we uncovered a novel mechanism of transcription regulation of the Sirt1 gene by the EGR1 (early growth response factor 1), which occurs in response to mechanical stretch. This stretch-responsive gene activation program is required to eliminate ROS generated during stretch.
EXPERIMENTAL PROCEDURES
Plasmids
pcDNA3-Egr1-FLAG (29), pTA-luc Sirt1 promoter (−202) (25), pCRUZ-Sirt1-HA, pYE-Sir2, and pYE-Sir2(H/Y) (23) were obtained from Addgene. pTA-luc was obtained from Clontech. pcDNA-Sirt1-FLAG (21) was a kind gift from Dr. Wei Gu.
Sirt1 Promoter Mutagenesis
Mutants of the Sirt1 promoter at the EGR1 canonical binding sites at −65 (MT1) and −122 (MT2) were obtained by replacing the GCGGGGGCG consensus sequence by GCAAGGACG. The pTA-luc Sirt1 promoter (−202) plasmid was mutagenized by site-directed mutagenesis by using the following primers: AGCCGAGCCGCAGGAACGCCAGTGCCGCGC for MT1 and GAGAGGCGCAGGAACGGGGGAGGGGCGGG for MT2. The QuikChange II site-directed mutagenesis kit (Stratagene) was used, and primers were designed following the manufacturer's instructions.
Cell Lines and Culture
C2C12 cells were obtained from ATCC. Myoblasts were grown in high glucose DMEM medium (Invitrogen) supplemented with 10% FBS and 0.5 mg/ml of minimal essential amino acids (growth medium, GM) in a CO2 incubator. Myotubes were obtained from myoblasts grown at subconfluency by replacing the medium by high glucose DMEM supplemented with 2% horse serum (differentiation medium, DM) after a wash with Dulbecco's phosphate base saline. 70–80% of myotubes are obtained at second day of culture on regular collagen-coated plates (Biocoat) or at the third day of culture on type I collagen-coated flexible bottomed plates (FlexCell Intl.).
Myotube Stretch Protocol
Myoblasts cultured onto type I collagen-coated flexible-bottom wells (Flex I plates, FlexCell Intl.) were subjected to differentiation. At third day in DM, myotubes were subjected to a 15% stretch protocol at 1 Hz (0.5 s of deformation alternating with 0.5 s of relaxation) with a Flexcell system. Unless otherwise specified, myotubes were stretched for 30 min and harvested at different intervals after stretch by scraping after two washes in Dulbecco's phosphate saline buffer.
Diaphragm ex Vivo Mechanical Stretch Protocol
This experimental protocol was performed essentially as described in our previous work (10). C57BL6 mice at 3–4 months of age were used. Mice were anesthetized, and the diaphragmatic muscle was excised from each animal and immediately immersed into a muscle bath containing a modified Krebs-Ringers solution bubbled with 95% O2 and 5% CO2 maintained at 25 °C. Right hemidiaphragm muscle sheets were kept in Krebs-Ringer solution to serve as nonstretched control muscle samples. Mechanical stretch was applied to the entire costal muscle of left hemidiaphragms by passively stretching the muscle in the direction of the muscle fibers. We applied passive tension of ∼0.4 n/cm, equivalent to a passive stress of 11 n/cm2. In these conditions, a mechanical stretch of ∼50% from the unstressed length was achieved, which corresponds to ∼120% of optimal muscle length.
RNA Extraction and Real-time PCR
Total RNA from cells was prepared with the RNeasy mini kit from Qiagen. Total RNA from diaphragm muscles was prepared with Trizol (Invitrogen). Reverse transcription was performed on 1 μg of total RNA. cDNA aliquots were amplified with the Brilliant SYBR Green Master Mix (Stratagene) in a MX3005P cycler (Stratagene). RNA contents were standardize for GAPDH with the formula 2−Ct×−Ct(GAPDH). Primer sequences will be provided upon request.
Protein Extraction
Cultured myotubes were washed in cold PBS and scraped. Cell pellets were resuspended in a cell lysis buffer (25 mm Tris-HCl, pH 7.5, 250 mm NaCl, 2 mm EDTA, and 0.3% Nonidet P-40) containing 0.5 mm PMSF, 1 mm DTT, and protease inhibitor mixtures (Pierce). After two cycles of freeze and thaw, extracts were microcentrifuged for 10 min at 10,000 rpm, and pellets were discarded. Total protein from diaphragm muscles was obtained as described previously (26). Protein concentration was determined with the Quick Start Bradford protein assay from Bio-Rad.
ChIP Assays
Hemidiaphragms subjected to a 30-min stretch as well as their unstretched counterparts were weighted immediately after stretch (30–35 mg) and placed in cold PBS containing 0.5 mm PMSF. The tissue was sliced in small pieces and incubated in 1% formaldehyde in PBS for 15 min at room temperature in a rotating platform. Cross-linking was stopped by adding glycine to a final concentration of 0.125 m followed by a 5-min incubation at room temperature. The cross-linked tissue was collected by centrifugation at 1250 rpm for 10 min followed by two washes with PMSF 0.05 mm in PBS. The tissue was first disrupted by mechanical grinding with a bouncer in PBS containing 0.5 mm PMSF and 2× protease inhibitor mixture. Cells were collected by centrifugation and lysed in 10 mm Hepes, pH 8, 85 mm KCl, 0.5% Nonidet P-40, and 2× protease inhibitor mixture. The nuclear fraction was collected by centrifugation at 5000 rpm for 10 min and lysed for 20 min in 50 mm Tris HCl, 10 mm EDTA, 1% SDS at 4 °C (10 μl/mg of tissue). Nuclear extracts were sonicated (eight strokes at 30% for 15 s followed by 15 s of rest) while kept in a ice-water bath. After centrifugation for 10 min at 14,000 rpm, chromatin fractions were frozen in liquid nitrogen and kept at −80 °C. Chromatin from two hemidiaphragms was pooled, and 200 μl were used for each immunoprecipitation assay. After a 5× dilution in ChIP dilution buffer, 10 μl were separated (input), and the remaining diluted chromatin was incubated with the indicated specific antibody or rabbit IgG (negative control) or anti-RNA polymerase (positive control). An EZ-CHIP kit (Upstate) was used from this step. For ChIP, assays performed on myotubes the EZ-CHIP kit was used following the manufacturer's instructions.
On EGR1 immunoprecipitates, primers with sequences GCGTGGGTGGCGGGAGAGG (forward) and CATCTTCCAACTGCCTCTCTGGCC (reverse) were used to detect the Sirt1 promoter. Binding of FOXO4 and SIRT1 to the Sod2 promoter, was detected by primers with the following sequences, TTATGGAAACATTTGATAGCCACTGCTTCTTAGAC (forward) and CGCGTGCTTGCTACAGCCACGC (reverse). Positive and negative controls were performed on immunoprecipitates with RNA polymerase and IgG antibodies. Detection of the immunoprecipitated DNA was performed by PCR followed by electrophoresis in 1% agarose gels. To determine promoter occupancy percentage, the immunoprecipitated DNA was quantified by real-time PCR and calculated using the formula 100 × 2−(Ct(input)−Ct(IPx)) − 100 × 2−(Ct(input)−Ct(IP IgG)), for the indicated antibodies or 100 × 2−(Ct(input)−Ct(IP RNA polymerase)) − 100 × 2−(Ct(input)−Ct(IP IgG)) for positive controls.
Luciferase Assays
C2C12 myoblasts were co-transfected with either pTA-Luc or pTA-Luc Sirt1 promoter (wild type or mutant versions) and a hRluc-TK vector (pGL4.74 from Promega) with a 10:1 ratio. When indicated, pcDNA-Egr1 and pcDNA-Sirt1 were co-transfected. Transfections were performed with FuGENE 2 (Roche Biochemicals). When assays were performed on myotubes, the transfected myoblasts were induced to differentiate 12 h after transfection. Assays were performed with the Dual Glo Luciferase kit (Promega). Results reflect the average of two experiments performed by quatriplicate.
RNA Silencing
C2C12 myoblasts were seeded on Flexcell plates in antibiotic-free media supplemented with FBS. Cells were incubated at 37 °C in a CO2 incubator until the cells were 70–80% confluent. For each well, 80 pmol of siRNA were diluted into siRNA transfection medium Lipofectamine RNAiMAX (Invitrogen) and transfected according to the manufacturer's instructions.
Lentiviral Infection
pKLO nontarget and five pKLO shEGR1 plasmids were obtained from Sigma-Aldrich. C2C12 myoblasts were transiently transfected with each of the pKLO.1 shEGR1 plasmids and the pKLO.1 nontarget and allowed to differentiate. On the second day of differentiation, EGR1 expression was assessed by Western blot on total protein. The sh-EGR1 pKLO.1 plasmid (TRCN00002128), which harbors the sequence CCGGGGATGAGCTTACCCGCCATATCTCGAGATATGGCGGGTAAGCTCATCCTTTTTG rendered higher suppression of EGR1 expression (∼50%) and was selected to produce lentivirus used in further experiments. Lentiviral production was performed at the C-BASS core from Baylor College of Medicine. C2C12 myoblasts growing on flexible bottomed plates were infected with a filtered crude lentiviral preparation (multiplicity of infection ≅ 1:1) obtained from nontarget and the selected shEGR1 pKLO.1 plasmid. The following day myoblasts were induced to differentiate and cells were harvested after 3 days in DM for further analysis.
Immunocytochemistry
C2C12 cells grown on Flexcell plates were infected with nontarget or shEGR1 lentiviral particles as explained above and allowed to differentiate. On the second day of differentiation, a set of cells was stretched for 30 min, and the other set was kept to use as no stretch control. After 3 h, the stretched and nonstretched myotubes were washed with PBS and fixed with paraformaldehyde 4% in PBS for 15 min. After two washes with PBS, cells were permeabilized with Triton 3% in PBS for 5 min and blocked with 5% albumin for 1 h. Myotubes were incubated overnight with a rabbit polyclonal SIRT1 antibody (Upstate) and a mouse monoclonal tubulin antibody (Sigma-Aldrich) diluted to 10 μg/ml in the blocking solution at 4°C. After three 15-min washes with PBS, cells were incubated with goat anti-rabbit Alexa Fluor 488 and goat anti-mouse Alexa Fluor 594 (1:250 dilution in PBS), washed with PBS (×3) and stained with DAPI (10 μg/ml) for 2 min. The flexible bottoms of the wells were excised with a scalpel and mounted on microscope glass with Prolong Gold mounting media (Invitrogen). Images of the slides were obtained in an Axioscop microscope with a 200× magnification.
Superoxide Anion and Mn-SOD Activity Determination
C2C12 myoblasts were grown on flexible-bottomed plates and allowed to differentiate. On the second day of differentiation (3 days on DM), myotubes were stretched for 30 min, and superoxide anion production was analyzed at 1, 2, 4, or 6 h after stretch. Nonstretched myotubes were used to measure basal superoxide production. The superoxide anion content was quantified with a chemiluminiscent assay based on luminol oxidation (Superoxide Anion Detection kit, Stratagene) following the manufacturer's instructions except that the NADPH oxidase activator was omitted. An aliquot from each sample was used to determine protein concentration. Mn-SOD activity was calculated by substracting CuZn activity from total SOD activity. Cell lysates were split in two, and half of the lysates were treated with 1% SDS for 20 min to inhibit Mn-SOD activity. SOD activity was determined by adding aliquots from the treated or untreated samples (20 μg) to 100 μl of a solution containing 20 μm xanthine, 0.1 units/μl xanthine oxidase aliquot and 100 μm luminol. Light intensity was measured every 10 s for 2 min. Initial reaction velocity was used to calculate the enzyme specific activity. Protein concentration from cell extracts was determined with Quick Start Bradford protein assay from Bio-Rad.
Statistics
Data from RNA, protein, CHIP assays, superoxide anion content, and Mn-Sod activity were means ± S.D. obtained from three independent experiments. Luciferase assays were performed by quadruplicate, and plotted data were means ± S.D. from two independent experiments. Statistical significance for two treatments assays (no stretch versus stretched) was calculated by Student's t test. Data from either time course analysis experiments or those involving different treatments were analyzed by ANOVA with α = 0.05.
RESULTS
Mechanical Stretch Induces Sirt1 Expression in Diaphragm Muscles
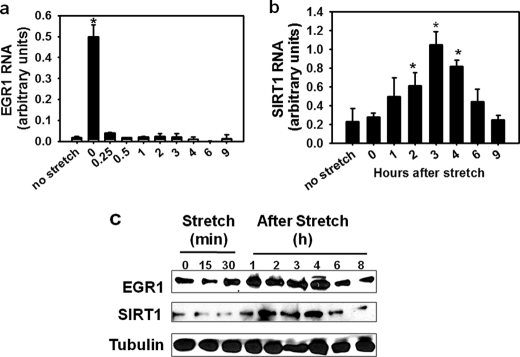
To evaluate the effect of stretch of skeletal muscles on SIRT1 expression, SIRT1 RNA and protein levels were measured in nonstretched and stretched diaphragms. SIRT1 mRNA content increased by 2.2-fold in stretched diaphragms 1 h after stretch (Fig. 1a) with a corresponding 100% increase in SIRT1 protein (Fig. 1, b and c). The stretch-dependent effect on SIRT1 RNA content correlates with an increase (3-fold) in the transcriptional activity of a minimal Sirt1 promoter (from −202 to +6) as measured by reporter assays performed in myotubes subjected to stretch (Fig. 1d).
FIGURE 1.
Mechanical stretch regulates SIRT1 expression in diaphragm muscles. a, SIRT1 RNA content was determined in excised mouse hemidiaphragm muscles (n = 3) subjected to a 30-min stretch (s) and the nonstretched (ns) hemidiaphragm by RT real-time PCR. b, SIRT1 protein was determined on whole protein (80 μg) from 30 min-stretched and nonstretched hemidiaphragms by Western blot. c, SIRT1 protein quantification was performed by densitometry from Western blots and corrected for tubulin (n = 5). Statistical difference between data from nonstretched and stretched diaphragm samples was determined by Student's t test; an asterisk represents p < 0.005. d, C2C12 myoblasts growing on FlexCell plates were transfected with pTA-luc (open boxes) or pTA-Sirt1 promoter (−212) (black boxes) Myoblasts were allowed to differentiate, and luciferase assays were performed on nonstretched (n/s) or stretched myotubes, 2 h (2h) or 4 h after stretch (4h). An asterisk indicates statistically significant difference from data obtained for pTA-luc nonstretched, p < 0.001. Bars indicate means, and error bars represent standard deviations.
EGR1 Is Involved in Stretch-dependent Transcriptional Activation of Sirt1
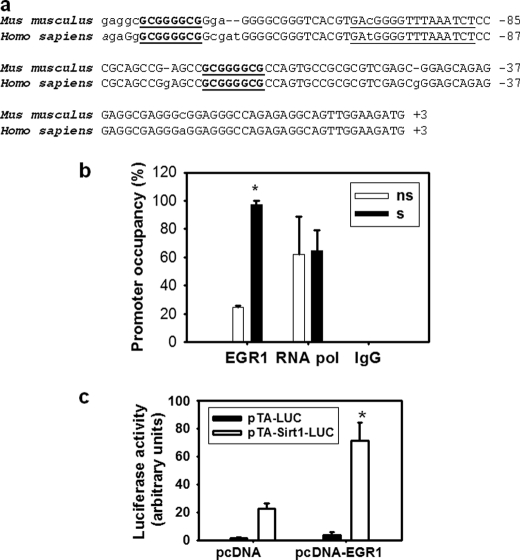
Analysis of the stretch-responsive region of the Sirt1 promoter revealed two EGR1 consensus binding sites at positions −65 and −122, which are conserved in the human promoter (Fig. 2a). Occupancy of the Sirt1 promoter region containing these regulatory elements was evaluated by ChIP experiments performed in nonstretched and stretched diaphragms. As seen in Fig. 2b, occupancy of these elements by EGR1 varied from 14% in nonstretched diaphragm muscles to 90% in stretched muscles. To evaluate the effect of EGR1 on Sirt1 transcription, the activity of a Sirt1 promoter reporter was measured under basal conditions or when EGR1 was overexpressed in C2C12 myoblasts. As shown in Fig. 2c, the activity of the Sirt1 promoter was increased by >3-fold by Egr1 overexpression. This result indicates that EGR1 has a stimulatory effect on Sirt1 transcription (Fig. 2c). To assess the effect of stretch on EGR1 and SIRT1 expression in C2C12 myotubes, the content of RNA and protein were determined before and at different times after stretch. EGR1 RNA content increased immediately after stretch (∼26-fold) (Fig. 3a). Changes in SIRT1 RNA content were observed between 1 and 6 h after stretch with maximal stimulation of ∼4-fold, 3 h after stretch (Fig. 3b). These changes were preceded by changes in EGR1 protein content that increased progressively in response to stretch between 1 and 4 h after stretch. Maximal stimulation occurred at 3 h after stretch followed by stimulation of SIRT1 protein expression occurring between 2 and 4 h after stretch (Fig. 3c and supplemental Fig. 1).
FIGURE 2.
EGR1 binds to the Sirt1 promoter and has a stimulatory effect on Sirt1 transcription. a, mouse and human Sirt1 promoter alignment. The initiator element consensus sequence is underlined, and EGR1 canonical binding sites are in boldface and underlined. b, ChIP assays were performed on 30 min stretched (s, black boxes) or nonstretched (ns, empty boxes) hemidiaphragms with an antibodies against EGR1 and RNA polymerase (positive control) and rabbit IgG (negative control). Immunoprecipitated DNA was quantified by real-time PCR, and promoter occupancy was calculated as described under “Experimental Procedures.” c, C2C12 myoblasts were transfected with pTA-luc (black boxes) or pTA-Sirt1 promoter (−212) (open boxes) in the presence of pcDNA or pcDNA-EGR1 (0.2 μg/well), and luciferase assays were performed with Dual Glo Luciferase (Promega) 24 h after transfection. Statistical difference between pcDNA and pcDNA-EGR1 transfected cells was calculated by t test. An asterisk indicates a statistically significant difference (p = 0.002). Bars indicate means, and error bars represent standard deviations.
FIGURE 3.
Mechanical stretch stimulates EGR1 and SIRT1 expression in C2C12 myotubes. C2C12 myotubes grown on flexible-bottomed plates were stretched for 30 min and collected before stretch and at the indicated times after stretch (a, b, and c). EGR1 (a) and SIRT1 (b) RNA content was determined by RT real-time PCR in stretched and nonstretched myotubes. Values were corrected for GAPDH RNA content. Error bars correspond to standard deviation, SD (n = 3). The overall statistically significant difference among groups was estimated by ANOVA, p < 0.001. An asterisk indicate values significant different from nonstretched myotubes by multiple comparison versus control analysis by Holman-Sidak method. c, 80 μg of protein from whole cell extracts obtained from nonstretched and stretched myotubes collected at the indicated times before or during stretch (Stretch) and after (After Stretch) were analyzed by Western blot with EGR1 and SIRT1 antibodies; a tubulin antibody was used to assess approximately equal loading.
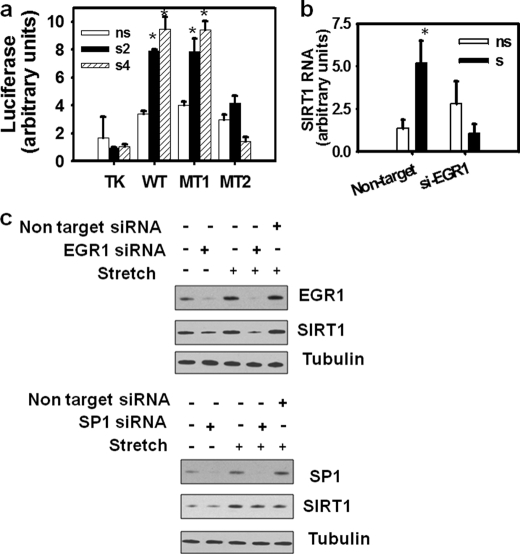
The involvement of EGR1 in the stretch-induced Sirt1 transcriptional activation was assessed by analyzing the effect of stretch of C2C12 myotubes, on the promoter activity of Sirt1 reporters containing mutations on the EGR1 binding sites. A Sirt1 promoter reporter harboring a mutation of the EGR1 binding site at −65 behaved as the wild type. (Fig. 4a, MT1). In contrast, a mutation on the EGR1 binding site at −122 abolished the effect of stretch on the Sirt1 promoter (Fig. 4a, MT2). These results indicate that the −122 EGR1 element is crucial to stimulate Sirt1 transcription in response to mechanical stretch.
FIGURE 4.
EGR1 is necessary for transcriptional activation of Sirt1 by stretch in skeletal muscle cells. a, C2C12 myoblasts growing on FlexCell plates were transfected with pTA-luc (Clontech) or pTA-Sirt1 promoter (−212) wild type or mutant versions, MT1 and MT2 obtained as described under “Experimental Procedures.” Myoblasts were allowed to differentiate for 3 days, and luciferase assays were performed on nonstretched (ns, open boxes) or stretched myotubes, 2 h (s2, black boxes), or 4 h after stretch (s4, dashed boxes). An asterisk indicates statistical significant difference from data obtained for pTA-luc nonstretched, p < 0.001. b, C2C12 myoblasts plated on FlexCell plates were infected with nontarget or shEGR1 lentiviral particles and allowed to differentiate for 3 days and subjected to a 30-min stretch (s, black boxes) or kept without being stretched (ns, open boxes); RNA content was determined in nonstretched myotubes and 3 h after stretch in the stretched myotubes by RT real-time PCR. An asterisk indicate statistical significant difference from data for nonstretched myotubes infected with nontarget viral particles, p = 0.004. Bars represent means, and error bars indicate standard deviations. c, cells were grown in growth medium (GM) for 24 h and transfected with 80 pmol of nonsilencing RNA or small interference RNA specific for EGR1 or SP1 (Santa Cruz Biotechnology) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions; after 8 h, media was replaced with differentiation medium (DM); on the second day of differentiation, EGR1, SP1, and SIRT1 protein were examined with specific antibodies by Western blot, and tubulin was used as a loading control.
EGR1 Silencing Abolishes Sirt1 Induction by Stretch in C2C12 Myotubes
To confirm the involvement of EGR1 on the stimulation of Sirt1 expression, we evaluated the effect of EGR1 silencing on SIRT1 expression in stretched myotubes. Knockdown of EGR1 abolished the stretch-dependent increase in SIRT1 RNA and SIRT1 protein in C2C12 myotubes (Fig. 4, b and c). Knockdown of SP1, a transcription factor with responsive elements (GGGCGG) that overlaps EGR1 elements, had no effect on the stretch-dependent increase of SIRT1 protein by stretch (Fig. 4c).
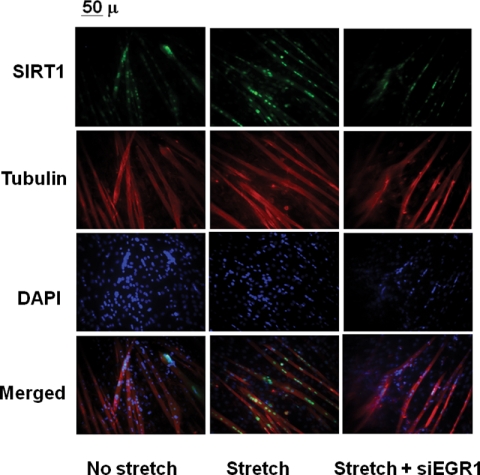
The effect of EGR1 silencing on Sirt1 protein expression was also evaluated by immunocytochemistry with a SIRT1 antibody (Fig. 5). Quantification of nuclei expressing detectable SIRT1 varied from 24 ± 1% in nonstretched myotubes to 54 ± 2% after stretch and 33 ± 7% in stretched myotubes when Egr1 was knocked down.
FIGURE 5.
EGR1 is necessary to increase nucler expression of SIRT1 in stretched skeletal muscle cells. C2C12 myotubes were infected with nontarget lentivirus (no stretch and stretch) or shEGR1 lentivirus, where indicated cells were stretched. Immunostaining was performed with a rabbit polyclonal anti-SIRT1, mouse monoclonal anti-tubulin, Alexa Fluor 488-conjugated anti-rabbit and Alexa Fluor 594 anti-mouse secondary antibodies, and DAPI staining. Images were collected in an Axioscop microscope with a 200× magnification.
Sirt1 Induction by Stretch Contributes to ROS Detoxification
SIRT1 promotes oxidative stress resistance by promoting a FOXO-dependent transcriptional activation of the mitochondrial superoxide dismutase Mn-SOD (3–5). Our previous work showed that, in contrast to FOXO1 and FOXO3a, FOXO4 is resistant to down-regulation by mechanical stretch in diaphragm muscles (26). Other authors reported that FOXO4 transcriptional activity was inhibited by acetylation in response to oxidative stress; instead, binding and deacetylation by SIRT1 prevented FOXO4 inhibition (3). We analyzed the dynamics of the association of SIRT1 and FOXO4 before and after stretch by coimmunoprecipitation and ChIP analysis. FOXO4 was immunoprecipitated from nonstretched and stretched myotubes, immediately after stretch or 3 h after stretch. An increase in acetylated FOXO4 was observed immediately after stretch, whereas interaction with SIRT1 was observed 3 h after stretch without a change in the state of FOXO4 acetylation (Fig. 6a). Results from ChIP assays performed in nonstretched or stretched myotubes showed that SIRT1 and FOXO4 binding to the Sod2 promoter at a FOXO element increased from ∼20% to full occupancy 3 h after stretch (Fig. 6b). These results point out that the stretch-dependent induction of SIRT1 promotes its association with FOXO4 and facilitates FOXO4 binding to the Sod2 promoter. The effect of stretch on SOD2 RNA content was measured in myotubes infected with nontarget or si-EGR1 lentiviral particles, before stretch and 4 h after stretch. As seen in Fig. 6c, SOD2 RNA increased by ∼3-fold after stretch and this stimulation was dependent on EGR1 expression.
FIGURE 6.
Stretch-induced SIRT1 promotes Mn-SOD expression and contributes to ROS scavenging. a, total protein (500 μg) from nonstretched (ns) or stretched myotubes collected immediately after (s0) or 3 h after stretch (s3) were immunoprecipitated with 5 μg of a goat polyclonal FOXO4 antibody (Santa Cruz Biotechnology). 80 μg of total protein (Input) from each sample were analyzed by Western blot with SIRT1 and FOXO4 antibodies. Immunoprecipitates (IP) from each sample (IP FOXO4) were subjected to Western blot with SIRT1 and acetyl-lysine antibodies. b, ChIP assays were performed with FOXO4 (black boxes) and SIRT1 (gray boxes) as specific antibodies and RNA polymerase (RNA pol) antibodies (positive control, open boxes) on nonstretched myotubes (ns), myotubes subjected to a 30-min stretch and harvested immediately (s0) or 3 h after (s3). Immunoprecipitated DNA was analyzed by real-time PCR, and promoter occupancy was estimated as described under “Experimental Procedures.” Statistical significance was calculated by ANOVA. An asterisk indicates data with significant difference from nonstretched samples, p ≤ 0.001. c, C2C12 myoblasts plated on FlexCell plates were infected with nontarget (black boxes) or shEGR1 (empty boxes) lentiviral particles and allowed to differentiate; on the third day of differentiation, myotubes were stretched or kept without being stretched; SOD2 RNA content was determined by RT real-time PCR in nonstretched myotubes (ns) or 3 h after stretch (s). Statistical significance was determined by ANOVA; an asterisk indicates statistical significant difference from data for nonstretched myotubes infected with nontarget viral particles, p < 0.001. d, Mn-SOD activity was determined as described under “Experimental Procedures” in whole cell extracts from nonstretched (ns), stretched myotubes collected immediately (s0) or 4 h after stretch (s4) from samples not treated (black boxes) or treated by addition of 10 mm nicotinamide to the media immediately after stretch (open boxes); an asterisk indicates data with a statistically significant difference from nontreated samples, p < 0.05. e, myoblasts previously infected with nontarget (non-si) or shEGR1 lentiviral particles (si-EGR1) were infected with pBaBe-Puro (non-si and si-EGR1) or pYE-hSir2 retroviral particles (non-si/Sirt1 and si-EGR1/SIRT1) and allowed to differentiate. On the third day of differentiation, cells were stretched by 30 min, and superoxide anion content was determined before stretch and at 1, 2, and 5 h after stretch. Analysis by two-way ANOVA reflected that the difference in mean values among the different treatments is greater than would be expected by chance after allowing for effects of differences by time with p = 0.003, pairwise versus non-si; si-EGR1, p = 0.0048 (significant); non-si/SIRT1, p = 0.0005 (significant); and si-EGR1/SIRT1, p = 0.0725 (not significant). Error bars indicate standard deviations (n = 3).
The effect of applying 30 min of mechanical stretch on Mn-SOD activity was studied in C2C12 myotubes. Mn-SOD activity was diminished 1 h after stretch, and the basal levels of Mn-SOD activity were recovered 4 h after stretch. Addition of nicotinamide, a SIRT1 inhibitor, to the media after stretch abolished the recovery in Mn-SOD activity (Fig. 6d). This suggests that SIRT1 activity is required to recover Mn-SOD activity after stretch.
To uncover the effect of stretch on superoxide production and the role played by EGR1-dependent induction of SIRT1, superoxide anion content was measured in myotubes expressing a si-EGR1 RNA in comparison with cells expressing a nonsilencing RNA (nontarget) (Fig. 6e). Maximal superoxide production was observed 1 h after stretch and then progressively decreased from 2 to 5 h after stretch in myotubes infected with a nontarget RNA lentivirus (Fig. 6e, non-si). When EGR1 was knocked down, myotubes failed to completely remove the excess in superoxide anion content produced after stretch (Fig. 6e, si-EGR1). Alternatively, constitutive SIRT1 expression lowered superoxide anion production of myotubes infected with nontarget silencing lentivirus (Fig. 6e, non-si/SIRT1), which is consistent with the role of SIRT1 in ROS detoxification. Moreover, SIRT1 expression rescued the ROS scavenging power of the EGR1 knocked down myotubes (Fig. 6e, siEGR1/SIRT1). These results strongly suggest that EGR1 contributes to ROS detoxification after stretch by inducing SIRT1 expression.
DISCUSSION
Muscle contractility leads to generation of ROS under physiological conditions. There are several lines of evidence that establish a nexus between exposure to ROS and diminished contractile function and fatigue during exercise (8). As part of muscle adaptation to exercise training, SOD activity has been shown to increase (27–30) mainly due to changes in Mn-SOD activity (29, 31). Rapid changes in SOD2 mRNA expression have been observed in rat skeletal muscles after an acute bout of treadmill running (32). However, the mechanisms underlying this antioxidative response are not fully understood (27). SIRT1 and FOXO are key players in the protection against oxidative stress. FOXO4 has been shown to activate the Sod2 promoter (5), and its transcriptional activity is enhanced by deacetylation by Sirt1 (3). In turn, SIRT1 expression is induced by FOXO3a in tissue under nutritional stress (25).
Our previous findings clearly indicated that a down-regulation of FOXO occurs upon stretch in diaphragms by exclusion of FOXO1 and FOXO3a from the nuclei by an Akt/Inhibitor of NFKappaB kinase-dependent mechanism. Alternatively, FOXO4 nuclear exclusion is prevented through JNK activation by stretch (26). In this study, we found that Sirt1 RNA and protein expression are induced by stretch of diaphragm muscles and myotubes. The activity of a minimal Sirt1 promoter containing the features responsible for FOXO-dependent transcriptional activation is stimulated by stretch despite decreased binding of FOXO3a to the Sirt1 promoter in stretched muscles (data not shown). Analysis of this region of the Sirt1 promoter revealed the existence of two EGR1 consensus binding sites, whose occupancy is enhanced by stretch of diaphragms. Sirt1 promoter mutants of the element at −122 failed to respond to stretch, which suggests this element is at least partly responsible for the stretch-dependent induction. The involvement of EGR1 in stretch-induced transcriptional activation of Sirt1 in skeletal muscle cells is suggested by the following: a) its stimulatory effect on the activity of the Sirt1 promoter in vitro; and b) Egr1 silencing suppressed the stretch-dependent induction of Sirt1 RNA and protein. Interestingly, silencing of Sp1, another mechanoresponsive transcription factor with overlapping binding sites with those of EGR1 on the Sirt1 promoter, had no effect on Sirt1 induction by stretch.
Our results suggest that the stretch-dependent induction of Sirt1 by EGR1 promotes FOXO4 deacetylation and FOXO4 binding to the Sod2 promoter. EGR1 silencing abolished the increase in SOD2 RNA expression triggered by stretch and affected the ability of myotubes to restore basal ROS levels. Such basal ROS levels are restored when SIRT1 is expressed artificially.
Our findings revealed a stretch-induced gene activation program that contributes to ROS detoxification in skeletal muscles. Our results demonstrated that mechanical stretch augments EGR1 expression and leads the transcriptional activation of Sirt1. Furthermore, SIRT1 stimulated FOXO-driven Sod2 expression and allowed ROS scavenging (see schematic in Fig. 7). In this way, an EGR1-SIRT1 interaction allows a transient increase in SIRT1 expression and elicits a timely response that contributes to ROS detoxification and protects skeletal muscles from oxidative damage.
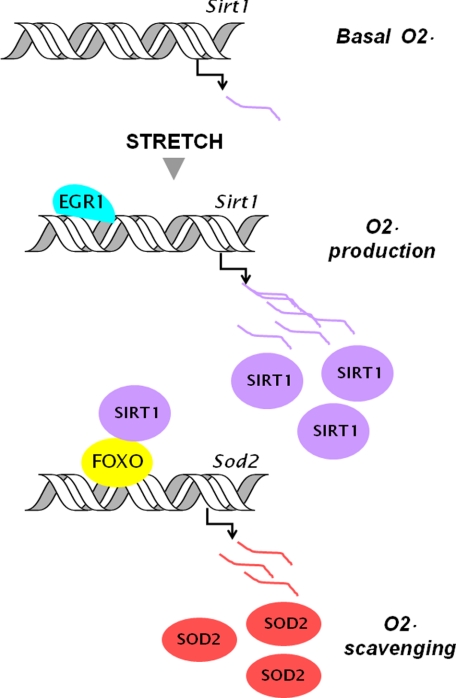
FIGURE 7.
EGR1 involvement in Sirt1 induction and ROS scavenging after stretch of skeletal muscle cells. Mechanical stretch of myotubes increases superoxide anion production and EGR1 expression. EGR1 binds to the Sirt1 promoter and activates SIRT1 expression. Induced SIRT1 binds to FOXO and promotes FOXO binding to the Sod2 promoter. Stimulation of SOD2 expression by SIRT1 contributes to ROS scavenging after stretch.
Several reports have identified that SIRT1 activation is beneficial for muscle function. Mice fed with resveratrol, a polyphenol that activates SIRT1, show increased muscle strength and resistance to muscle fatigue (33). On the other hand, SIRT1 expression has been reported to increase in rats and human skeletal muscle after exercise training (34, 35). Whether the stretch-dependent induction of Sirt1 driven by EGR1 in skeletal muscles, in vitro, is responsible for changes in SIRT1 expression in vivo and whether it plays a role in the antioxidative response that follows exercise training is an open question that needs to be addressed.
Supplementary Material
Acknowledgments
We thank Tyler Merceron for invaluable technical assistance, Haying Lu and Fred Pereira by the use of the Axioscop Microscope, and Cristina Velazco for help with the editing of graphs in this manuscript.
This work was supported in part by National Institutes of Health Grant HL63134. This work was also supported by a grant from the National Science Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ROS
- reactive oxygen species
- Mn-SOD
- manganese superoxide dismutase
- Luc
- luciferase
- ANOVA
- analysis of variance
- FOXO
- Forkhead box type.
REFERENCES
- 1. Reid M. B., Durham W. J. (2002) Ann. N.Y. Acad. Sci. 959, 108–116 [DOI] [PubMed] [Google Scholar]
- 2. Jackson M. J. (2008) Free Radic. Biol. Med. 44, 132–141 [DOI] [PubMed] [Google Scholar]
- 3. van der Horst A., Tertoolen L. G., de Vries-Smits L. M., Frye R. A., Medema R. H., Burgering B. M. (2004) J. Biol. Chem. 279, 28873–28879 [DOI] [PubMed] [Google Scholar]
- 4. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 5. Essers M. A., Weijzen S., de Vries-Smits A. M., Saarloos I., de Ruiter N. D., Bos J. L., Burgering B. M. (2004) EMBO J. 23, 4802–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nethery D., Callahan L. A., Stofan D., Mattera R., DiMarco A., Supinski G. (2000) J. Appl. Physiol. 89, 72–80 [DOI] [PubMed] [Google Scholar]
- 7. Javesghani D., Magder S. A., Barreiro E., Quinn M. T., Hussain S. N. (2002) Am. J. Respir. Crit. Care Med. 165, 412–418 [DOI] [PubMed] [Google Scholar]
- 8. Reid M. B. (2008) Free Radic. Biol. Med. 44, 169–179 [DOI] [PubMed] [Google Scholar]
- 9. Kumar A., Chaudhry I., Reid M. B., Boriek A. M. (2002) J. Biol. Chem. 277, 46493–46503 [DOI] [PubMed] [Google Scholar]
- 10. Kumar A., Boriek A. M. (2003) FASEB J. 17, 386–396 [DOI] [PubMed] [Google Scholar]
- 11. Kumar A., Khandelwal N., Malya R., Reid M. B., Boriek A. M. (2004) FASEB J. 18, 102–113 [DOI] [PubMed] [Google Scholar]
- 12. Kaeberlein M., McVey M., Guarente L. (1999) Genes Dev. 13, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tissenbaum H. A., Guarente L. (2001) Nature 410, 227–230 [DOI] [PubMed] [Google Scholar]
- 14. Rogina B., Helfand S. L. (2004) Proc. Natl. Acad. Sci. 101, 15998–16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. (2004) Nature 429, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 17. Bordone L., Motta M. C., Picard F., Robinson A., Jhala U. S., Apfeld J., McDonagh T., Lemieux M., McBurney M., Szilvasi A., Easlon E. J., Lin S. J., Guarente L. (2006) PLoS Biol. 4, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prozorovski T., Schulze-Topphoff U., Glumm R., Baumgart J., Schröter F., Ninnemann O., Siegert E., Bendix I., Brüstle O., Nitsch R., Zipp F., Aktas O. (2008) Nat. Cell Biol. 10, 385–394 [DOI] [PubMed] [Google Scholar]
- 19. McBurney M. W., Yang X., Jardine K., Hixon M., Boekelheide K., Webb J. R., Lansdorp P. M., Lemieux M. (2003) Mol. Cell. Biol. 23, 38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lavu S., Boss O., Elliott P. J., Lambert P. D. (2008) Nat. Rev. Drug Discov. 7, 841–853 [DOI] [PubMed] [Google Scholar]
- 21. Alcendor R. R., Gao S., Zhai P., Zablocki D., Holle E., Yu X., Tian B., Wagner T., Vatner S. F., Sadoshima J. (2007) Circ. Res. 100, 1512–1521 [DOI] [PubMed] [Google Scholar]
- 22. Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 23. Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 24. Wang C., Chen L., Hou X., Li Z., Kabra N., Ma Y., Nemoto S., Finkel T., Gu W., Cress W. D., Chen J. (2006) Nat. Cell Biol. 8, 1025–1031 [DOI] [PubMed] [Google Scholar]
- 25. Nemoto S., Fergusson M. M., Finkel T. (2004) Science 306, 2105–2108 [DOI] [PubMed] [Google Scholar]
- 26. Pardo P. S., Lopez M. A., Boriek A. M. (2008) Am. J. Physiol. Cell. Physiol. 294, C1056–1066 [DOI] [PubMed] [Google Scholar]
- 27. Ji L. L. (2008) Free. Radic. Biol. Med. 44, 142–152 [DOI] [PubMed] [Google Scholar]
- 28. Leeuwenburgh C., Fiebig R., Chandwaney R., Ji L. L. (1994) Am. J. Physiol. 267, R439–445 [DOI] [PubMed] [Google Scholar]
- 29. Powers S. K., Criswell D., Lawler J., Ji L. L., Martin D., Herb R. A., Dudley G. (1994) Am. J. Physiol. 266, R375–380 [DOI] [PubMed] [Google Scholar]
- 30. Higuchi M., Cartier L. J., Chen M., Holloszy J. O. (1985) J. Gerontol. 40, 281–286 [DOI] [PubMed] [Google Scholar]
- 31. Ji L. L., Stratman F. W., Lardy H. A. (1988) Arch. Biochem. Biophys. 263, 150–160 [DOI] [PubMed] [Google Scholar]
- 32. Hollander J., Fiebig R., Gore M., Ookawara T., Ohno H., Ji L. L. (2001) Pflugers Arch. 442, 426–434 [DOI] [PubMed] [Google Scholar]
- 33. Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 34. Suwa M., Nakano H., Radak Z., Kumagai S. (2008) Metabolism 57, 986–998 [DOI] [PubMed] [Google Scholar]
- 35. Marfe G., Tafani M., Pucci B., Di Stefano C., Indelicato M., Andreoli A., Russo M. A., Manzi V., Sinibaldi-Salimei P. (2010) BMC Physiol. 10:7, doi:10.1186/1472-6793-10-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.