Abstract
The human cytidine deaminase APOBEC3G (A3G) is an innate restriction factor that inhibits human immunodeficiency virus, type 1 (HIV-1) replication. Regulation of A3G gene expression plays an important role in this suppression. Currently, an understanding of the mechanism of this gene regulation is largely unknown. Here, we have identified and characterized a TATA-less core promoter with an NFAT/IRF-4 composite binding site that confers cell type-specific transcriptional regulation. We found that A3G expression is critically dependent on NFATc1/NFATc2 and IRF-4. When either NFATc1 or NFATc2 and IRF-4 were co-expressed, A3G promoter activity was observed in cells that normally lack A3G expression and expression was not detected in the presence of the individual factors. This induced A3G expression allowed normally permissive CEMss cells to adopt a nonpermissive state, able to resist an HIV-1Δvif challenge. This represents the first reporting of manipulating the restrictive state of a cell type via gene regulation. Identification of NFAT and IRF family members as critical regulators of A3G expression offers important insight into the transcriptional control mechanisms that regulate innate immune responses and identifies specific targets for therapeutic intervention aimed at effectively boosting our natural immunity, in the form of a host defensive factor, against HIV-1.
Keywords: Gene Regulation, HIV, NFAT Transcription Factor, Tissue-specific Transcription Factors, Transcription Promoter, APOBEC3G, Cellular Restriction Factors, IRF Transcription Factor
Introduction
Human A3G is an innate immune resistance factor for a broad range of retroviruses. The gene is expressed in hematopoietic cell populations, lymphoid tissues, and selected established T cell lines (1). The A3G protein restricts human immunodeficiency virus, type 1 (HIV-1)2 infection by accessing the budding virion and disrupting the reverse transcription of HIV-1 RNA in target cells. The post-entry block impairs initiation (2), inhibits transcript elongation (3), and induces G-to-A hypermutation in the nascent viral cDNA through cytidine deamination (4–7). HIV-1 Vif potently counteracts this restriction in the producer cell by targeting A3G to the proteasome, thereby preventing incorporation of A3G into the virus particle (8–11). The mechanism for producer cell mediated A3G restriction and HIV-1 Vif counteraction have been extensively studied and characterized (12) and the Vif:A3G regulatory circuit is one of the most interesting examples of how cellular restriction factors participate in the exertion of a powerful intracellular defense mechanism.
A3G mRNA and protein levels vary across both developmental and differentiation transitions, and these differences influence the restrictive capacity of the particular cell type. Decreased A3G expression and susceptibility to HIV-1 occurs during differentiation of human monocytes to monocyte-derived macrophages (13). Conversely, increased A3G expression with enhanced resistance to HIV-1 occurs during dendritic cell maturation (14). Finally, decreased levels of A3G have been noted in the CD4+ T helper 2 (Th2) subset, compared with T helper 1 (Th1) cells (15). Taken together, these observations suggest that A3G is an important part of an effective innate immune response (16–20).
The molecular pathways responsible for the spatial and temporal regulation of A3G gene expression have not been defined. Several studies have shown that interferon, cytokine, and mitogenic stimulation can induce A3G gene expression and potential binding sites for transcription factors in the putative promoter region have been tentatively identified (21–24). Many of these transcription factors (e.g. Ets-1, c-Myc, and IRF-1) are ubiquitously expressed and therefore are not likely to influence A3G expression across the developmental and differentiation transitions. Accordingly, the transcription factors governing promoter selectivity and cell type specificity have yet to be established.
Here, we performed a detailed analysis to define a minimal cell type-specific promoter element that contains neither a TATA nor CAAT box in their usual upstream location but contains a critical composite binding site for the NFAT and IRF families of transcription factors. When NFATc1 (also known as NFAT2) or NFATc2 (also known as NFAT1) and IRF-4 were expressed in tandem, A3G expression was strongly induced in nonimmune cells and T cell lines that do not usually express A3G. This manipulated expression correlated with the ability of these cells to potently resist an HIV-1Δvif challenge. Identifying the transcription factor families and member proteins that control cell type-specific A3G expression is thus important for understanding how the virus and antiviral innate immune responses intersect.
EXPERIMENTAL PROCEDURES
5′ Rapid Amplification of cDNA Ends (RACE)
RACE clones were generated using First Choice RLM-RACE (Ambion) with poly(A)+ RNA from selected CD4+ T cell subsets. The RNA was reverse transcribed at 50 °C with Thermoscript RT-PCR (Invitrogen) and A3G specific reverse primer 792R (5′-CAG GTG ACC TCA TAC TCC TGG T-3′). The RLM-RACE clones were amplified by nested PCR from the cDNA using the 5′-RACE outer (5′-GCT GAT GGC GAT GAA TGA ACA CTG-3′) and inner (5′-CGC GGA TCC GAA CAC TGC GTT TGC TGG CTT TGA TG-3′) and 3′-A3G-specific primers 792R (above) and 799R (5′-CTC TGG GTG GTA CTT AAG TTC GGA A-3′). Cycling conditions were as follows: 95 °C at 12 min; cycle, 94 °C at 30 s, 60 °C at 30 s, 72 °C at 2 min, and 72 °C at 10 min. The PCR product DNA was inserted into pCR2.1 TOPO (Invitrogen) and then sequenced.
Generation of pGL4-A3G Upstream Luciferase Reporter Constructs
The A3G upstream truncation series of −402 nt, −298 nt, and −240 nt were amplified from BAC clones (Stratagene, CTD-3214F20, genomic alignment of BAC ends AQ18854 and AQ212961) and cloned into the pGL4.10 luciferase reporter construct (Promega). The reverse primer for all constructs was 5′-CCT TGG CCG GCT AGT CCC GA-3′. The forward primer sequences are as follows: −402 nt, 5′-ATG GTG GAG TGG CGG CTC-3′; 298 nt, 5′-ACT TTC TCT TTC CCT TTG-3′; and 240 nt, 5′-ACA GAG CGG CCT GTC TTT-3′. Mutations in the putative IRF (5′-GGC GCT GGC TGC AAT GAC GCG AGA TTT CCC TTT G-3′), NFAT (5′-CTG CAA TGA CTT TCT CTT GAA CTT TGC AAT T-3′), and Sp1 (5′-GGA GAG GAG GCT CCA GCT ATA CAT GAC CAC CAG G-3′) sites were introduced into the 402-nt construct by site-directed mutagenesis PCR (GeneTailor, Invitrogen) (bold-faced letters are sequence in which subsequent mutations were introduced). Mutations were confirmed by sequencing.
Cell Lines and Culture and Stimulation Conditions
All cell lines were maintained under standard conditions. 293T and HeLa cells were cultured in DMEM (10% serum, 100 units/ml penicillin/streptomycin), whereas the CEM-A, CEM, and CEMss cell lines were grown in RPMI (10% serum, 100 units/ml penicillin/streptomycin). The A3G-negative 293T and HeLa cell lines as well as the A3G-expressing CEM-A cell line are adherent. Where designated, cells were stimulated with ionomycin (500 ng/ml, Santa Cruz Biotechnology, catalog no. sc-3592), cyclosporin A (CsA; 10 μg/ml, Santa Cruz Biotechnology, catalog no. sc-3503), or DMSO for the indicated times. Cells under chronic ionomycin or CsA treatment were washed and resuspended in fresh, ionomycin-containing media every 48 h or CsA-containing media every 24 h.
Cell Transfection
HeLa and CEM-A cells were transfected with Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions. 293T cells were transfected with polyethyleneimine (25). Constructs were co-transfected with the internal Renilla reporter construct pGL4.74 at a 2:1 luciferase:Renilla ratio, or with pcDNA3.1, IRF-4, or IRF-8 and GFP at a 6:3:1 IRF:luciferase:GFP ratio. An SV40-driven luciferase reporter, pGL4.13, was included as the positive control (pGL4 series, Promega). Each transfection was performed in triplicate, and a minimum of four independent experiments is shown. NFATc1, IRF-4, and IRF-8 cDNAs (Open Biosystems 8327711, 4861223, and 8991963, respectively) were subcloned into the pcDNA3.1 (NFATc1 and IRF-4) or pCMV4 (IRF-8) expression vector. The pcDNA2-NFATc2 expression vector was kindly provided by Laurie Glimcher (Harvard Medical School, Boston, MA).
Luciferase Reporter Assay
Cells were processed 36–48 h post-transfection with the Dual-Glo luciferase assay kit according to the manufacturer's instructions (Promega). Signal was acquired on the PerkinElmer 1420 luminescence counter Victor Light luminometer. Transfections were done in triplicate and normalized to the Renilla internal control (pGL4.74), and the average of the three transfections was compared with the pGL4.13 for each assay. In the transient transfections of the IRF-4 and IRF-8 cDNAs into 293T, fluorescence and luminescence were acquired on a Synergy 4. Luciferase was normalized to GFP and the pGL4.10 background was subtracted, and the relative promoter activity compared with the pcDNA3.1+pGL4.57 normalized signal. Statistical analysis was done using the Student's t test under the conditions of two-tailed distribution and two-sample equal variance.
Bioinformatic Analyses
To characterize the putative A3G promoter, the ALGGEN PROMO, which runs TRANSFAC (version 8.3), was utilized. We used the combined string and weight matrix searching of the Transcription Element Search System (TESS) to predict transcription factor binding sites (TFBS) in the DNA sequence of the 402-nt construct. The statistically significant factors that were predicted to bind to the DNA sequence by the combined search query were validated by independent analyses with WWW Promoter Scan and WWW Signal Scan.
Cell Fractionation and Immunoblot Analysis
Cells were harvested and fractionated into nuclear and cytoplasmic fractions as described previously (26). Whole cell lysates were prepared by harvesting cells and resuspending in lysis buffer (0.5 m Tris, 10% glycerol, 3% SDS, 5% β-mercaptoethanol). Lysates were resolved on an 8% SDS-PAGE gel and probed with monoclonal α-NFATc1 (7A6; antibody sc-7294X, Santa Cruz Biotechnology) or α-NFATc2 (4G6-G5, antibody sc-7296X, Santa Cruz Biotechnology), or a 12% SDS-PAGE gel and probed with a polyclonal α-A3G (kindly provided by Michael Malim, King's College London, London, UK), polyclonal α-IRF-4 (H-140, antibody sc-28696X, Santa Cruz Biotechnology), or monoclonal α-actin (CP-01, Calbiochem).
ChIP
Chromatin was prepared according to the Abcam ChIP protocol. Prior to immunoprecipitation, samples were precleared with 80 μl salmon sperm DNA/protein A/G PLUS-agarose (Santa Cruz Biotechnology, antibody sc-2003), and 100 μl of the supernatant was retained as the input control sample. The remaining supernatant was divided into four fractions, each receiving 5 μg antibody-Ig control (control IgG, antibody sc-2025, Santa Cruz Biotechnology), α-NFATc1, α-NFATc2, or α-IRF-4 (as used above) and incubated overnight. Following incubation with antibody, samples were processed according to the Millipore ChIP protocol with the following modifications; IP samples were captured with 30 μl of salmon sperm DNA/protein A/G PLUS-agarose for 1 h on a rotating platform in a cold room, beads were washed with 1× TE buffer twice, and DNA was purified using the QIAquick PCR purification kit (Qiagen).
Quantitative Real-time PCR
Real-time PCR analysis was performed using a TaqMan assay (Applied Biosystems). A3G primers and probe were as follows: A3G_ChIP_F-369, 5′-GGG GAG GGG CTT GTG C-3′; A3G_ChIP_R-293, 5′-AAG GCA ATT GCA AAG GGA A-3′; and A3G_ChIP_P-319, 5′-6FAM-GGG CGC TGG CTG CAA TGA CTT TC-TAMRA-3′. Cycling conditions were 95 °C for 10 min followed by 45 cycles of 95 °C for 15 s/60 °C for 1 min. The expected product length for A3G is 95 nt, which was confirmed by gel electrophoresis of the products. For each run, samples for each ChIP antibody sample were assayed in triplicate, and the percent input was determined as described previously (27). Student's t test under the conditions of two-tailed distribution and two-sample equal variance was used to determine statistical significance.
Immunofluorescence
293T cells were grown on coverslips and transfected by Lipofectamine LTX. 48 h post-transfection, cells were fixed with 4% formaldehyde, permeabilized with 0.2% Triton X-100, washed with PBS, and blocked in PBSAT (PBS/1% BSA/0.1% Tween 20). Coverslips were then incubated with α-A3G, incubated with an α-rabbit conjugated to Alexa Fluor 568 (Molecular Probes), incubated with DAPI, and mounted. Image acquisition was done using a ZEISS LSM700 point scanning confocal system, mounted to an Axio Observer Z1 running off of ZEN2009. CEM-A cells were grown on coverslips and treated with 10 μg/ml CsA for 3 days. Slides were fixed and stained for A3G as above. Images were acquired using a Nikon Eclipse TS100, with a 60× objective and Spot RT camera. Image analysis was done with Spot Advanced software.
HIV-1 Virus Challenge and Replication Assay
CEM and CEMss cells were pretreated as indicated and inoculated with 5 ng of p24Gag for 24 h. Cells were then washed and resuspended in medium containing DMSO or ionomycin at ∼1 × 106 cells/ml. Supernatants were harvested every 48 h. Replication kinetics were monitored with an HIV-1 p24Gag ELISA assay (PerkinElmer Life Sciences).
RESULTS
Identification of Alternative A3G Transcription Start Sites
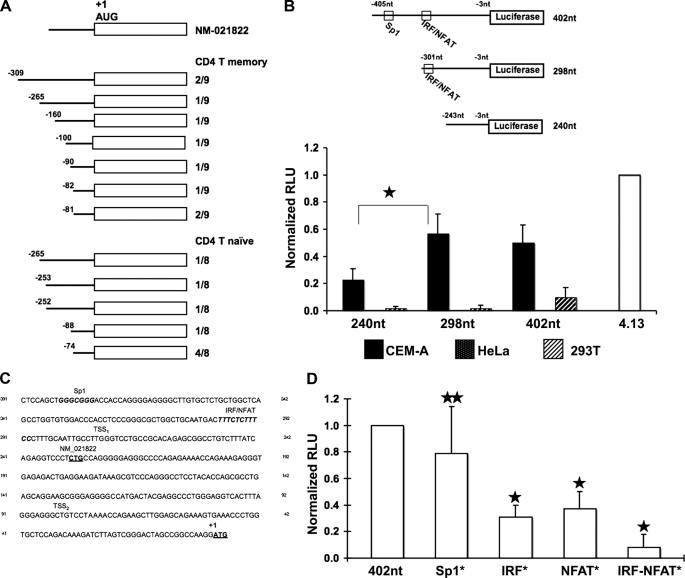
Using a combination of 5′ RACE and high density resolution tiling arrays, the ENCyclopedia Of DNA Elements (ENCODE) Project has found that transcription start sites (TSSs) are somewhat promiscuous but nonrandom (28). Specifically, in regard to the regulatory region of the A3G gene, TSSs have previously been reported −58 nt to −361 nt relative to the translation start site using the A3.01 T cell line (29). To address the possibility that T cell lines may exhibit an aberrant transcription profile, we employed nonexhaustive 5′ RACE analysis to map TSSs using cDNA derived from CD4+ T cells (naïve and memory cell populations) and cultured T cell lines; all cell types constitutively expressed A3G. The results obtained from the primary CD4+ T cells showed a clustering of TSSs ∼ 275 nt (6/17 clones; referred to as TSS1) and ∼80 nt (10/17 clones; referred to as TSS2) upstream of the translation start site (+1 AUG; Fig. 1A). There was no bias toward detection of the upstream or downstream TSS in naïve or memory CD4+ T cells. Notably, we observed the same variability and clustering in cDNAs derived from the CEM and HUT78 T cell lines (data not shown).
FIGURE 1.
Delineation and analysis of the A3G minimal promoter. A, 5′ RACE was performed using RNA isolated from CD4+ T cell naïve and memory subsets, and 17 individual clones were analyzed. NM_021822 represents a previously defined TSS. The TSSs recorded are tabulated. The AUG start codon is indicated. B, the upper schematic depicts the luciferase reporter constructs with numbering relative to the initiating ATG, and the construct length shown. Highlighted are a previously described Sp1 binding site and a putative composite IRF/NFAT binding site that was delineated via TRANSFAC® analysis (see C below). Luciferase activity was measured for each construct in CEM-A (black bars), HeLa (speckled bars), and 293T (hatched bars). The activity of each construct was normalized against the 4.13 SV40-luciferase positive control (normalized relative light units), M p value<0.00001. C, TRANSFAC® analysis of the region upstream of the translation start site revealed several putative transcription factor binding sites including a predicted IRF/NFAT composite site (consensus IRF binding site, TTTCNNTT; consensus NFAT binding site, TTTCC). The previously reported Sp1 site (29) is shown for comparison. The TSS1 and TSS2 clusters defined by RACE in A are indicated, as is the previously reported transcription start site (NM_021822). D, the Sp1, IRF, and NFAT binding sites predicted in C were mutated individually or in combination and assayed in the luciferase reporter system. p values were determined by comparing activity of the mutant construct to parental construct (star, p value > 0.05; two stars, p value < 0.00000001.
Differential Cell Type-specific A3G Gene Expression
The different A3G expression patterns in human tissue and hematopoietic cell populations suggest a cell type-specific regulation (13, 30–33). To characterize such differential regulation, we determined the transcriptional activity of the putative A3G promoter in two different cell types: cells that normally express A3G (termed nonpermissive cells because they restrict an HIV-1Δvif infection) and cells that do not express A3G (dubbed permissive as they are vulnerable to an HIV-1Δvif infection). Constructs were created linking putative A3G promoter fragments to a firefly luciferase reporter gene. The 402-nt construct and the 298-nt encompass the TSS1 and TSS2 sequences (Fig. 1A). In contrast, the 240-nt construct includes the TSS2 only. Each construct was expressed transiently, along with a Renilla luciferase internal control, in CEM-A, HeLa, and 293T cells. These lines have distinct tissue origins, but only the CEM-A T cell line expresses detectable endogenous levels of A3G.
Transcriptional activity of individual constructs in each cell line was measured with a Dual-Luciferase assay system (Promega). When transfected into CEM-A cells, luciferase expression from the 240-nt construct was significantly less than that from the 298-nt and 402-nt constructs (Fig. 1B) Luciferase activity was undetectable when the constructs were transfected into HeLa cells, and although a low level of activity was detected in transfected 293T cells, none of the constructs exhibited statistically differential activity. These results suggest that a major regulator for the cell type-specific regulation of A3G likely resides in this 298-nt construct.
Transcription Factor Binding Sites in Putative A3G Promoter
TFBSs with cell type specificity were identified with in silico mapping algorithms (34, 35). The computationally identified TFBS with high conservation and log likelihood scores located distal to TSS1 and TSS2 include a previously identified Sp1 (−383), as well as a novel IRF/NFAT (−301), numbered according to the +1 ATG (Fig. 1C). No recognizable TATA or CAAT elements were detected. Interestingly, Sp1 is a ubiquitously expressed protein that appears to play an important role in the binding of the RNA polymerase II complex to the initiation site; it is particularly critical in exerting this function in promoters that lack canonical TATA or CAAT elements. In contrast to Sp1, IRF and NFAT proteins display significant cell and tissue type-specific expression. The IRF/NFAT composite site identified here is of particular note as cooperative binding of NFAT and IRF to such a composite site has already been shown to critically activate the IL-4, IL-10, and IL-12 promoters in a strikingly tissue-specific manner (36–39). IRF-4 has also been shown to be essential for the cell type-specific regulation of activation-induced deaminase, a member of the activation-induced deaminase/APOBEC family of cytidine deaminases (40).
To determine the functional role that these identified TFBS play, we performed site-directed mutagenesis of the NFAT, IRF, and Sp1 binding sites (alone and in tandem) in the 402-nt construct and measured the effect(s) on transcriptional activity in CEM-A cells. The NFAT and IRF composite binding site exerted significant control over promoter activity (Fig. 1D). When compared with the parental construct, mutation of either binding site resulted in a 3–4-fold decrease. Mutation of both NFAT and IRF binding sites caused an almost complete loss of promoter activity. Mutation of the Sp1 site had surprisingly little effect, suggesting that the Sp1 site is not critical for cell type-specific transcriptional regulation of the A3G promoter.
Endogenous NFAT and IRF Proteins Bind to A3G Regulatory Sequences
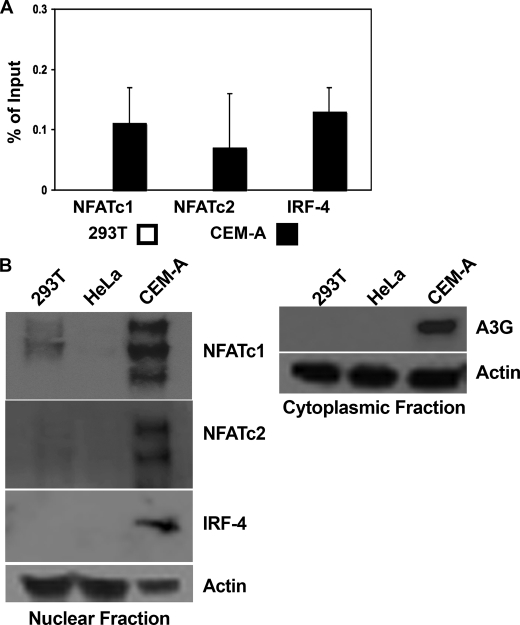
To determine whether NFAT and IRF proteins in fact occupy the IRF/NFAT composite site of the A3G core promoter, we performed ChIP experiments, comparing the CEM-A and 293T cell lines. We began these studies with a primary focus on two of the five known NFAT family members, NFATc1 and NFATc2. Both proteins are expressed at high levels in primary CD4+ T cells, the major target cells for HIV-1 and both have been shown to bind the same DNA target sequence(s). They are functionally redundant, but their expression patterns oscillate during T cell development and activation (41). Of note, NFAT expression is also seen in nonimmune cells, suggesting that any strict cell type-specific control of the A3G promoter is likely conferred by an additional factor or factors. IRF-4, a lymphoid/myeloid restricted family member (42), is one such candidate transcription factor. It has been previously shown to associate with both NFATc1 and NFATc2 to modulate cell type-specific cytokine expression in T cells (36–38). As seen in Fig. 2A, neither NFAT nor IRF-4 protein binding is detected within the core promoter sequence in 293T cells, whereas protein-specific binding to the same fragment within the CEM-A cell line is readily detectable.
FIGURE 2.
Binding of the minimal promoter fragment and differential expression patterns of IRF and NFAT. A, ChIP assays were performed on 293T and CEM-A cell lysates using antibodies against NFATc1, NFATc2, or IRF-4. Pulldown was detected by quantitative real-time PCR and percent of input determined by comparison with an Ig control pulldown. Average cycle threshold of GAPDH for input samples was used as an internal control for quality of input DNA, 293T Ct 29.0 ± 1.47, CEM-A Ct 28.6 ± .86. B, 293T, HeLa, and CEM-A cell lysates were subcellularly fractionated and probed for the presence of NFAT and IRF family members. Immunoblots probed with an antibody specific for actin served to monitor loading equivalency. Ct, threshold cycle.
We next correlated the relative amounts of nuclear localized NFATc1, NFATc2, and IRF-4 in our cell lines with the results of the ChIP experiments. Multiple NFATc1 and NFATc2 isoforms were present in CEM-A cells, whereas only low levels of NFATc1 were detected in 293T cells (Fig. 2B). These low levels of NFATc1 are consistent with the minimal core promoter activity detected previously (Fig. 1B). Not surprisingly, the immune cell-restricted IRF-4 was only observed in the CEM-A cells (Fig. 2A). HeLa cells did not express NFATc1, NFATc2, or IRF-4. A3G protein expression was detected only in CEM-A cells, its expression correlating with the presence of both NFAT and IRF.
NFAT and IRF-4 Are Necessary and Sufficient for A3G Expression
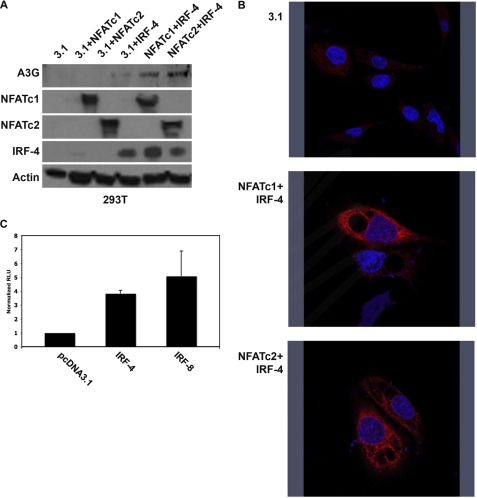
To further define the roles of NFATc1, NFATc2, and IRF-4 in this gene regulation, we performed transient transfections, expressing these transcription factors, individually and in tandem, in 293T cells. Neither NFATc1 nor NFATc2 expression alone was sufficient for A3G promoter activity. Weak gene expression was seen with the ectopic expression of IRF-4, but robust expression was detected only when IRF-4 and NFAT (either NFATc1 or NFATc2) were co-transfected (Fig. 3A). A3G expression detected in the IRF-4 transfection suggests that the low levels of endogenous NFAT expression observed in the 293T cell line (Fig. 2B) can function with IRF-4 to drive A3G expression. High levels of A3G expression, however, require both NFAT and IRF family members. Immunofluorescence of the transiently transfected 293T cells showed that the induced A3G exhibited a cytoplasmic localization, similar to the reported subcellular localization of endogenous A3G (Fig. 3B). It is important to note that these transfections likely result in the overexpression of the transcription factors of interest. This overexpression and the complexity of the NFAT family makes it difficult to conclude with certainty the identity of the specific NFAT family members involved in endogenous A3G gene expression. However, it is clear that the simultaneous, exogenous expression of the NFAT and IRF proteins can drive A3G expression in the 293T cell line, which does not normally express this gene.
FIGURE 3.
NFAT and IRF are necessary and sufficient for induction of A3G expression. A, 293T cells were co-transfected with the indicated expression plasmids. Whole cell lysates were probed for expression of transfected plasmids and A3G. Immunoblots probed for actin expression are also shown (loading control). B, 293T cells were transfected with the indicated plasmids, and cells were stained for A3G. Nuclei were counterstained with DAPI. C, 293T cells were transfected with pcDNA3.1, IRF-4, or IRF-8 plus reporter construct. Relative promoter activity was determined by comparing the IRF-4 and IRF-8 signal with the pcDNA3.1 signal.
Initially, IRF-4 was assayed because its expression is restricted to the lymphoid lineage, and it is expressed in the CEM-A T cell line. However, in vivo, A3G expression is also detected in cells of the myeloid lineage, and IRF-8 expression dominates in this subpopulation of cells. To compare the ability of IRF-4 and IRF-8 to transactivate the A3G promoter, luciferase reporter assays were performed. The A3G minimal promoter construct with either IRF-4 or IRF-8 was co-transfected into 293T cells. The expression of either IRF-4 or IRF-8 induced a 4–5-fold increase in A3G promoter activity when compared with the vector control transfection (Fig. 3C). Following up this result, we also observed IRF-8 was capable of inducing A3G protein expression when overexpressed in 293T cells (data not shown), similar to the A3G induction observed in the expression of IRF-4 via transient transfection (Fig. 3A). The activity of these related IRF family members in cells of distinct lineages may explain the restricted expression pattern of A3G in immune cells in vivo. Taken together, these experiments show that ectopic expression of NFAT and IRF proteins is sufficient for induction of A3G gene expression in cells that do not naturally express A3G.
Nuclear Translocation of NFAT Induces A3G Expression in Permissive Cells
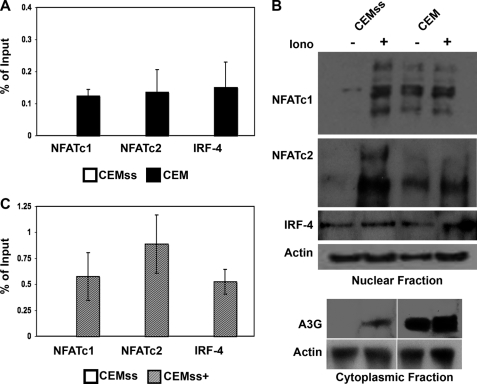
We next probed the question of NFAT and IRF-4 protein occupancy of the A3G promoter in genetically related, but phenotypically distinct, T cells. CEMss cells do not normally express A3G and succumb to an HIV-1Δvif challenge, whereas CEM cells express levels of A3G comparable with that observed in primary T cells and potently suppress an HIV-1Δvif infection (1). ChIP experiments showed endogenous NFATc1, NFATc2, and IRF-4 from CEM nuclear extracts were fully capable of binding the TFBS, supporting the established paradigm that A3G gene expression confers upon cells the ability to restrict HIV-1Δvif. In contrast, binding of this regulatory sequence in cells vulnerable to an HIV-1Δvif infection, such as CEMss T cells, was not detectable (Fig. 4A).
FIGURE 4.
Enrichment of nuclear NFAT induces transactivation of the A3G promoter in T cells. A, antibodies against NFATc1, NFATc2, IRF-4, and an Ig control were used in a ChIP assay. Immunoprecipitated samples from CEMss and CEM were analyzed in a TaqMan real-time PCR assay using primers directed to the A3G minimal promoter region. The average cycle threshold of GAPDH for input samples was used as an internal control for quality of input DNA, CEMss Ct 27.4 ± .87, CEM Ct 27.3 ± .78. B, Mock-treated (−) and ionomycin (Iono)-stimulated (+) CEMss and CEM cell lysates were subcellularly fractionated and probed for NFATc1, NFATc2, or IRF-4 expression in the nuclear fraction. A3G expression was assayed in the cytoplasmic fraction. Actin expression is shown as a loading control. C, CEMss and ionomycin stimulated CEMss (CEMss+) cells were assayed for NFATc1, NFATc2, or IRF-4 occupation of the A3G promoter as in A. Average cycle threshold of GAPDH for input samples was used as an internal control for quality of input DNA; CEMss Ct 27.4 ± .87, CEMss+ Ct 28.2 ± .78. Ct, threshold cycle.
Examination of the expression profile of the NFAT and IRF-4 proteins in the CEM and CEMss cell lines supported the ChIP results. In unstimulated T cells, NFAT is found in the cytoplasm, and only under conditions that activate calcineurin is NFAT-dephosphorylated, allowing its translocation to the nucleus where it can act as a transcription factor. Interestingly, although the overall cellular level of expression of NFAT in both CEM and CEMss cells was comparable (data not shown), neither NFATc1 nor NFATc2 were found in the nuclei of unstimulated CEMss cells (Fig. 4B). Stimulation of these cells with ionomycin, a pleiotropic activator that results in calcineurin activation, led to the appearance of both NFATc2 and NFATc1 in the nucleus. Concomitant with this relocalization of NFAT was the appearance of cytoplasmic A3G expression in the CEMss cells (Fig. 4B). CEM cells, in contrast, contained NFATc1 and NFATc2 in the nucleus under both unstimulated and ionomycin-treated conditions; ionomycin did not affect additional mobilization of NFAT. As would be predicted from such an observation, levels of A3G in these cells were altered little in response to ionomycin treatment. IRF-4, in contrast to NFAT, is normally found within the nucleus of T cells, and this was observed in both the CEM and CEMss lines. Expression levels were comparable and unresponsive to ionomycin.
To follow up the NFAT mobilization observation, ChIP analysis was carried out with untreated and ionomycin-stimulated CEMss cells. Ionomycin treatment of CEMss cells resulted in detectable binding of NFATc1, NFATc2, and IRF-4 to the A3G promoter, whereas untreated cells displayed no occupancy of the promoter (Fig. 4C). Based on these observations, we propose that NFAT and IRF binding to the A3G promoter is dependent on the nuclear localization of NFAT, and this binding represents an important regulatory step in the gene expression of A3G.
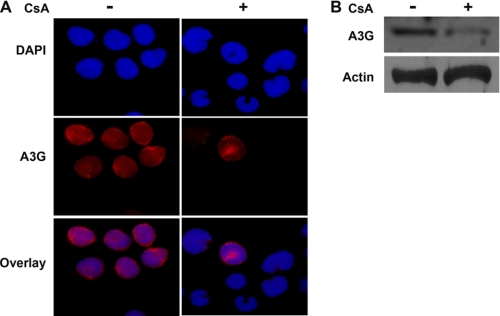
To further verify the significance of the regulation of A3G by the binding of NFAT and IRF-4 to the A3G promoter, we inhibited NFAT occupancy of the A3G promoter using cyclosporin A. Cyclosporin A directly inhibits calcineurin activation and, thereby, albeit indirectly, NFAT nuclear translocation. CEM-A cells, which normally express A3G, were treated with cyclosporin A and A3G cellular expression examined by immunofluorescence (Fig. 5A) and Western blotting (Fig. 5B). Cyclosporin A treatment decreased the abundance of the A3G protein in CEM-A cells, consistent with the previous results showing that nuclear localization of NFAT is critical for A3G promoter activity (Fig. 5A). This observation was confirmed by Western blot of treated and untreated cells (Fig. 5B). From these experiments, it is clear that enriching NFAT levels in the nucleus induces expression of A3G.
FIGURE 5.
Inhibition of NFAT nuclear translocation decreases A3G expression. A, CEM-A cells were mock-treated or treated with cyclosporin A, and A3G expression was detected by immunofluorescence. B, mock-treated or cyclosporin-treated CEM-A cells were lysed, and A3G expression was examined (upper panel). Actin expression is shown as a loading control.
NFAT-mediated Induction of A3G in CEMss Cells Renders Them Resistant to HIV-1Δvif Infection
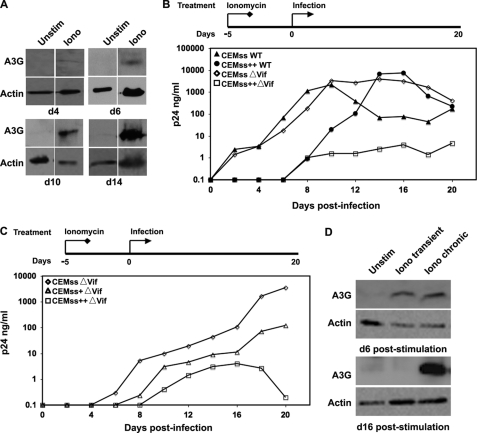
Ultimately, the most important consideration of manipulating A3G expression levels is to determine the biological significance of this regulation. We infected ionomycin-treated CEMss cells with both wild-type and HIV-1Δvif to examine whether artificial induction of this protein could confer upon cells an ability to resist a viral challenge (Fig. 6). Initially, we verified that A3G gene expression was stably induced by treatment of CEMss cells with ionomycin (Fig. 6A). We then examined whether this stimulated gene expression was active in a functional assay. Untreated CEMss cells, not unexpectedly, were equally susceptible to both wild-type and HIV-1Δvif (1) (Fig. 6B). Peak viremia was recorded at approximately the same level, on day 10 post-challenge for both infections. Interestingly, treatment of CEMss cells with ionomycin resulted in a kinetic delay of wild-type infection, peak viremia was not seen until day 14. Most impressively, when these cells were stimulated with ionomycin and challenged with HIV-1Δvif, they potently resisted a HIV-1Δvif infection over the entire viral challenge time course. Peak viremia was 400–2000-fold lower than that seen in all other experimental conditions, essentially recapitulating the phenotype of the parental CEM cells, which are the standard for nonpermissivity in these replication assays. This phenotypic reversion has been observed previously (1), but only when A3G itself was directly expressed in the CEMss line. This is the first reporting of recapitulating the phenotypic conversion via gene regulation.
FIGURE 6.
Induced A3G protein is stable and exerts an antiviral effect. A. The stability of induced A3G expression in CEMss cells was determined by treating cells with ionomycin (Iono) over a 14-day time course. The kinetics of A3G expression induction were examined by immunoblotting for expression on days 4, 6, 10, and 14. B, mock and ionomycin-stimulated (Stim) CEMss (CEMss++) cells were treated for 5 days prior to viral challenge with wild-type HIV-1 (filled symbols) or HIV-1Δvif (open symbols). Subsequent viral replication was monitored by HIV-1 p24Gag ELISA. C, CEMss cells were mock or ionomycin-stimulated for 5 days prior to infection. Ionomycin- treated cells then either continued to receive stimulation (++) or ionomycin was washed out of the medium after a 24-h incubation with HIV-1Δvif virus (+). Spreading infection was assayed by p24Gag ELISA. D, whole cell lysates were prepared from mock, transient ionomycin-, and chronic ionomycin-stimulated cultures on d1 of the replication curve, corresponding to day 6 of the stimulation time course. Additional whole cell lysates were prepared on day 10 of the replication curve, corresponding to day 16 of the stimulation protocol. Whole cell lysates from both days were probed for A3G and actin expression by Western blot. Unstim, unstimulated.
To further dissect the importance of the timing of stimulated A3G expression, we developed a transient ionomycin stimulation protocol designed to examine induced A3G expression in the initial exposure phase of infection. Based on previous expression induction kinetics (Fig. 6A), CEMss cells were pretreated with ionomycin for 5 days prior to viral challenge. Upon challenge, ionomycin treatment was discontinued in a subset of cells to create transiently stimulated CEMss cells, whereas a second population continued to be treated. At the time of infection, A3G expression levels were the same under both the transient and chronic stimulation conditions (Fig. 6D, top panels). As seen previously, chronic ionomycin stimulation led to a significant suppression of viral replication and even more interesting, a simple transient ionomycin stimulation of CEMss cells with allowed them to effectively restrict an HIV-1Δvif challenge (Fig. 6C). In fact, this transient stimulation resulted in not only the kinetic delay in viral replication, but peak viremia was also suppressed almost 100-fold when compared with untreated cells infected with HIV-1Δvif (Fig. 6C). This acquired resistance to a viral challenge may have important implications for understanding early events in acute infection.
DISCUSSION
In this study, we identified the minimal core promoter fragment and critical transcription factors responsible for functional A3G gene expression. Using this data, we then successfully induced the expression of the A3G gene in two different cell lines, which do not normally express endogenous A3G. Examination of primary T cells for transcriptional start sites revealed that transcription initiates from two distinct regions, a common feature of TATA-less promoters. The region encompassing TSS1 confers the striking cell type specificity. A DNA fragment containing this sequence acts as a promoter in a T cell line but lacks detectable function in either the 293T or HeLa cell line. Although the defined Sp1 site in this sequence is likely to be important for in vivo A3G promoter activity and has been shown to function in reporter assays (29), and there are certainly other key DNA motifs within this core promoter, it is the overlapping NFAT and IRF binding sequence that is essential for the observed cell type specificity. This suggests a regulated model whereby basal transcription machinery and the composite NFAT/IRF site coordinately regulate the critical cell type-specific activation of the promoter.
NFATc1 and NFATc2 are expressed in T cells at various stages of development and states of activation and have been implicated in several essential regulatory mechanisms that exhibit a cell type specificity (41). However, both are also expressed in a variety of cell types, suggesting that additional transcription factors participate in establishing the cell type-specific regulation seen with the A3G promoter. The IRF family of DNA-binding factors regulates interferon-inducible genes. Within this family, the proteins IRF-4 and IRF-8 exhibit a strict restriction of expression in cells of the lymphoid and myeloid lineages. IRF-4 is the predominant form expressed in T cells and the IFN-inducible IRF-8 is found in macrophages and dendritic cells (42). Both cooperate with other transcription factors to exert a cell type-specific regulation (39, 40, 43–45). Regarding, the NFAT/IRF partnering specifically, IRF-4 and IRF-8 have been shown to individually cooperate with NFAT proteins to regulate cytokine production in cells of the immune system, in response to a variety of stimuli (36–39). IRF-4 expression is induced upon T cell activation and, along with NFATc1/c2, is critically involved in the genetic regulation of interleukin-2 and -4. In contrast, interferon-γ treatment of monocytes stimulates IRF-8, which regulates the transcription of interleukin-12 (46). This differential responsiveness of IRF-4 and IRF-8, accounts for the finer points of cell type-specific regulation within different cell types of the immune compartment. In accordance with these findings, we consider it likely that IRF-4 critically participates in the regulation of A3G promoter activity in T cells, whereas IRF-8 likely adopts that role in macrophages; A3G expression is detected in both cell types, whereas IRF-4 expression is restricted to lymphocytes, and IRF-8 is primarily detected in cells of myeloid lineage (42). Furthermore, we propose an association between the pattern of NFATc1/NFATc2 and IRF-4/IRF-8 expression and induction and the regulation of the innate immune response (24). Understanding the regulation of the innate immune response, which must also include consideration of cellular restriction factors such as A3G upon exposure to HIV-1 is essential.
We verified that NFATc1/NFATc2 and IRF-4 binding to the A3G promoter was correlative as measured by ChIP experiments. Neither NFATc1/c2 nor IRF-4 binding was individually detected. In either case, when examining CEM versus CEMss cells, or stimulated versus unstimulated CEMss cells, the increased nuclear localization of the NFAT family member was associated with increased IRF-4 binding. The ionomycin stimulation of CEMss cells led to significant NFAT relocalization to the nucleus resulting in the detection of A3G promoter occupancy at a higher level than even in CEM cells, which endogenously express A3G. Although this is intriguing, it cannot unequivocally be stated whether this increased occupancy is solely at the A3G promoter. It must be noted that, by alignment, the promoter region of APOBEC3F (A3F) is virtually identical to A3G. The distinguishing of A3G versus A3F expression is certainly interesting, but it does not affect the interpretation of these results as both A3G and A3F are potent HIV-1 restriction factors. The primary focus of this work was the induction of A3G, but the parallel induction of other APOBEC3 family members is likely. The A3 genes are the result of segmental duplication and homologous sets of conserved TFBS are expected among the related family members. Alignment of upstream regions of the A3 family members reveals that APOBEC3D/E, A3F, and A3G are virtually identical in putative promoter regions. Each of these proteins exhibits potent anti-HIV-1 activity, and a simultaneous elevation of all three proteins may prove highly beneficial in a therapeutic setting.
NFAT activity can be modulated via a “threshold phenomenon” whereby NFAT-controlled expression is mediated by regulating the amount of nuclear-localized protein. In neither 293T nor CEMss cells, were the nuclear levels of NFAT sufficient to potentiate A3G promoter activation. However, by manipulating each cell type using different methods, we affected A3G expression in a similar fashion, confirming the importance of the identified transcription factors. In 293T cells, robust A3G gene expression was seen when NFAT and IRF-4 were co-transfected. This data suggest that co-expression of IRF-4 and NFAT was both necessary and sufficient to induce A3G expression. In CEMss cells, which express IRF-4, nuclear mobilization of NFAT was accomplished via ionomycin. This translocation then resulted in binding of NFAT and IRF-4 to the A3G promoter, followed by the appearance of A3G protein. Finally, in CEM-A cells cyclosporin A treatment inhibited NFAT nuclear translocation, resulting in a detectable decrease of A3G expression. These observations indicate that the level of nuclear NFAT regulates the expression of A3G. Additional experiments in CEMss confirmed that the ionomycin-induced A3G allowed the cells to restrict an HIV-1Δvif challenge. This conferment of viral restriction was identical to that observed when the A3G was overexpressed via the stable transduction of the cDNA in the CEMss cell line (1). This reporting of the ability to suppress HIV-1 infection via transcriptional control of a restriction factor is novel and opens an exciting line of investigation examining the potential of this type of approach.
Although the stimulated A3G expression in CEMss cells did not dramatically alter the replication kinetics of wild-type HIV-1, it is intriguing and significant that partial protection was observed as indicated by the altered kinetics of wild-type HIV-1 growth in these cells. The delay itself is relatively moderate (∼4 days), but it is consistently observed and may suggest an important in vivo relevance when considering events of acute infection, such as the critical establishment of viral set point. A shift in the balance between viral replication and innate defense exerted during the period of acute infection may have long lasting and important consequences for disease outcome.
Ultimately, the importance of understanding the cellular regulation of A3G gene expression is underscored by clinical observations that correlate elevated A3G expression with either the ability to resist HIV-1 exposure or successfully control infection (18, 20, 47). These novel insights into regulation of A3G expression may lay the molecular foundation for development of therapeutics aimed at exploiting and manipulating the A3G regulatory circuit allowing for modulation of A3G to protective levels in vivo.
Acknowledgments
We thank Jennifer Stanton, Matthew Stoltz, and Robert Bellin for technical assistance.
This work was supported, in whole or in part, by the Richard and Susan Smith Family Foundation (to A. M. S.), by Grant P30AI42845 from the Center for AIDS Research at the University of Massachusetts, by National Institutes of Health Grants AI071766-01 (to A. M. S.) and AI-3070072 (to S. M. W.), and by Elizabeth Glaser Pediatric AIDS Foundation Grant PF-77521 (to M. A. F.).
- HIV-1
- human immunodeficiency virus, type 1
- CsA
- cyclosporin A
- TFBS.
- transcription factor binding site(s)
- TSS
- transcription start site
- RACE
- rapid amplification of cDNA ends.
REFERENCES
- 1. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Nature 418, 646–650 [DOI] [PubMed] [Google Scholar]
- 2. Guo F., Cen S., Niu M., Saadatmand J., Kleiman L. (2006) J. Virol. 80, 11710–11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bishop K. N., Verma M., Kim E. Y., Wolinsky S. M., Malim M. H. (2008) PLoS Pathog. 4, e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. (2003) Cell 113, 803–809 [DOI] [PubMed] [Google Scholar]
- 5. Lecossier D., Bouchonnet F., Clavel F., Hance A. J. (2003) Science 300, 1112. [DOI] [PubMed] [Google Scholar]
- 6. Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. (2003) Nature 424, 99–103 [DOI] [PubMed] [Google Scholar]
- 7. Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. (2003) Nature 424, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marin M., Rose K. M., Kozak S. L., Kabat D. (2003) Nat. Med. 9, 1398–1403 [DOI] [PubMed] [Google Scholar]
- 9. Sheehy A. M., Gaddis N. C., Malim M. H. (2003) Nat. Med. 9, 1404–1407 [DOI] [PubMed] [Google Scholar]
- 10. Stopak K., de Noronha C., Yonemoto W., Greene W. C. (2003) Mol. Cell 12, 591–601 [DOI] [PubMed] [Google Scholar]
- 11. Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003) Science 302, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 12. Farrow M. A., Sheehy A. M. (2008) Future Microbiol. 3, 145–154 [DOI] [PubMed] [Google Scholar]
- 13. Peng G., Greenwell-Wild T., Nares S., Jin W., Lei K. J., Rangel Z. G., Munson P. J., Wahl S. M. (2007) Blood 110, 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pion M., Granelli-Piperno A., Mangeat B., Stalder R., Correa R., Steinman R. M., Piguet V. (2006) J. Exp. Med. 203, 2887–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vetter M. L., Johnson M. E., Antons A. K., Unutmaz D., D'Aquila R. T. (2009) PLoS Pathog. 5, e1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Biasin M., Piacentini L., Lo Caputo S., Kanari Y., Magri G., Trabattoni D., Naddeo V., Lopalco L., Clivio A., Cesana E., Fasano F., Bergamaschi C., Mazzotta F., Miyazawa M., Clerici M. (2007) J. Infect. Dis. 195, 960–964 [DOI] [PubMed] [Google Scholar]
- 17. Vázquez-Pérez J. A., Ormsby C. E., Hernández-Juan R., Torres K. J., Reyes-Terán G. (2009) Retrovirology 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin X., Brooks A., Chen H., Bennett R., Reichman R., Smith H. (2005) J. Virol. 79, 11513–11516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Land A. M., Ball T. B., Luo M., Pilon R., Sandstrom P., Embree J. E., Wachihi C., Kimani J., Plummer F. A. (2008) J. Virol. 82, 8172–8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pace C., Keller J., Nolan D., James I., Gaudieri S., Moore C., Mallal S. (2006) J. Virol. 80, 9259–9269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peng G., Lei K. J., Jin W., Greenwell-Wild T., Wahl S. M. (2006) J. Exp. Med. 203, 41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tanaka Y., Marusawa H., Seno H., Matsumoto Y., Ueda Y., Kodama Y., Endo Y., Yamauchi J., Matsumoto T., Takaori-Kondo A., Ikai I., Chiba T. (2006) Biochem. Biophys. Res. Commun. 341, 314–319 [DOI] [PubMed] [Google Scholar]
- 23. Rose K. M., Marin M., Kozak S. L., Kabat D. (2004) J. Biol. Chem. 279, 41744–41749 [DOI] [PubMed] [Google Scholar]
- 24. Stopak K. S., Chiu Y. L., Kropp J., Grant R. M., Greene W. C. (2007) J. Biol. Chem. 282, 3539–3546 [DOI] [PubMed] [Google Scholar]
- 25. Durocher Y., Perret S., Kamen A. (2002) Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Badran B. M., Wolinsky S. M., Burny A., Willard-Gallo K. E. (2002) J. Biol. Chem. 277, 47136–47148 [DOI] [PubMed] [Google Scholar]
- 27. Ahyi A. N., Chang H. C., Dent A. L., Nutt S. L., Kaplan M. H. (2009) J. Immunol. 183, 1598–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Birney E., Stamatoyannopoulos J. A., Dutta A., Guigó R., Gingeras T. R., Margulies E. H., Weng Z., Snyder M., Dermitzakis E. T., Thurman R. E., Kuehn M. S., Taylor C. M., Neph S., Koch C. M., Asthana S., Malhotra A., Adzhubei I., Greenbaum J. A., Andrews R. M., Flicek P., Boyle P. J., Cao H., Carter N. P., Clelland G. K., Davis S., Day N., Dhami P., Dillon S. C., Dorschner M. O., Fiegler H., Giresi P. G., Goldy J., Hawrylycz M., Haydock A., Humbert R., James K. D., Johnson B. E., Johnson E. M., Frum T. T., Rosenzweig E. R., Karnani N., Lee K., Lefebvre G. C., Navas P. A., Neri F., Parker S. C., Sabo P. J., Sandstrom R., Shafer A., Vetrie D., Weaver M., Wilcox S., Yu M., Collins F. S., Dekker J., Lieb J. D., Tullius T. D., Crawford G. E., Sunyaev S., Noble W. S., Dunham I., Denoeud F., Reymond A., Kapranov P., Rozowsky J., Zheng D., Castelo R., Frankish A., Harrow J., Ghosh S., Sandelin A., Hofacker I. L., Baertsch R., Keefe D., Dike S., Cheng J., Hirsch H. A., Sekinger E. A., Lagarde J., Abril J. F., Shahab A., Flamm C., Fried C., Hackermüller J., Hertel J., Lindemeyer M., Missal K., Tanzer A., Washietl S., Korbel J., Emanuelsson O., Pedersen J. S., Holroyd N., Taylor R., Swarbreck D., Matthews N., Dickson M. C., Thomas D. J., Weirauch M. T., Gilbert J., Drenkow J., Bell I., Zhao X., Srinivasan K. G., Sung W. K., Ooi H. S., Chiu K. P., Foissac S., Alioto T., Brent M., Pachter L., Tress M. L., Valencia A., Choo S. W., Choo C. Y., Ucla C., Manzano C., Wyss C., Cheung E., Clark T. G., Brown J. B., Ganesh M., Patel S., Tammana H., Chrast J., Henrichsen C. N., Kai C., Kawai J., Nagalakshmi U., Wu J., Lian Z., Lian J., Newburger P., Zhang X., Bickel P., Mattick J. S., Carninci P., Hayashizaki Y., Weissman S., Hubbard T., Myers R. M., Rogers J., Stadler P. F., Lowe T. M., Wei C. L., Ruan Y., Struhl K., Gerstein M., Antonarakis S. E., Fu Y., Green E. D., Karaöz U., Siepel A., Taylor J., Liefer L. A., Wetterstrand K. A., Good P. J., Feingold E. A., Guyer M. S., Cooper G. M., Asimenos G., Dewey C. N., Hou M., Nikolaev S., Montoya-Burgos J. I., Löytynoja A., Whelan S., Pardi F., Massingham T., Huang H., Zhang N. R., Holmes I., Mullikin J. C., Ureta-Vidal A., Paten B., Seringhaus M., Church D., Rosenbloom K., Kent W. J., Stone E. A., Batzoglou S., Goldman N., Hardison R. C., Haussler D., Miller W., Sidow A., Trinklein N. D., Zhang Z. D., Barrera L., Stuart R., King D. C., Ameur A., Enroth S., Bieda M. C., Kim J., Bhinge A. A., Jiang N., Liu J., Yao F., Vega V. B., Lee C. W., Ng P., Shahab A., Yang A., Moqtaderi Z., Zhu Z., Xu X., Squazzo S., Oberley M. J., Inman D., Singer M. A., Richmond T. A., Munn K. J., Rada-Iglesias A., Wallerman O., Komorowski J., Fowler J. C., Couttet P., Bruce A. W., Dovey O. M., Ellis P. D., Langford C. F., Nix D. A., Euskirchen G., Hartman S., Urban A. E., Kraus P., Van Calcar S., Heintzman N., Kim T. H., Wang K., Qu C., Hon G., Luna R., Glass C. K., Rosenfeld M. G., Aldred S. F., Cooper S. J., Halees A., Lin J. M., Shulha H. P., Zhang X., Xu M., Haidar J. N., Yu Y., Ruan Y., Iyer V. R., Green R. D., Wadelius C., Farnham P. J., Ren B., Harte R. A., Hinrichs A. S., Trumbower H., Clawson H., Hillman-Jackson J., Zweig A. S., Smith K., Thakkapallayil A., Barber G., Kuhn R. M., Karolchik D., Armengol L., Bird C. P., de Bakker P. I., Kern A. D., Lopez-Bigas N., Martin J. D., Stranger B. E., Woodroffe A., Davydov E., Dimas A., Eyras E., Hallgrímsdóttir I. B., Huppert J., Zody M. C., Abecasis G. R., Estivill X., Bouffard G. G., Guan X., Hansen N. F., Idol J. R., Maduro V. V., Maskeri B., McDowell J. C., Park M., Thomas P. J., Young A. C., Blakesley R. W., Muzny D. M., Sodergren E., Wheeler D. A., Worley K. C., Jiang H., Weinstock G. M., Gibbs R. A., Graves T., Fulton R., Mardis E. R., Wilson R. K., Clamp M., Cuff J., Gnerre S., Jaffe D. B., Chang J. L., Lindblad-Toh K., Lander E. S., Koriabine M., Nefedov M., Osoegawa K., Yoshinaga Y., Zhu B., de Jong P. J. (2007) Nature 447, 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muckenfuss H., Kaiser J. K., Krebil E., Battenberg M., Schwer C., Cichutek K., Münk C., Flory E. (2007) Nucleic Acids Res. 35, 3784–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. (2002) Genomics 79, 285–296 [DOI] [PubMed] [Google Scholar]
- 31. Pido-Lopez J., Whittall T., Wang Y., Bergmeier L. A., Babaahmady K., Singh M., Lehner T. (2007) J. Immunol. 178, 1671–1679 [DOI] [PubMed] [Google Scholar]
- 32. Wang Y. J., Wang X., Zhang H., Zhou L., Liu S., Kolson D. L., Song L., Ye L., Ho W. Z. (2009) J. Neuroimmunol. 206, 14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ying S., Zhang X., Sarkis P. T., Xu R., Yu X. (2007) Acta Biochim. Biophys. Sin. 39, 297–304 [DOI] [PubMed] [Google Scholar]
- 34. Farré D., Roset R., Huerta M., Adsuara J. E., Roselló L., Albà M. M., Messeguer X. (2003) Nucleic Acids Res. 31, 3651–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Messeguer X., Escudero R., Farré D., Núñez O., Martínez J., Albà M. M. (2002) Bioinformatics 18, 333–334 [DOI] [PubMed] [Google Scholar]
- 36. Hu C. M., Jang S. Y., Fanzo J. C., Pernis A. B. (2002) J. Biol. Chem. 277, 49238–49246 [DOI] [PubMed] [Google Scholar]
- 37. Lee C. G., Kang K. H., So J. S., Kwon H. K., Son J. S., Song M. K., Sahoo A., Yi H. J., Hwang K. C., Matsuyama T., Yui K., Im S. H. (2009) Mol. Immunol. 46, 613–621 [DOI] [PubMed] [Google Scholar]
- 38. Rengarajan J., Mowen K. A., McBride K. D., Smith E. D., Singh H., Glimcher L. H. (2002) J. Exp. Med. 195, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu C., Rao K., Xiong H., Gagnidze K., Li F., Horvath C., Plevy S. (2003) J. Biol. Chem. 278, 39372–39382 [DOI] [PubMed] [Google Scholar]
- 40. Luo H., Tian M. (2010) Mol. Immunol. 47, 1383–1395 [DOI] [PubMed] [Google Scholar]
- 41. Rao A., Luo C., Hogan P. G. (1997) Annu. Rev. Immunol. 15, 707–747 [DOI] [PubMed] [Google Scholar]
- 42. Ozato K., Tailor P., Kubota T. (2007) J. Biol. Chem. 282, 20065–20069 [DOI] [PubMed] [Google Scholar]
- 43. Lazorchak A. S., Schlissel M. S., Zhuang Y. (2006) Mol. Cell Biol. 26, 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagulapalli S., Goheer A., Pitt L., McIntosh L. P., Atchison M. L. (2002) Mol. Cell Biol. 22, 7337–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van der Stoep N., Quinten E., Marcondes Rezende M., van den Elsen P. J. (2004) Blood 104, 2849–2857 [DOI] [PubMed] [Google Scholar]
- 46. Mamane Y., Heylbroeck C., Génin P., Algarté M., Servant M. J., LePage C., DeLuca C., Kwon H., Lin R., Hiscott J. (1999) Gene 237, 1–14 [DOI] [PubMed] [Google Scholar]
- 47. An P., Bleiber G., Duggal P., Nelson G., May M., Mangeat B., Alobwede I., Trono D., Vlahov D., Donfield S., Goedert J. J., Phair J., Buchbinder S., O'Brien S. J., Telenti A., Winkler C. A. (2004) J. Virol. 78, 11070–11076 [DOI] [PMC free article] [PubMed] [Google Scholar]