Abstract
In eukaryotes, ribosome assembly requires hundreds of conserved essential proteins not present in the mature particle. Despite their importance, the function of most factors remains unknown. This is because protein deletion often affects the composition of the entire particle. Additionally, many proteins are present in assembling ribosomes for extended times, which makes it difficult to pinpoint their role to a particular step. Here we have combined classical yeast biochemistry with experiments using recombinant proteins and RNA to study the role of Dim2 and its interaction with Nob1, the nuclease that generates the 3′-end of 18 S rRNA. Analysis of Dim2 mutants in which the interaction with Nob1 is disrupted demonstrates that this interaction between Dim2 and Nob1 is essential for optimal growth, and RNA binding experiments show that Dim2 increases Nob1 RNA affinity. Furthermore, our data indicate that Dim2 helps regulate Nob1 cleavage activity at the 3′-end of 18 S rRNA, as point mutants where this interaction is abolished in vitro accumulate pre-ribosomes containing Nob1 and 20 S rRNA in vivo. Interestingly, the site of interaction with Nob1 is mapped to the canonical RNA binding surface of a KH-like domain in Dim2, providing another example where an RNA-binding domain can be repurposed for protein interactions.
Keywords: Ribosomal RNA (rRNA), Ribosomal RNA Processing, Ribosome Assembly, Ribosome Structure, RNA-Protein Interaction
Introduction
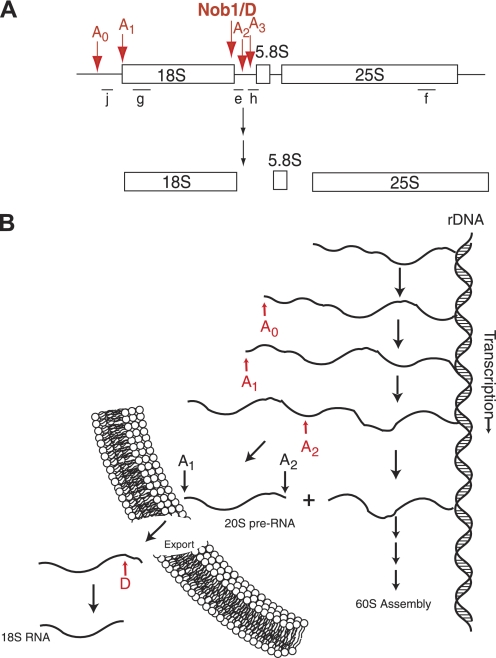
Ribosome assembly in eukaryotes is a complex and highly regulated process that consumes much of the energy of an actively growing cell (1). Ribosome assembly begins with the transcription of the ribosomal RNAs (rRNAs),3 three of which (18 S, 5.8 S, and 25 S) are produced as a single transcript (Fig. 1). During transcription, this RNA is extensively methylated at 2′-OH groups (2), and initial cleavages separate the rRNAs destined for the small and large ribosomal subunit (3). Data in the literature indicate that 18 S rRNA processing is initiated in the nucleolus by a non-essential cleavage step (termed A0) in the region 5′ to 18 S rRNA (5′-ETS), followed by the essential cleavages at sites A1 (to generate the 5′-end of 18 S rRNA) and A2 (to separate 18 S rRNA from 5.8 S and 25 S rRNA (4), Fig. 1). The final step of 18 S rRNA maturation is the cytoplasmic cleavage at site D to form the mature 3′-end of 18 S rRNA. This cleavage step is carried out by the PIN-domain-containing nuclease Nob1 (5–8).
FIGURE 1.
Pre-18 S rRNA processing in yeast. A, schematic overview of the rRNA transcript and cleavage sites for 18 S production. The locations of the Northern probes used in Fig. 5 are shown with bars and letters under the RNA. B, order of early rRNA processing and transcription steps in yeast. For simplicity steps required for 60 S assembly are excluded.
Modification and cleavage of pre-rRNAs is integrated with rRNA folding and binding of ribosomal proteins via a large machinery. In yeast, this essential machinery comprises ∼200 proteins, assembly factors that bind transiently to pre-ribosomal complexes. Despite their importance for assembling this essential biomolecule, in many cases their specific roles in ribosome biogenesis remain unknown. The function of enzymes can be dissected relatively easily, resulting in the identification of all rRNA-modifying enzymes as well as many of the nucleolytic enzymes, particularly in the 60 S pathway. In contrast, the function of the majority of the assembly factors is harder to study, as by sequence analysis many proteins only contain either RNA or protein interaction motifs, making it hard to propose initial models for protein function.
To address the role of these proteins, Tollervey and colleagues have recently developed an elegant technique to map the RNA-binding sites of RNA-binding ribosome assembly factors by crosslinking (9). Whereas this method has been fruitful in many cases, not all ribosome assembly factors with RNA-binding motifs have produced crosslinks to rRNA (10, 11). One such elusive protein is Dim2 (10).
Baserga and co-workers (12, 13) have used yeast 2-hybrid screens to systematically map the protein-protein interaction networks within the previously defined UtpA and UtpB complexes. A subpopulation of the proposed interactions has also been further studied using truncations or point mutations. Together, these maps have yielded detailed insight into a so-far intractable large RNA-protein complex. However, a detailed dissection of the role of these protein interactions, as well as how they interact with the transcriptional apparatus or the rRNA processing machinery, awaits further study.
To address the role of Dim2 in ribosome assembly, and perhaps develop a generally applicable method for dissecting the role of ribosome assembly factors without enzymatic functions, we focused on the protein-binding partner of Dim2, a protein required for 40 S ribosome assembly (14–16). Here we have tested the existence of a proposed interaction between Dim2 and the D-site nuclease Nob1. Our data show that this interaction is required for optimal ribosome biogenesis and provide evidence for a role for Dim2 in recruiting and activating Nob1 for cleavage at the 3′-end of 18 S rRNA. In addition, our data reveal that this interaction makes use of a KH-like domain, a motif typically associated with RNA binding.
EXPERIMENTAL PROCEDURES
Yeast Strains and Yeast Expression Plasmids
Yeast strain YKK192, created from the Open Biosystems Nob1-TAP strain, has a galactose-inducible promoter inserted in front of the Dim2 open reading frame. It was generated using PCR-based recombination (17), and correct insertion was verified by PCR and Western analysis. The Dim2 open reading frame, including an N-terminal HA tag, was cloned into pRS416TEF (18) using oligonucleotides a and b listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
For Northern probes, the location of the probe (e.g., D-A2, between cleavage sites D and A2) or mature RNAs recognized (e.g., 18 S) are also indicated. See Fig. 1 for schematic of probe locations and cleavage sites.
| Primer | Use/Location | Sequence |
|---|---|---|
| a | 5′-HA | GCCGCCACTAGTATGTATCCTTATGACGTGCCTGACTATGCCAGCCTGGGGACCTATGGTTGCGCCTACTGCTTTG |
| b | 3′ | TCAGACAAGCTTTTAGTAGCGTTCTTTTAATCTAG |
| c | 5′ | GATCGAGGCGCCATGGTTGCGCCTA |
| d | 3′ΔC | CAAGTAAAGCTTTCAATCTAGAATCAATAGTGAGATAGAATCG |
| e | d-A2 | GCTCTCATGCTCTTGCC |
| f | 25 S | GGGCAGGCTGCAGCTTCCTACCAG |
| g | 18 S | CATGGCTTAATCTTTGAGAC |
| h | A2-A3 | ATGAAAACTCCACAGTG |
| i | U2 | CAGATACTACACTTG |
| j | A0-A1 | GGAAACAGCTGAAATTCC |
Protein Expression Plasmids
The Dim2 open reading frame, as well as deletion constructs, were cloned into pSV272 for protein expression using oligonucleotides b, c, and d listed in Table 1. Dim2 mutants were generated using the QuikChange protocol.
Expression and Purification of Dim2
His-MBP-tagged Dim2 plasmid DNA was freshly transformed into Rosetta(DE3) Escherichia coli cells (Novagen). Protein expression was carried out as previously described for Bms1 (19). Cells were lysed in 50 mm NaPO4 (pH 8.0), 150 mm NaCl, and 0.5 mm PMSF and purified via Ni-NTA chromatography using 1 ml HisTrap columns (GE Healthcare). Ni-eluate fractions containing Dim2 were pooled and dialyzed at least 3 h into 50 mm Tris pH 8.0, 150 mm NaCl, 1 mm DTT (Buffer A) at 4 °C. Dialyzed protein was loaded onto a MonoS column equilibrated in Buffer A and eluted in a salt gradient to a final concentration of 40% of Buffer B (50 mm Tris, pH 8.0, 2 m NaCl, 1 mm DTT) over 12 column volumes. Fractions containing Dim2 were pooled and concentrated and further purified over a Superdex200 column in 50 mm HEPES pH 8.0, 200 mm NaCl, and 1 mm DTT. Dim2-containing fractions were pooled, concentrated, glycerol was added to 10%, and protein flash-frozen for long-term storage at −80 °C. Dim2-MBP protein concentration was determined with a nanodrop spectrophotometer using ϵ = 76,180 m−1 cm−1. To obtain untagged Dim2, TEV protease was added during the dialysis step. All subsequent steps were carried out identically. Dim2 protein concentrations were determined using ϵ = 11,460 m−1 cm−1. Dim2 mutant proteins were purified using the same protocol as the wild-type protein, except that a MonoQ ion-exchange column was used.
Transcriptions
rDNA fragments were cloned into pUC19 between XmaI and PstI sites, and then transcribed as described (7).
RNA Binding Assay
RNA binding experiments were carried out using electromobility shift assays. Radioactively labeled RNA was folded, and binding reactions were carried out as described (7). 10-μl binding reactions were incubated at 30 °C for 2 h and then immediately loaded onto a 6% acrylamide/THEM pH 7.5 gels (33 mm Tris, 67 mm HEPES, 1 mm EDTA, 10 mm MgCl2) after addition of 10 μl of RNA loading dye (0.1% xylene cyanol, 0.1% bromphenol blue, 45% glycerol). Gels were run at 10 watts for ∼2 h, dried, and exposed to a phosphoscreen overnight. Signal for protein-bound and free RNA was detected using Typhoon scanner and quantification was performed using ImageQuant software (Molecular Dynamics). Data were fit to the Hill equation.
In Vitro Pull-down Assays
10 μm recombinant MBP-tagged Dim2 (or MBP-tagged Nob1) was incubated with 10 μm untagged Nob1 (or untagged Dim2) for 30 min at room temperature. Pre-equilibrated amylose resin was added to the protein mixture and incubated on a shaker for 30 min at room temperature. The mixture was applied to a disposable column and the flow through was collected by brief centrifugation at <2 g. The column was washed with 1 ml of gel filtration buffer and briefly centrifuged to collect all residual buffer. Bound proteins were eluted with a 50 mm maltose in gel filtration buffer and analyzed via SDS-PAGE.
Northern Analysis
YKK192 was transformed with Dim2 constructs in pRS416TEF (or empty plasmid vector). Transformants were grown in galactose in midlog phase for at least 12 h before being grown in YPD for sample collection. 10 OD units of cells were collected at 0, 8, and 12 h after the switch to glucose media. Cells were pelleted and washed twice in water before being stored at −80 °C. For RNA extraction, cells were resuspended in 400 μl of TES buffer (10 mm Tris, pH 7.5, 10 mm EDTA, 0.5% (w/v) SDS). 400 μl of acidic phenol was added, and samples were incubated at 65 °C for 45 min. The acid phenol extraction was repeated, followed by a phenol/chloroform extraction. RNA was ethanol precipitated at −80 °C for an hour and resuspended in nuclease-free water and stored at −20 °C. RNAs were separated on a 1% agarose/formaldehyde gel, transferred to Amersham Biosciences Hybond-N membrane (GE Healthcare) by passive transfer, and crosslinked at high efficiency using optimal conditions of the UV-crosslinker (FB-UV XL-1000; Fisher Scientific). To detect transferred RNAs, 10 μm of the Northern probe (IDT, Table 1) was end-labeled using T4 polynucleotide kinase (NEB) and [γ-P32]ATP. The 5-μl reaction was incubated at 37 °C for 1 h, diluted with 25 μl of water, and purified using G-50 columns (GE Healthcare). Membranes were washed twice with 0.1% SDS for 10 min at 65 °C followed by a 1–4 h of incubation at 37 °C in prehybridization buffer (7.5× Denhardt's Solution, 5× SSPE, 0.75 m NaCl, 75 mm Na-citrate, pH 7.0, 0.1% SDS, 0.1 mg/ml single-stranded DNA). Then, 5 μl of the probe (in 500 μl prehybridization buffer) was denatured at 95 °C for 3 min and added to the membrane. Membranes were incubated with probes at 37 °C overnight. Membranes were washed in 5× SSPE, 0.1% SDS for 10 min at 37 °C followed by a wash in 0.5× SSPE, 0.1% SDS for 10 min at 37 °C. Membranes were exposed to a phospho-screen or film for 2–6 h (depending on the probe used), and analyzed using ImageQuant software.
Sucrose Gradients
YKK192 was transformed with Dim2 constructs in pRS416TEF (or empty vector). Overnight cultures were shifted to YPD for 8 h prior to sample collection. After 7.5 h, 100 OD units of cells were incubated with 100 μg/μl cycloheximide at 30 °C while shaking and subsequently pelleted and resuspended in gradient lysis buffer containing 100 μg/μl cycloheximide (20 mm Tris 8.0, 150 mm NaCl, 5 mm MgCl2) before being flash frozen. Cells were lysed under liquid nitrogen by grinding with mortar and pestle before being thawed and loaded onto 10–50% 5 ml sucrose gradients containing 100 μg/μl cycloheximide. Gradients were made by layering five sucrose-containing solutions, with each layer being frozen at −80 °C for 15 min before the next one was added. Frozen gradients were thawed at 4 °C for 1 h prior to sample loading to allow for thawing and smoothing of the gradient. Gradients were centrifuged at 40,000 rpm for 2 h. 375-μl fractions were collected and half of the volume was TCA-precipitated and analyzed by Western blots using IgG antibody to detect the TAP tag on Nob1.
RESULTS
Prior work suggested that Dim2 and Nob1 might interact with each other, as an interaction was found in a yeast 2-hybrid assay (20). Furthermore, Nob1 was also co-precipitated with Dim2 in yeast. We wanted to confirm that this interaction is direct and not mediated by concomitant association with the same pre-ribosomal particle(s). In addition, we wanted to dissect the role of this interaction in ribosome assembly. To test if these proteins interacted directly, we overexpressed and purified recombinant Nob1 from E. coli, as previously described (7). Dim2 was also overexpressed and purified from E. coli as described under “Experimental Procedures.” The identity of both proteins was confirmed by mass spectrometry, which also revealed that full-length Dim2 was obtained.
Dim2 Binds Directly to Nob1
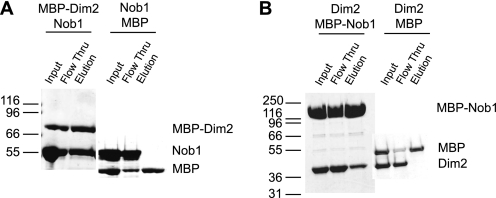
To test if Nob1 and Dim2 bind directly we performed pull-down experiments. MBP-tagged Dim2 was incubated with untagged Nob1, bound to amylose resin, washed extensively, and then eluted with maltose. The data in Fig. 2A show that Nob1 is retained in the presence of Dim2-MBP, but not MBP alone. Furthermore, the reverse pull-down using MBP-tagged Nob1 and untagged Dim2 verifies this result (Fig. 2B). These data show that Dim2 and Nob1 directly interact with each other.
FIGURE 2.
Dim2 interacts directly with Nob1. A, in vitro pulldowns with recombinant Nob1 and MBP-tagged Dim2. B, reverse in vitro pulldowns with recombinant MBP-Nob1 and Dim2. Control reactions show that Nob1 and Dim2 do not interact with MBP alone.
Mutations in Dim2 Central KH-like Domain Abolish Nob1 Binding in Vitro
To understand the role of the Dim2·Nob1 interaction in ribosome assembly, we wanted to create point mutants that disrupt this interaction and dissect their effects on ribosome assembly in vivo. To map the Nob1 binding site, we created Dim2 truncations that removed either the C-terminal KH-domain or the N-terminal domain. Both of these Dim2 fragments retain Nob1 binding, indicating that the Nob1-binding site resides in the central domain (supplemental Fig. S1B and data not shown). Sequence analysis indicates that this central domain contains conserved features of canonical KH-domains, including the predicted βααββα secondary structure elements, as well as conserved hydrophobic residues that help in packing of the sheets typically found in KH-domains (supplemental Fig. S2A). However, the Dim2 central domain does not contain the GXXG signature loop between the first and second α-helices. As a result, certain structure prediction programs do not annotate it as a KH-domain. Because this region of Dim2 (aa 87–178) has strong similarity but lacks the GXXG loop, it is called the central “KH-like” domain hereafter.
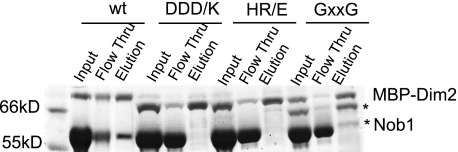
To further confirm that the central KH-like domain is involved in Nob1 binding, we created several point mutants in this region and tested the recombinant mutant proteins for Nob1 binding. The first mutant recreates a GXXG loop by converting the NSWT sequence in Dim2's central KH-like domain to the GKDG in Dim2 C-terminal KH-domain (GXXG mutant). Furthermore, we noticed that several conserved, charged residues were surface-exposed, according a structure prediction for Saccharomyces cerevisiae Dim2, based on an archeal Dim2 structure (supplemental Fig. S2B and Ref. 21). In the predicted structure, these residues surround the NSWT loop. Because the nature of the charge (positive or negative) was strictly conserved on these residues, suggesting that they may be involved in a conserved interaction, we mutated these residues (His-104, Arg-105; Asp-167, Asp-169, and Asp-170) to reverse the charge. The resulting mutants are referred to as HR/E and DDD/K, respectively.
All mutants were expressed in E. coli as MBP fusion proteins and incubated with Nob1 as described for wild-type protein. Interestingly, the GXXG, HR/E, and DDD/K mutations abolish Nob1 binding in vitro (Fig. 3). Fig. 3 also shows that for the mutant proteins, little full-length protein was obtained. Instead, most of the protein was truncated at the C terminus, resulting in a product similar, or identical, to one observed by Vanrobays et al. (15).4 However, removing the entire C-terminal domain does not affect Nob1 binding as described above (supplemental Fig. S1B), indicating that the truncation in the mutants does not cause the loss in Nob1 binding. Furthermore, the mutant, truncated proteins still bound RNA (supplemental Fig. S3) showing that the mutant truncated proteins are properly folded. Taken together, these results indicate that the Dim2 central KH-like domain interacts with Nob1.
FIGURE 3.
Mutations in the Dim2 central KH-like domain abolish its interaction with Nob1. Nob1 interacts with MBP-tagged wild-type Dim2 but not the DDD/K, HR/E, or GXXG mutants. Asterisks denote Dim2 breakdown products, not Nob1, as confirmed by Western blotting (data not shown).
Mutations in the Dim2 Central KH-like Domain Are Lethal
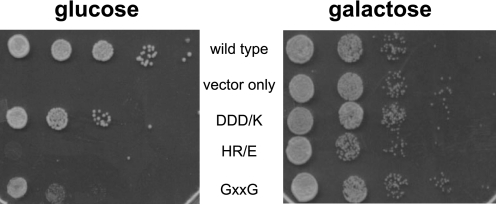
To test if the interaction between Nob1 and Dim2 was important for cell viability, we tested if the Dim2 mutants that abolish the Nob1·Dim2 interaction in vitro affect ribosome assembly in vivo. Because Dim2 is an essential protein and cells cannot grow in its absence, we first created a yeast strain in which Dim2 was under the control of the inducible galactose promoter. As expected, this strain requires galactose and does not grow in the presence of glucose (Fig. 4). Viability in glucose is restored when this yeast strain is transformed with a plasmid encoding full-length wild-type Dim2 under the control of the constitutive TEF promoter (Ref. 18 and Fig. 4). In contrast, plasmids encoding the HR/E or GXXG mutants do not provide for growth in glucose, and the DDD/K mutant gives a distinct slow growth phenotype. Importantly, Western blotting shows that all proteins are stable (data not shown).
FIGURE 4.
Mutations in the Dim2 central KH-like domain are lethal. A yeast strain with Dim2 under a galactose-inducible promoter was transformed with pRS416TEF plasmids encoding wild type or Dim2 mutants and then grown on glucose- or galactose-containing plates.
Mutations in the Dim2 Central KH-like Domain Result in 20 S Accumulation
Prior data in the literature indicate that Dim2 binds to pre-ribosomes prior to Nob1. This conclusion is based on the observation that deletion of Dim2 or its C-terminal KH-domain inhibits co-transcriptional cleavage at site A2, while deletion or mutations in Nob1 inhibit cytoplasmic cleavage at site D (5, 6, 14, 15). In addition, Dim2 co-purifies with ribosome assembly factors involved in both late and early steps, indicating that Dim2 is associated with late as well as early pre-ribosomal particles (22–25). In contrast, Nob1 mostly co-purifies with late ribosome assembly factors (23, 24, 26). Because Dim2 directly binds Nob1, we wanted to know if the Dim2·Nob1 interaction was required to recruit Nob1 to pre-ribosomal particles. Because Nob1 is the endonuclease required for cleavage at site D (5–8) and its depletion leads to accumulation of 20 S rRNA (5, 6), one might be able to observe 20 S accumulation (the Nob1 phenotype) if the Dim2 mutations only affect the binding of Nob1 to Dim2. In contrast, if the Dim2 mutations also affect processes other than Nob1 binding, inhibition of cleavage at site A2 may be observed, as seen in the absence of Dim2.
We used the glucose-repressible Dim2 strain transformed with plasmids encoding wild type or mutant Dim2 and depleted endogenous Dim2 for 0, 8, or 12 h. Total RNA was extracted from these cells, fractionated on agarose gels, and probed by Northern blotting. A probe directed against the spliceosomal U2 snRNA was used as a loading control. The data in Fig. 5 show that depletion of Dim2 leads to a loss of 20 S rRNA and accumulation of the 21 S rRNA, indicating that cleavage at site A2 is inhibited, which is consistent with prior data (14–16).
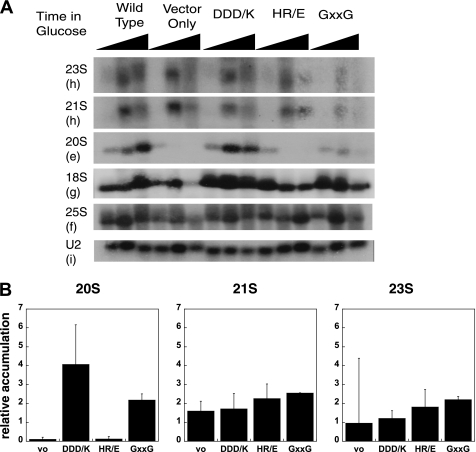
FIGURE 5.
Northern analysis of pre-rRNA processing in strains containing Dim2 mutants. A, Nob1TAP; Dim2::Gal strains supplemented with plasmids encoding wild type or mutant Dim2 under the constitutive TEF promoter were grown in glucose-containing medium for 0, 8, or 12 h prior to harvest. Total RNA was extracted and separated on an agarose-formaldehyde gel. The gel was transferred onto a membrane and probed with the indicated oligonucleotides. The sequences and locations of these probes are listed in Table 1. B, accumulation of 20 S, 21 S, and 23 S pre-rRNAs after 8 h in glucose relative to the strain with plasmid-encoded wild type Dim2 (adjusted for loading differences by normalization to the U2 probe). These averages are obtained from three or more independent experiments.
We next analyzed the Dim2 mutants and compared the accumulation of the 20 S pre-rRNA as well as the non-productive 21 S and 23 S intermediates to that observed in cells containing wild-type Dim2. Importantly, the DDD/K mutant shows a 4-fold accumulation of 20 S rRNA, consistent with an effect on Nob1 binding to pre-ribosomes (Fig. 5B). This effect is larger at 8 h than at 12 h after shut-off of endogenous Dim2, indicating that secondary events happen at later times. In contrast, the pre-rRNA processing phenotype of the HR/E mutant resembles that of the Dim2 depletion with accumulation of the 21 S and 23 S rRNAs and depletion of the productive 20 S intermediate. The GXXG mutant has an intermediate phenotype, as it shows a 2-fold accumulation of the 20 S rRNA as well as an accumulation of 21 S and 23 S rRNAs.
Together, these data show that Dim2 is required for cleavage at site D in addition to the previously documented requirement for cleavage at site A2. Furthermore, our data also provide evidence that, in addition to the C-terminal RNA-binding domain, (15), the central KH-like domain is also required for cleavage at site A2. We do not think that the ability to bind Nob1 is required for cleavage at site A2, as the DDD/K mutant is deficient in Nob1 binding but only minimally affects cleavage at site A2.
Mutations in the Central KH-like Domain of Dim2 Reduce Nob1 Binding to Pre-ribosomes in Vivo
To further test if the Dim2 mutations affect the Nob1 interaction with pre-ribosomes, we also performed sucrose gradient centrifugation followed by Western analysis to detect free Nob1 and Nob1 associated with pre-ribosomal particles. Lysates were prepared from yeast cells expressing no Dim2, or wild type or mutant Dim2, with endogenous Dim2 depleted for 8 h. After clarification, lysates were fractionated on a 10–50% sucrose gradient to separate free proteins from ribosomes or pre-ribosomal particles.
As expected and consistent with prior data (5, 27, 28), in cells containing wild type Dim2, Nob1 is bound to 43 S pre-ribosomes, 90 S pre-ribosomes, as well as in polysomes, where it is associated with 43 S pre-ribosomes (Fig. 6, data not shown and Ref. 28). In cells lacking Dim2, some Nob1 is found free at the top of the gradient, not associated with pre-ribosomal particles. Furthermore, because no 20 S rRNA is made in these cells (Ref. 14, 15 and Fig. 6) there is no association with polysomes. However, association with a 40 S-like particle remains. Given that there is no 20 S rRNA (Fig. 6 and data not shown) but instead 21 S rRNA, it seems likely that this represents a 40 S-like particle containing 21 S rRNA that does not mature to 18 S rRNA (18 S levels are depleted). Alternatively, Nob1 could be bound to mature 40 S ribosomes, a protein complex containing ribosome assembly factors akin to one recently found in the absence of rRNA transcription (29), or, finally, Nob1 might be bound to entirely unrelated particles, including the proteasome as previously suggested (20). Nevertheless, the increased amount of free Nob1 in the absence of Dim2 is consistent with a role for Dim2 in recruiting Nob1.
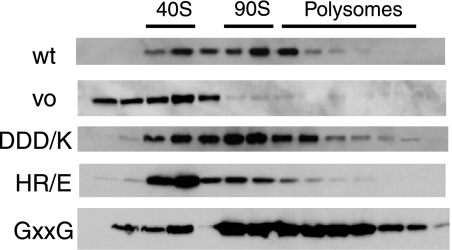
FIGURE 6.
Nob1 binding to pre-ribosomes in strains containing Dim2 mutants. Nob1TAP; Dim2::Gal strains supplemented with plasmids encoding wild type or mutant Dim2 were grown in glucose-containing medium for 8 h prior to harvest. Lysates were then fractionated on 10–50% sucrose gradients. Proteins were TCA-precipitated, run on an SDS-PAGE gel, and probed for Nob1.
We next analyzed association of Nob1 with pre-ribosomes in cells containing mutant forms of Dim2. All three mutants show a slightly increased amount of free Nob1, which can be detected in fraction 2 in all Dim2 mutants but not wild-type Dim2. Furthermore, the amount of Nob1 in fraction 3 is increased. However, the remaining substantial ribosome association of Nob1 also shows that Nob1 binds independently of Dim2, likely held via its own RNA binding activity as well as by protein-protein interactions (7). These results are consistent with a non-essential role for Dim2 in helping to recruit Nob1.
In addition, the HR/E mutant shows decreased association of Nob1 with polysomal fractions, consistent with the observation that no 20 S rRNA is made. Northern analysis confirms that fractions 6–8 contain 35 S rRNA, and indicates that the 40 S-like particles in the HR/E mutant contain 21 S rRNA (data not shown), as expected from the bulk Northern analysis in Fig. 5 that shows that no 20 S rRNA is made.
In contrast, Nob1 is bound to 40 S particles and polysomal fractions in the GXXG and DDD/K mutants, consistent with the formation of 20 S rRNA in these particles (Fig. 6). Notably, both mutants show increased association of Nob1 in polysomal particles as previously observed upon accumulation of 20 S rRNA (28).
The observation that Nob1 bound to 20 S rRNA accumulates in 40 S and polysomal fractions in the DDD/K and GXXG mutants suggests that the defect from these mutations does not simply arise because Nob1 is no longer bound to polysomal fractions. Instead, the Nob1 function in D-site cleavage is somehow impaired in these mutants.
Dim2 Binding Strengthens the Nob1 RNA Binding Affinity
Nob1 is the nuclease for cleavage at site D (5–8), and correspondingly, it binds pre-rRNAs containing cleavage site D (7). The Northern data and polysome gradients described above suggest that mutations in Dim2 that abolish the interaction with Nob1 stall ribosome assembly at the Nob1-dependent D-site cleavage step, resulting in accumulation of a particle that has Nob1 bound but is unable to cleave. Thus, we wanted to test if Dim2 affected Nob1 interaction with rRNA or its cleavage activity.
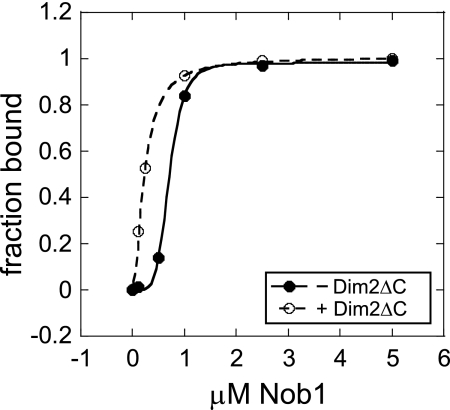
Because the Dim2 C-terminal KH-domain binds rRNA (15), complicating the analysis of an effect from Dim2 binding on Nob1 RNA binding activity, we made a C-terminal truncation of Dim2 (Dim2ΔC). As expected, Dim2ΔC does not bind rRNA (supplemental Fig. S1A and Ref. 15), but it retains the ability to bind Nob1 (supplemental Fig. S1B). Addition of Dim2ΔC increases the Nob1 RNA binding affinity (Fig. 7). Because this effect is independent of the Dim2 RNA binding activity it must be an allosteric effect on the Nob1 RNA binding site. This finding suggests that within Nob1 the Dim2 and RNA-binding sites communicate and that the presence of Dim2 changes the mode of the Nob1 interaction with rRNA to increase its RNA binding affinity. No effect from Dim2 on the Nob1 rRNA cleavage activity was observed (data not shown).
FIGURE 7.
Binding of Dim2ΔC strengthens the Nob1 RNA binding affinity. Data were fit to the Hill equation and yield K1/2 values of 0.14 μm2 and 0.26 μm4 in the presence and absence of Dim2ΔC, respectively. Different Hill coefficients were used because independent experiments indicate that the addition of Dim2 changes the stoichiometry of the Nob1·rRNA interaction from 4:1 to 2:1 (A. C. Lamanna and K. Karbstein, unpublished data). The different Hill coefficients preclude direct comparison of K1/2 values. However, the different Hill coefficients do not change the finding that Dim2 increases Nob1 RNA binding affinity, as all data points in the presence of Dim2 are to the left of the corresponding ones in the absence.
DISCUSSION
A Direct Interaction between Dim2 and Nob1 Is Essential for Ribosome Assembly
Prior data in the literature had indicated that Dim2 might interact directly with Nob1, as an interaction was found in a yeast 2-hybrid screen as well as in vivo pull-downs (20). However, yeast 2-hybrid screens suffer from a large number of false positives, and because Dim2 and Nob1 are known to bind the same ribosome assembly intermediate (23, 24, 26, 30), it is possible this mediated their interaction in vivo. Thus, we tested for binding of recombinant Dim2 and Nob1 using in vitro pull-downs. Our data show that Nob1 and Dim2 directly interact. Mapping of the protein complex interface by truncation analysis indicated that the central KH-like domain in Dim2 is responsible for this interaction. This was confirmed by the analysis of point mutants around the NSWT loop that replaces the GXXG loop found in canonical KH-domains. These point mutations in Dim2 abolish the interaction with Nob1 and provide a tool to study the role of the Nob1-Dim2 interaction in vivo. Analysis of the consequences of these mutations on ribosome assembly reveals multiple functions of Dim2 as described below.
Roles for Dim2 in Ribosome Assembly
Dim2 Is Required for Cleavage at Site D
Northern analysis of the DDD/K (and to a lesser extent the GXXG) mutant shows that this mutation inhibits cleavage at site D, as expected from a mutation that disrupts the interaction with Nob1. However, inspection of the Nob1 association with pre-ribosomes also indicates that Nob1 remains fully bound to 43 S pre-ribosomes. This suggests that (a) the Nob1 RNA binding affinity is sufficient for binding to 43 S pre-ribosomes (although binding is reduced in the absence of Dim2) and (b) that Dim2 must affect Nob1 activity in a more subtle manner. Our data show that Dim2 modulates the way in which Nob1 interacts with rRNA, as binding of truncated Dim2, which cannot bind RNA by itself, strengthens the interaction of Nob1 with RNA via an allosteric effect. It is possible that this modification of the rRNA binding interface in Nob1 is required for efficient Nob1-dependent cleavage at site D. However, in a simple recombinant system to study Nob1-dependent cleavage that includes Nob1 and pre-rRNA fragments, Dim2 has no effect on Nob1-dependent cleavage (data not shown). Nevertheless, we note that rRNA cleavage in that system is slow (31), suggesting it may lack elements of the in vivo cleavage reaction. A direct effect from Dim2 on Nob1-dependent cleavage could either require these other elements or the requirement for Dim2 might be mediated by other factors. Comprehensive analysis of all assembly factors stably bound to late 43 S pre-ribosomes (Tsr1, Rio2, Enp1, Ltv1, Dim1, and Nob1) shows that Dim2 is part of an extensive network of interactions between Nob1 and the essential kinase Rio2 (41), rendering Rio2 a possible candidate for such a role. In addition, it is possible that any effect from Dim2 on Nob1-dependent cleavage involves either ribosomal proteins or assembly factors not stably bound to the pre-43 S ribosome, such as Fap7 or Rio1. Additional experiments will be required to distinguish between these possibilities.
Dim2 Is Required for Cleavage at Site A2
Northern analysis of the HR/E mutant suggests that this mutation affects an earlier step of ribosome assembly (that is probably independent of Nob1 binding), as the HR/E mutation inhibits cleavage at site A2 (and also, to a lesser extent, at A0 and A1) This finding suggests that the central KH-like domain in Dim2 forms interactions with additional ribosome assembly factors early during assembly. We tested if Dim2 interacts with Rcl1, the suggested nuclease for cleavage at site A2, but were not able to detect an interaction (data not shown). The observation that different mutations in the KH-like central domain disrupt different steps in ribosome assembly suggests that Dim2 is part of a highly dynamic region in pre-ribosomes that is being remodeled during assembly. It also suggests that there might be multiple ways to create a protein binding interface with a KH-like domain, such that different regions of the KH-like domain are used for different interactions. This latter conclusion is consistent with previous structural analysis that shows that canonical KH-domains, which bind RNA, can also pack against each other on the other side of the domain (32, 33).
Dim2 Contributes to Nob1 Binding to Pre-ribosomes
The gradient centrifugation data presented herein suggest that Dim2 contributes to binding of Nob1 to pre-ribosomal particles, as Nob1 accumulates in unbound form on top of the gradient in the absence of Dim2 and is increased in free fractions with Dim2 mutants that disrupt Nob1 binding. However, they also show that Nob1 can bind to pre-ribosomes independently of Dim2, as each strain showed remaining binding of Nob1 to pre-ribosomal particles.
Furthermore, we observe that the amount of Nob1 in free particles is greater when no 20 S rRNA but 21 S or 23 S rRNA is made (no Dim2 and the HR/E mutant). This suggests that Nob1 binding to 20 S-containing 43 S pre-ribosomes is stronger than binding to the 21 S-containing pre-40 S-like particle. Two possible models could explain this observation. It is possible that the Dim2·Nob1 interaction is remodeled during ribosome assembly, such that it is more important early in assembly. Alternatively, and perhaps more likely, the difference could arise from Nob1 interactions with pre-ribosomes being remodeled, such that they are stronger later in assembly compared with earlier. Thus, the Dim2 interaction would be relatively more important early on. In that context, we have recently shown that particles that are not yet cleaved at site A2 are in a different structure from particles cleaved at that site (31). These different structures result in differences in the mode of interaction between Nob1 and pre-rRNA, which could explain the observed differences.
Dim2 Is Required for 40 S Export and SSU Processosome Assembly
Previous work from the Lafontaine laboratory has shown that Dim2 has a nuclear export sequence (NES), which is required for efficient export of the 40 S subunit (15). In addition, long time courses of Dim2 depletion also inhibit SSU (small subunit) processosome assembly, pointing to additional roles of Dim2 in early processing steps.
A Role for Eukaryotic Dim2 in Translation Initiation?
The observation of a complex between archeal Dim2 (aDim2) and aIF2α has led to suggestions for a role for aDim2 in translation initiation (21). We note however that the Dim2/IF2α interface observed in the crystal is not conserved in eukaryotic Dim2. Furthermore, the archeal Dim2 has a bona-fide RNA-binding domain in place of the KH-like central domain found in eukaryotic Dim2. In the structure, this KH-domain interacts with the anti-Shine-Dalgarno sequence, which is not conserved in eukaryotes. Finally, archeal Dim2 lacks the N-terminal domain found in eukaryotic Dim2. Our data suggest that deletion of this domain is detrimental in vivo.5 Together, these observations suggest that there are substantial differences between eukaryotic and archeal Dim2, leading us to be cautious with making inferences about the function of Dim2 from one kingdom to the next.
A KH-like Domain Provides a Protein-Protein Interaction Interface
KH-domains are well-characterized RNA-binding domains in which a three-stranded antiparallel β-sheet packs against three α-helices (34). The RNA binding surface is provided by a conserved loop between the first two α-helices. The first and last residue of this 4-residue loop are conserved as glycines, hence the name GXXG loop. In addition, the first two helices and the second strand of the β-sheet are also typically part of the RNA binding surface. We denote the Dim2 central domain as a KH-like domain because it contains all the conserved secondary structure elements as well as conserved hydrophobic residues to stabilize the KH-fold, yet it does not contain the GXXG loop, which is instead replaced with the sequence NSWT (supplemental Fig. S2A).
Our data show that the central KH-like domain of Dim2 provides the interface for interaction with Nob1 and possibly other ribosome assembly factors. Previous structural work with proteins containing two tandem KH-domains has shown that in a few cases two KH-domains can interact with each other “on the back site”, i.e. away from the RNA binding motif. While different modes of packing of these KH-domains have been observed (32, 33, 35), none of them involve the RNA binding surface and all are consistent with concomitant binding of both KH-domains to RNA. In contrast, the data herein imply that the surface that usually is used for RNA binding is used for the interaction of the KH-like domain with Nob1 (supplemental Fig. S2C). Our data suggest that both the NSWT loop, which replaces the canonical GXXG loop, and the first α-helix, which contains the HR motif, interact with Nob1. Both of these structural elements are part of the RNA binding site in canonical KH-domains. The DDD motif is part of a helix not found in all KH-domains (both the KH and KH-like domain in Dim2 as well as all 15 KH-domains in vigilin have this helix (36)). Interestingly, the location of the DDD sequence in the predicted structure is close to the HR motif and the NSWT loop (supplemental Fig. S2B), and it is also similar to the location of an extension to the last α-helix found in the Nova proteins. This helix is known to contribute to RNA binding via specific hydrogen bonds (37, 38).
This analysis indicates that the mode of interaction between the Dim2 KH-like domain and Nob1 appears to be very similar to the mode of interaction between canonical KH-domains and RNA, and suggests that this is an example of the canonical RNA binding surface of a KH-type domain used for protein-protein, not RNA-protein, interactions. Similar suggestions have been made for individual KH-domains in multi-KH-domain-containing proteins. For example, the last two KH-domains in human vigilin have been shown to interact with a histone-methyltransferase even in the absence of RNA, suggesting strongly that at least one of these KH-domains is a protein-interacting module, although the mode of interaction (via the RNA binding surface or other surfaces) is not clear. It is also worth pointing out that not all the 15 KH-domains in vigilin contain the conserved GXXG loop. In all vigilin proteins the first KH-domain is divergent, and so are at least two additional ones (although their locations are not conserved), although S. cerevisiae vigilin contains only six canonical GXXG-loop containing KH-domains, and eight or nine divergent ones. Similarly, the first KH-domain in the alternative splicing factor PSI has a mutation in the GXXG loop. Nevertheless, in the case of PSI it is known that the first KH-domain contributes to RNA binding (39), and in the case of human vigilin, the last two KH-domains that are known to interact with proteins are canonical KH-domains. Thus, there does not seem to be a strict correlation between mutations in the GXXG loop and the ability of KH-domains to bind proteins instead of RNA.
The finding that the “RNA-binding” surface of a KH-like domain is used for protein-protein interactions is reminiscent of another RNA-binding domain, the RNA-recognition-motif (RRM), which has recently been shown to bind proteins (40). Perhaps this is another remnant of the evolution from the RNA to the protein world, which likely started with small peptides that bound RNA. Once proteins became more prevalent, protein-protein interaction domains had to be evolved. It seems plausible that some of these motifs would be directly descended from ancestral RNA-binding domains, as suggested by data herein.
Supplementary Material
Acknowledgments
We thank members of our laboratory for comments on the manuscript and J. Maddock for hosting H. A. W. in the final stages of this project.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM086451.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
We have used the 55 and 66 kDa markers on the SDS-PAGE gel to obtain an estimate of 2 kDa for the size difference between truncated and full-length Dim2. Using an average of 110 Da/amino acid, we thus estimate the loss of about 18 amino acids. That would suggest that the KH-domain is truncated between α-helices 3 and 4 (supplemental Fig. S2). Indeed, α-helix 4 is not present in many KH-domains, consistent with the observation that the truncated proteins still bind RNA.
H. A. Woolls and K. Karbstein, unpublished data.
- rRNA
- ribosomal RNA
- RRM
- RNA recognition motif
- TCA
- trichloroacetic acid
- aa
- amino acids
- MBP
- maltose binding protein.
REFERENCES
- 1. Warner J. R. (1999) Trends Biochem. Sci. 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 2. Kos M., Tollervey D. (2010) Mol. Cell 37, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Osheim Y. N., French S. L., Keck K. M., Champion E. A., Spasov K., Dragon F., Baserga S. J., Beyer A. L. (2004) Mol. Cell 16, 943–954 [DOI] [PubMed] [Google Scholar]
- 4. Venema J., Tollervey D. (1999) Annu. Rev. Gen. 33, 261–311 [DOI] [PubMed] [Google Scholar]
- 5. Fatica A., Oeffinger M., Dlakić M., Tollervey D. (2003) Mol. Cell Biol. 23, 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fatica A., Tollervey D., Dlakić M. (2004) RNA 10, 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamanna A. C., Karbstein K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14259–14264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pertschy B., Schneider C., Gnädig M., Schäfer T., Tollervey D., Hurt E. (2009) J. Biol. Chem. 284, 35079–35091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Granneman S., Kudla G., Petfalski E., Tollervey D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9613–9618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Granneman S., Petfalski E., Swiatkowska A., Tollervey D. (2010) EMBO J. 29, 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bohnsack M. T., Martin R., Granneman S., Ruprecht M., Schleiff E., Tollervey D. (2009) Mol. Cell 36, 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freed E. F., Baserga S. J. (2010) Nucleic Acids Res. 38, 4798–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Champion E. A., Lane B. H., Jackrel M. E., Regan L., Baserga S. J. (2008) Mol. Cell Biol. 28, 6547–6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vanrobays E., Gélugne J. P., Caizergues-Ferrer M., Lafontaine D. L. (2004) RNA 10, 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanrobays E., Leplus A., Osheim Y. N., Beyer A. L., Wacheul L., Lafontaine D. L. (2008) RNA 14, 2061–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Senapin S., Clark-Walker G. D., Chen X. J., Séraphin B., Daugeron M. C. (2003) Nucleic Acids Res. 31, 2524–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 18. Mumberg D., Müller R., Funk M. (1995) Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 19. Karbstein K., Jonas S., Doudna J. A. (2005) Mol. Cell 20, 633–643 [DOI] [PubMed] [Google Scholar]
- 20. Tone Y., Toh-E A. (2002) Genes Dev. 16, 314231–314257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jia M. Z., Horita S., Nagata K., Tanokura M. (2010) J. Mol. Biol. 398, 774–785 [DOI] [PubMed] [Google Scholar]
- 22. Collins S. R., Kemmeren P., Zhao X. C., Greenblatt J. F., Spencer F., Holstege F. C., Weissman J. S., Krogan N. J. (2007) Mol. Cell Proteomics 6, 439–450 [DOI] [PubMed] [Google Scholar]
- 23. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 24. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Hofert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 25. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 26. Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. (2003) EMBO J. 22, 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Granneman S., Nandineni M. R., Baserga S. J. (2005) Mol. Cell Biol. 25, 10352–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soudet J., Gélugne J. P., Belhabich-Baumas K., Caizergues-Ferrer M., Mougin A. (2010) EMBO J. 29, 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Merl J., Jakob S., Ridinger K., Hierlmeier T., Deutzmann R., Milkereit P., Tschochner H. (2010) Nucleic Acids Res. 38, 3068–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., Hurt E. (2006) Nature 441, 651–655 [DOI] [PubMed] [Google Scholar]
- 31. Lamanna A. C., Karbstein K. (2010) J. Mol. Biol. 305, 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Díaz-Moreno I., Hollingworth D., Kelly G., Martin S., Garcia-Mayoral M., Briata P., Gherzi R., Ramos A. (2010) Nucleic Acids Res. 38, 5193–5205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beuth B., Pennell S., Arnvig K. B., Martin S. R., Taylor I. A. (2005) EMBO J. 24, 3576–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valverde R., Edwards L., Regan L. (2008) FEBS J 275, 2712–2726 [DOI] [PubMed] [Google Scholar]
- 35. Valverde R., Pozdnyakova I., Kajander T., Venkatraman J., Regan L. (2007) Structure 15, 1090–1098 [DOI] [PubMed] [Google Scholar]
- 36. Musco G., Stier G., Joseph C., Castiglione Morelli M. A., Nilges M., Gibson T. J., Pastore A. (1996) Cell 85, 237–245 [DOI] [PubMed] [Google Scholar]
- 37. Jensen K. B., Musunuru K., Lewis H. A., Burley S. K., Darnell R. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5740–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lewis H. A., Musunuru K., Jensen K. B., Edo C., Chen H., Darnell R. B., Burley S. K. (2000) Cell 100, 323–332 [DOI] [PubMed] [Google Scholar]
- 39. Chmiel N. H., Rio D. C., Doudna J. A. (2006) RNA 12, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kielkopf C. L., Lücke S., Green M. R. (2004) Genes Dev. 18, 1513–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Campbell M. G., Karbstein K. (2011) PLoS One, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.