Abstract
The amyloid precursor protein (APP) is cleaved by β- and γ-secretases to generate the β-amyloid (Aβ) peptides, which are present in large amounts in the amyloid plaques of Alzheimer disease (AD) patient brains. Non-amyloidogenic processing of APP by α-secretases leads to proteolytic cleavage within the Aβ peptide sequence and shedding of the soluble APP ectodomain (sAPPα), which has been reported to be endowed with neuroprotective properties. In this work, we have shown that activation of the purinergic receptor P2X7 (P2X7R) stimulates sAPPα release from mouse neuroblastoma cells expressing human APP, from human neuroblastoma cells and from mouse primary astrocytes or neural progenitor cells. sAPPα shedding is inhibited by P2X7R antagonists or knockdown of P2X7R with specific small interfering RNA (siRNA) and is not observed in neural cells from P2X7R-deficient mice. P2X7R-dependent APP-cleavage is independent of extracellular calcium and strongly inhibited by hydroxamate-based metalloprotease inhibitors, TAPI-2 and GM6001. However, knockdown of a disintegrin and metalloproteinase-9 (ADAM9), ADAM10 and ADAM17 by specific siRNA, known to have α-secretase activity, does not block the P2X7R-dependent non-amyloidogenic pathway. Using several specific pharmacological inhibitors, we demonstrate that the mitogen-activated protein kinase modules Erk1/2 and JNK are involved in P2X7R-dependent α-secretase activity. Our study suggests that P2X7R, which is expressed in hippocampal neurons and glial cells, is a potential therapeutic target in AD.
Keywords: Alzheimer Disease, ATP, MAP Kinases (MAPKs), Purinergic Receptor, Secretases
Introduction
The amyloid precursor protein (APP)3 is a transmembrane protein expressed by neurons, astrocytes, microglia, and neural progenitor cells (NPCs) in the central nervous system (CNS). APP can be cleaved by three different secretases called: α, β, and γ, depending on their cleavage sites on the APP (1, 2). The amyloid-β (Aβ) peptides generated by β- and γ-secretases are found in senile plaques of patients with Alzheimer Disease (AD). The α-secretase cleaves APP within the Aβ peptide sequence, precluding the formation of neurotoxic Aβ peptides. The soluble fragment of APP, sAPPα produced after cleavage by the α-secretase, has been shown to have neurotrophic and neuroprotective properties (3–5). Several enzymes capable of mediating non-amyloidogenic α-processing of APP have been identified: ADAM9, -10, and -17 (6). However, several proteases may cooperate in the physiological α-cleavage of APP in CNS. Various reports have shown that ADAM activation is induced by G protein-coupled receptor (GPCR) agonists. Ca2+ increase, ROS production, and phosphorylation of ADAMs by protein kinases C (PKC) or mitogen-activated protein kinases (MAP kinases), are involved in ADAM activation following stimulation of GPCR (7, 8). However, receptors other than GPCR are able to activate PKC and MAP kinases and/or to induce Ca2+ increase and ROS production. Indeed, the purinergic receptor P2X7 (P2X7R) is endowed with such properties (9).
P2X7R is a non-selective ATP-gated cation channel, expressed by various cells of neural or hematopoietic origin (10). Upon ligand binding, it enables an influx of Ca2+ and Na+ and an efflux of K+. Prolonged activation of P2X7R results in the formation of non-selective macromolecular pores and ultimately cell death (11–13). Brief stimulation induces the non-cytotoxic release of leaderless pro-inflammatory cytokines, such as IL1-β and IL-18, from TLR-ligand-activated macrophages, microglia and Schwann cells (14).
Moreover, activation of P2X7R results in the ecto-domain shedding of many cell surface proteins by metalloproteases: l-Selectin, CD23, TNFα, CD27, and matrix metalloproteinase-9 (MMP-9) (15–18).
As P2X7R is up-regulated around senile plaques in animal models of AD and in patients with AD (19, 20) and can trigger the proteolytic cleavage of membrane proteins, we examined whether activation of P2X7R may affect APP processing. In this work, we demonstrate, in human and mouse neuroblastoma cell lines, as well as in primary astrocytes and NPCs, that the specific activation of P2X7R induces the release of the fragment sAPPα. We show that this proteolytic cleavage is mediated by a metalloprotease different from those previously described for sAPPα release (6). As P2X7R is expressed in neurons from the hippocampus and in glial cells, its capacity to trigger non-amyloidogenic APP processing suggests that it may have a neuroprotective effect in AD.
EXPERIMENTAL PROCEDURES
Animals
4–8-week-old C57BL/6 mice were purchased from Charles River Laboratories. P2X7R-deficient mice (21), backcrossed to C57BL/6 mice for 7 generations, were from the Jackson Laboratory (Bar Harbor, ME).
Reagents and Antibodies
Fibroblast growth factor (FGF) was obtained from Peprotech (Rocky Hill, NJ), B27 supplement and DMEM/F12 medium from Invitrogen (Carlsbad, CA). Epidermal Growth Factor (EGF), insulin, ATP, benzoylbenzoyl ATP (Bz-ATP), oxidized-ATP (oATP), and Brilliant Blue G (BBG) were purchased from Sigma-Aldrich. Z-VAD-FMK, GM6001, TAPI-2, U0126, U0124, SP600125, SB203580, and thapsigargin were from Calbiochem (San Diego, CA). A438079, a selective antagonist of P2X7R, was obtained from Tocris Bioscience (Bristol, UK).
The following antibodies (Ab) were used for immunofluorescence and Western blotting: unconjugated mouse monoclonal antibody (mAb) anti-human APP clone 22C11 and clone 6E10 (Chemicon, Temecula, CA), anti-V5 (Invitrogen), goat anti-actin (I-19), affinity-purified rabbit anti-P2X7R antibodies recognizing residues 576 to 595 of P2X7R (Alomone Labs, Jerusalem, Israel), rabbit anti-ADAM10 IgG (eBiosciences, Hatfield, UK), rabbit anti-ADAM17 IgG (eBiosciences), goat anti-ADAM9 (R&D Systems, Minneapolis, MN), rabbit anti-Erk1/2 (pTpY202/204), anti-JNK/SAPK(pTpY183/185), anti-p38 (pTpY180/182), phosphospecific antibodies, rabbit anti-Erk1/2, anti-JNK/SAPK, anti-p38 antibodies (Cell Signaling, Danvers, MA). Affinity-purified goat anti-rabbit IgG Abs coupled to peroxidase (Rockland Immunochemicals, Gilbertsville, PA), goat anti-mouse IgG Abs coupled to peroxidase, mouse mAb anti-goat/sheep IgG conjugated to peroxidase (Sigma-Aldrich), were used as secondary Ab for Western blot analyses and goat anti-mouse IgG Abs coupled to Alexa 488 (Invitrogen) were used as secondary Ab for immunofluorescence.
Cell Cultures
Neuro2a mouse neuroblastoma cells were maintained in DMEM containing 10% fetal calf serum (FCS). Primary cultures of astrocytes were generated from hemispheres of 1–3-day-old mice. Cells were grown in DMEM containing 10% FCS. NPC cultures were prepared from striatum of mice at E14.5 day. Spheres of NPCs were grown in DMEM/F12 supplemented with B27, EGF (20 ng/ml), FGF (10 ng/ml), and insulin (20 μg/ml). SKNBE human neuroblastoma cells were maintained in DMEM/F12 medium-containing 10% FCS.
Western Blot Analyses
Proteins from cell lysates were separated by SDS-PAGE and transferred to nitrocellulose membranes, blocked with 4% nonfat milk or 5% BSA in Tris-buffered saline containing 0.2% Tween 20. Blots were immunostained with primary Ab at 4 °C overnight and probed with secondary Ab conjugated to horseradish peroxidase. Specific bands were visualized by enhanced chemiluminescence (PerkinElmer, Boston, MA).
Detergent-resistant Membrane (DRM) Preparation
Cells were submitted to surface biotinylation using sulfo-NHS-LC-Biotin (Pierce). Five million cells were resuspended in Tris-buffered saline (TBS) (Tris 50 mm, NaCl 150 mm, EDTA 1 mm, pH 7.4) containing 0.05% Triton X-100 and protease inhibitors (Roche, Basel, Switzerland) and passed 6 times through a 26G3/8 needle. After 20 min of incubation on ice, lysates were adjusted to 45% sucrose and placed at the bottom of a SW41 centrifuge tube. A sucrose step gradient was performed by layering 6 ml of 36% and 3.5 ml of 5% (w/v) sucrose in TBS. Sucrose percentages were assessed by refractometry. After centrifugation at 270,000 × g at 4 °C for 15 h, 2-ml aliquots of the gradients were collected from the top and submitted to precipitation with neutravidin-agarose beads (Pierce) at 4 °C overnight. Neutravidin-bound proteins from each sucrose gradient fraction were separated by SDS-PAGE and transferred to nitrocellulose membranes. Blots were immunostained with rabbit anti-P2X7R Abs.
Calcium Experiments
Cells were loaded with 2 μm Fura-2 AM (Molecular Probes, Invitrogen) in DMEM containing 10% FCS (1 mm Ca2+) for 30 min at 37 °C. Then, cells were washed and suspended in modified Krebs-HEPES medium (128 mm NaCl, 2.5 mm KCl, 2.7 mm CaCl2, 16 mm glucose, 20 mm HEPES, pH 7.4). We measured the Ca2+ increase in cells by dual excitation spectrofluorimetric analysis at 340 and 380 nm (ratio of A340/A380). Bz-ATP (300 μm) or thapsigargin (1 μm) were added after 50 s and Triton X-100 (0.1%) after 200 s.
Measurement of LDH Release
Cell lysis was quantified by measuring the release of lactate dehydrogenase (LDH). Cells (1 × 105) were incubated in 100 μl of DMEM for various lengths of time. Cells were centrifuged at 200 × g for 10 min, and supernatants were tested for LDH release using the oxidation reaction of β-NADH in the presence of pyruvate (Sigma kit for LDH).
Plasmids and Transfections
The cDNA encoding the human APP was cloned by RT-PCR from SHSY5Y, sequenced, and introduced into pcDNA3.1D/V5-His (Invitrogen).
Neuro2a cells were transfected with the APP construct, using GeneJuice according to the manufacturer's instructions (Novagen, Darmstadt, Germany). A stable cell line was established and a clone Neuro2a-hAPP was selected.
Immunofluorescence
Neuro2a cells, seeded on poly-lysine coated glass coverslips, were fixed with 4% paraformaldehyde. Nonspecific sites were blocked using PBS containing 0.5% FCS. Permeabilization was carried out using 0.2% Triton X-100 in blocking buffer for 10 min. Ab were applied for 1 h at room temperature. Cells were then washed three times with PBS, and secondary Ab applied for 1 h at room temperature. Cells were counterstained with Hoechst, washed, mounted with Fluoromount-G (Southernbiotech, Birmingham, AL) and observed with a fluorescent DBM Leica microscope (Leica, Wetzlar, Germany).
Stimulation of sAPP Release
Cells were grown in 24-well plates to near confluency. Medium was replaced by DMEM containing 0.5% BSA, and cells were stimulated or not with Bz-ATP or ATP. For experiments with pharmacological inhibitors, cells were treated or not for 30 min at 37 °C with inhibitor. Then the medium was replaced with DMEM 0.5%BSA with or without inhibitor, and cells were stimulated or not with Bz-ATP. The supernatants were collected and analyzed for sAPP release by Western blot. The bands corresponding to sAPP were quantified with Quantity One Software (Bio-Rad). sAPP release is expressed as a percentage of specific sAPP release in control condition. Specific sAPP release corresponds to sAPP release after 1 mm Bz-ATP stimulation minus the amount of spontaneous sAPP release. % of sAPP release = (specific sAPPα release/specific sAPPα release in control condition) × 100.
Quantification of APP and APP Fragments
The levels of APP or APP fragments were quantified in cell supernatants or cell lysates using ELISAs kits according to the manufacturer's protocols. To quantify the human APP and sAPPα, we used the PhosphoQuestTM APP human kit (DiscoverX corporation Ltd, Birmingham, UK) and for sAPPβ the human sAPPβ-w (highly sensitive) assay kit (Immuno-Biological Laboratories Co., Ltd, Hambourg, Germany). The human Aβ 1–40 and 1–42 peptide levels were determined with the PhophoQuestTM human amyloid 1–40 or 1–42 kit (DiscoverX).
RNA Interference
Small interfering RNAs (siRNA) targeting mouse P2X7R, ADAM9, ADAM10, ADAM17, and siRNA controls were from Dharmacon (Cramlington, UK). Neuro2a cells were transfected with 1 μm siRNA using the cell line Nucleofector kit V (Amaxa, Gaithersburg, MD) and seeded in flat bottom 24-well plates. Inhibition of targeted gene expression was confirmed by Western blot 48 h after transfection.
Statistical Analysis
Data are expressed as a mean value ± S.D. Data were analyzed by using Student's t test and *, p < 0.05 (at least) was considered statistically significant. When data involve more than one variable, statistical significance was estimated with one-way ANOVA followed by Student-Newman-Keuls Method or Tukey Test using SigmaStat software.
RESULTS
Characterization of Neuro2a Neuroblastoma Cells
To determine whether Neuro2a cells express P2X7R, we labeled the cell surface with biotin and analyzed the biotinylated proteins by Western blotting with the anti-P2X7R antibody after fractionation on a sucrose gradient. We observed that P2X7R is expressed at the plasma membrane and a fraction of the receptor is associated with DRMs in Neuro2a cells as observed in different cell types (22–24) (Fig. 1A). In contrast, the majority of the plasma membrane APP is outside the DRMs (Fig. 1A).
FIGURE 1.
Characterization of Neuro2-hAPP cells. A, Neuro2a cells were surface-labeled with sulfo-NHS-LC-biotin and lysed in 0.05% Triton X-100 before fractionation on a 36%/5% sucrose density gradient. Neutravidin-bound proteins from fraction 1 to 5 were analyzed by Western blot using rabbit anti-P2X7R and mouse anti-APP Abs (22C11). The low density fraction (fraction 2) contains DRMs. The heavy density fractions (fractions 4–5) contain the bulk of solubilized membrane proteins. B, Neuro2a cells were incubated with (□) or without 10 μm A438079 (■) and loaded with 2 μm Fura-2 AM. They were stimulated with 300 μm Bz-ATP in modified Krebs HEPES at 37 °C, and maximal increase was obtained after addition of 0.1% Triton X-100. The Ca2+ increase was determined as described under “Experimental Procedures.” C, LDH assay was performed on Neuro2a cells treated with 0 mm (□), 1 mm (■), or 3 mm (▴) Bz-ATP for increasing lengths of time. The results are representative of two experiments performed on different days. D, permeabilized Neuro2a-hAPP cells were labeled with 22C11, 6E10, and anti-V5 mAbs and then with Alexa488-conjugated anti-mouse IgG. Cells were counterstained with Hoechst. Control represents cells stained in absence of primary Ab. E, immunoblots were performed with rabbit anti-P2X7R antibodies on Neuro2a and Neuro2a-hAPP cells (upper panel). The same blots were stripped and probed for actin to show equal loading (lower panel). F, Neuro2a (■) and Neuro2a-hAPP (□) cells were loaded with 2 μm Fura-2 AM. Then, these cells were stimulated with 300 μm Bz-ATP in modified Krebs HEPES at 37 °C and maximal increase was obtained by addition of 0.1% Triton X-100. The increase in Ca2+ was determined as described under “Experimental Procedures.”
We then analyzed the functional activity of the surface-expressed P2X7R. Bz-ATP stimulation of Neuro2a cells induces a rapid increase in intracellular Ca2+ characteristic of channel opening (Fig. 1B). Pretreatment of Neuro2a cells with two inhibitors of P2X7R, oATP (data not shown) and A438079 (Fig. 1B), inhibited Bz-ATP-induced Ca2+ influx, confirming the functionality of P2X7R in Neuro2a cells. To determine if sustained stimulation of P2X7R induces Neuro2a cell death, we treated the cells with various Bz-ATP concentrations for increasing lengths of time. We observed the release of LDH, a marker for plasma membrane disruption, following treatment with 3 mm Bz-ATP, but no cell death was observed with 1 mm Bz-ATP even after 5 h of stimulation (Fig. 1C). Altogether, these results indicate that Neuro2a cells express a functional P2X7R.
As excellent mAbs capable of detecting human APP and its fragments have been described, we expressed the human APP in Neuro2a cells by transfecting them with a plasmid encoding the V5-tagged human APP cDNA (Fig. 1D). Neuro2a-hAPP cells, stably transfected with this vector, express the human APP protein as observed by immunocytochemistry using mAbs reacting against the V5 tag or specific for human APP (mAb 6E10) (Fig. 1D). In addition, we have shown that Neuro2a and Neuro2a-hAPP cells express comparable levels of P2X7R (Fig. 1E) and that P2X7R-stimulation induces identical increases in intracellular Ca2+in both cell lines (Fig. 1F).
ATP and Bz-ATP Induce APP Release
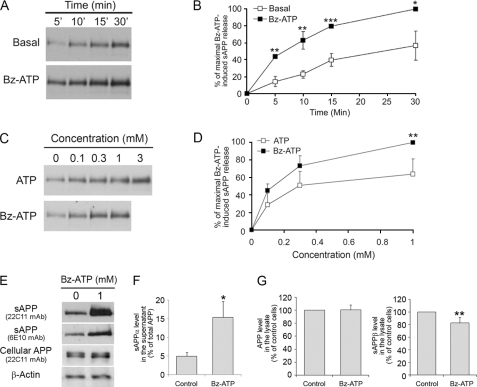
As P2X7R has been involved in the proteolytic cleavage and ecto-domain shedding of several plasma membrane proteins, we examined the effect of P2X7R stimulation on APP processing. As 1 mm Bz-ATP does not induce cell death (Fig. 1C), we treated Neuro2a-hAPP with this concentration for increasing lengths of time. P2X7R activation induced significant APP release as early as 5 min following stimulation, compared with basal release (n = 3; p < 0.01) (Fig. 2, A and B). We observed APP shedding after treatment of Neuro2a-hAPP with concentrations of 1 mm ATP and 0.3 mm Bz-ATP (Fig. 2, C and D). Thus, Bz-ATP stimulation is more efficient than ATP activation, which is characteristic of P2X7R. These results show that ATP or Bz-ATP treatment induces the release of the soluble fragment of APP (sAPP), suggesting that P2X7R is involved in this phenomenon.
FIGURE 2.
ATP and Bz-ATP stimulations trigger sAPP shedding. A, supernatants from Neuro2a-hAPP cells stimulated with or without Bz-ATP (1 mm) at 37 °C for the length of time indicated were analyzed by Western blot using anti-APP mAb (22C11). Blot shown is representative of three independent experiments. B, time course of basal (□) and Bz-ATP-stimulated (■) sAPP secretion. Each point corresponds to the amount of sAPP released divided by the maximal amount of sAPP released after 1 mm Bz-ATP stimulation for 30 min × 100. Data points represent the means ± S.E. of three independent experiments (one-way ANOVA followed by Student-Newman-Keuls Method, H = 21.7, *, p < 0.05; **, p < 0.01; ***, p < 0.001). C, supernatants from Neuro2a-hAPP cells stimulated with increasing concentrations of ATP (□) or Bz-ATP (■) during 15 min at 37 °C were analyzed by Western blot using anti-APP mAb (22C11). Blot shown is representative of four independent experiments. D, ATP (□) and Bz-ATP (■) dose-dependent release of sAPP. Each point corresponds to the amount of sAPP released divided by the maximal amount of sAPP released after 1 mm Bz-ATP stimulation for 30 min × 100. Data points represent the means ± S.E. of four independent experiments (one-way ANOVA followed by Tukey Test, F = 14.7, **, p < 0.01). E, Neuro2a-hAPP cells were stimulated with or without Bz-ATP (1 mm) for 15 min at 37 °C. Supernatants and cell lysates were analyzed by Western blot using two anti-APP mAbs: 22C11 which recognizes the N-terminal region of APP or 6E10 that reacts with the sAPPα fragment only or anti-actin Ab. These data are representative of three independent experiments. F, Neuro2a-hAPP cells were stimulated with or without Bz-ATP (1 mm) for 15 min at 37 °C. Supernatants were subjected to sandwich ELISA to quantify sAPPα (n = 3). Values are the means ± S.E. of three independent experiments (Student's t test, *, p < 0.05). G, Neuro2a-hAPP cells were treated as in F, and cell lysates were subjected to sandwich ELISA to quantify sAPPα and APP (n = 3), and sAPPβ (n = 4) were quantified by sandwich ELISA. Values are the means ± S.E. of at least three independent experiments (Student's t test, ** p < 0.01).
Bz-ATP Treatment Induces the Release of the sAPP-α Fragment
Two soluble forms of APP have been described that result from the proteolytic cleavage of APP by two secretases: α or β (1, 2). To determine which APP fragment is released following Bz-ATP stimulation, we used two mAbs: one reacting with the APP N-terminal region (mAb 22C11) and one which only recognizes the sAPPα fragment (mAb 6E10). As shown in Fig. 2E, both antibodies react with sAPP in the supernatant from Neuro2a-hAPP cells treated with Bz-ATP, demonstrating that Bz-ATP stimulation induces an α-cleavage of APP (Fig. 2E). To determine whether P2X7R activation also affects the β-secretase cleavage of APP, we compared the amounts of sAPPα and sAPPβ fragments using specific ELISA. As expected, we found a 3-fold increase of sAPPα in the supernatant of Bz-ATP-stimulated Neuro2a-hAPP cells as compared with controls (Fig. 2F) (specific increase in sAPPα released = 28 ± 9 ng/ml, n = 3 independent experiments). In contrast, we did not detect sAPPβ in the supernatants of cells stimulated or not with Bz-ATP i.e. the levels of sAPPβ were below the detection threshold 0.4 ng/ml. In addition, the amount of APP remained constant in cell lysates of stimulated or control cells (120 ± 18 ng/ml for 105 Bz-ATP-stimulated Neuro2a cells versus 120 ng/ml ± 10 ng/ml for 105 control Neuro2a cells) while sAPPβ decreased significantly in lysates of Bz-ATP-stimulated cells as compared with controls (Fig. 2G). We also titrated the amounts of peptides Aβ1–40 and Aβ1–42 in supernatants and cell lysates. We did not find these peptides using ELISAs that detect 10 pg/ml of Aβ peptides. In summary, these results show that Bz-ATP stimulation triggers the release of sAPPα and simultaneously decreases the amount of sAPPβ suggesting strongly that P2X7R activates the α-secretase pathway predominantly.
Bz-ATP-induced APP Release Is Mediated by the P2X7R
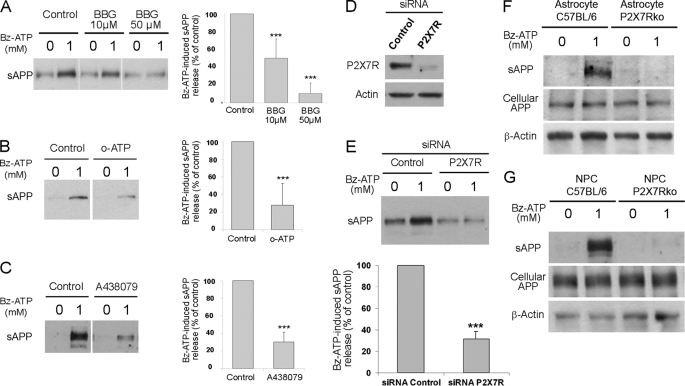
We next used two sets of experiments with pharmacological inhibitors and siRNA to demonstrate that P2X7R stimulation by Bz-ATP triggers the release of sAPPα. Pre-treatment of Neuro2a-hAPP cells with pharmacological inhibitors of P2X7R, BBG, oATP, or the more selective one, A438079 (25, 26), inhibits sAPPα release (Fig. 3, A–C) (n = 3; p < 0.001 for BBG; n = 5; p < 0.001 for oATP and A438079). Transfection of Neuro2a-hAPP cells with siRNA specific for P2X7R inhibited the expression of the P2X7R protein (Fig. 3D) and abolished Bz-ATP induced APP cleavage (Fig. 3E) (n = 4; p < 0.001), while non-target control siRNA had no effect.
FIGURE 3.
Bz-ATP stimulation induces sAPP release via activation of P2X7R. Neuro2a-hAPP cells were preincubated at 37 °C for 1 h with 10 μm and 50 μm BBG (A), for 2 h with 300 μm oATP (B), and for 30 min with 10 μm A438079 (C), and stimulated with or without Bz-ATP (1 mm) for 15 min at 37 °C. Supernatants were analyzed by Western blot using anti-APP mAb (22C11). Histograms correspond to the densitometric analysis of sAPP release as described under “Experimental Procedures.” Values are the means ± S.E. of at least three independent experiments (one-way ANOVA followed by Tukey Test, F = 36.3 for BBG and Student's t test for oATP and A438079, **, p < 0.01; ***, p < 0.001). D, cell lysates of Neuro2a-hAPP cells transfected with control or P2X7R siRNA were analyzed, using anti-P2X7R or anti-actin Abs, by Western blot (representative of four independent experiments). E, Neuro2a-hAPP cells were transfected with control or P2X7R siRNA and stimulated with or without Bz-ATP (1 mm) for 15 min at 37 °C. Supernatants were analyzed by Western blot using anti-APP mAb (22C11). Histograms correspond to the densitometric analysis of sAPP release as described under “Experimental Procedures.” Values are the means ± S.E. of four independent experiments (Student's t test, ***, p < 0.001). F, supernatants and cell lysates from C57BL/6 or P2X7Rko astrocytes stimulated without or with Bz-ATP (1 mm) for 15 min at 37 °C were analyzed by Western blot using anti-APP mAb (22C11) or anti-actin Ab. Blots shown are representative of four independent experiments. G, supernatants and cell lysates from C57BL/6 or P2X7Rko NPCs stimulated with or without Bz-ATP (1 mm) for 15 min at 37 °C were analyzed by Western blot using anti-APP mAb (22C11) or anti-actin Ab. Blots shown are representative of three independent experiments.
P2X7R-induced APP Release Is Also Observed in Primary Neural Cells
The involvement of P2X7R in ATP, endocannabinoid, and excitatory amino acid release has been previously shown in astrocytes (27–29). When we treated astrocytes from wild-type and P2X7R-deficient mice with Bz-ATP for 15 min, we observed the release of sAPPα in the supernatant from wild-type cells but not in the supernatant from P2X7R-deficient cells (Fig. 3F).
As NPCs also express a functional P2X7R (13) and APP can induce their proliferation (4, 5), we determined whether P2X7R activation induces the cleavage of APP in these cells. As shown in Fig. 3G, Bz-ATP treatment triggers the release of sAPPα in the supernatants from wild-type NPCs but not in the supernatant from P2X7R-deficient NPCs. These results demonstrate that P2X7R-activation induces specific sAPPα release in mouse primary astrocytes as well as in neural progenitor cells.
P2X7R Activation Also Induces sAPP Release in Human Neuroblastoma Cells, SKNBE
As human P2X7R has different pharmacological properties from mouse P2X7R, we tested whether human P2X7R activation mediates sAPP release.
We tested several neuroblastoma cell lines and found that SKNBE neuroblastoma express P2X7R mRNA transcripts containing none of the previously described loss-of-function polymorphic mutations (30, 31). Bz-ATP stimulation of SKNBE cells induced a rapid increase in intracellular Ca2+ (Fig. 4A) as well as sAPP release in the supernatants (Fig. 4B). Pretreatment of neuroblastoma cells with two P2X7R inhibitors, oATP and A438079, blocked Bz-ATP-induced intracellular Ca2+ increase (Fig. 4A) and sAPP release (Fig. 4, B and C) (n = 3; p < 0.001 for oATP and A438079). These results show that P2X7R activation induces sAPPα release in human neuroblastoma cells.
FIGURE 4.
Bz-ATP induces sAPP release in human SKNBE neuroblastoma cells. A, SKNBE cells incubated with (□) or without 10 μm A438079 (■) and HEK293 cells (●) were loaded with 2 μm Fura-2 AM for 30 min and stimulated with 300 μm Bz-ATP and 0.1% Triton X-100 in modified Krebs-HEPES at 37 °C. The Ca2+ increase was determined by dual excitation spectrofluorimetric analysis at 340 and 380 nm (ratio of OD340/OD380). B, SKNBE cells were preincubated for 2 h with 300 μm oATP and for 30 min with 10 μm A438079 and then stimulated with or without Bz-ATP (1 mm) for 15 min at 37 °C. Supernatants were analyzed by Western blot using anti-APP mAb (22C11). C, histograms correspond to the densitometric analysis of sAPP release as described under “Experimental Procedures.” Values are the means ± S.E. of three independent experiments (Student's t test, ***, p < 0.001).
Metalloproteases Are Involved in APP Processing Induced by P2X7R Stimulation
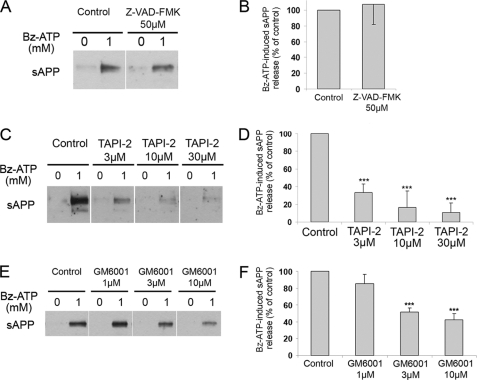
Proteolytic cleavage mediated by P2X7R involves two types of proteases: caspases and metalloproteases (14, 16, 32). To determine whether caspases are involved in P2X7R-induced APP release, we treated Neuro2a-hAPP cells with the pan-caspase inhibitor, Z-VAD-FMK and found no inhibition of P2X7R-dependent APP release by this inhibitor (Fig. 5, A and B).
FIGURE 5.
Activation of the P2X7R induces APP cleavage by a metalloprotease. Neuro2a-hAPP cells preincubated for 1 h at 37 °C with 50 μm Z-VAD-FMK (A and B) or increasing concentrations of GM6001 (C and D) or TAPI-2 (E and F), and stimulated with or without Bz-ATP (1 mm) for 15 min at 37 °C. Supernatants were analyzed by Western blot using anti-APP mAb (22C11). Histograms correspond to the densitometric analysis of sAPP release as described under “Experimental Procedures.” Values are the means ± S.E. of three independent experiments (one-way ANOVA followed by Tukey Test, F = 37.4 for GM6001 and F = 42.7 for TAPI-2, ***, p < 0.001).
In contrast, treatment with two different metalloprotease inhibitors, TAPI-2 and GM6001, resulted in significant inhibition of P2X7R-stimulated sAPP production compared with untreated cells (n = 3; p <0.001 for 3 μm TAPI-2 and 10 μm GM6001) (Fig. 5, C–F). Therefore, APP cleavage induced by P2X7R stimulation involves a metalloprotease.
APP Cleavage Stimulated by Bz-ATP Is Independent of ADAM9, -10, and -17
Different candidates for α-secretase activity have been proposed, including the metalloproteases ADAM9, -10, and -17 (6).
Using siRNA against ADAM9, -10, and -17, we inhibited the expression of the respective proteins (Fig. 6, A and C). However, while ADAM expression was strongly inhibited, Bz-ATP treatment still induced sAPPα release (Fig. 7, B, D, E). To avoid compensatory effects between ADAMs, we transfected Neuro2a-hAPP cells with siRNAs specific for the three ADAMs. As shown in Fig. 6, D and E, strong inhibition of ADAMs did not block sAPPα release induced by P2X7R stimulation. These findings strongly suggest that APP cleavage induced by P2X7R activation is mediated by a metalloprotease distinct from the well-established α-secretases.
FIGURE 6.
P2X7R-mediated APP processing is independent of ADAM9, ADAM10, and ADAM17. A, cell lysates of Neuro2a-hAPP cells transfected with control, ADAM10 or ADAM17 siRNA were analyzed by Western blot using anti-ADAM10 or anti-ADAM17 Abs. B, Neuro2a-hAPP cells were transfected with control, ADAM10 or ADAM17 siRNA and then stimulated with or without Bz-ATP (1 mm) for 15 min at 37 °C. Supernatants were analyzed by Western blot using anti-APP mAb (22C11). C, Neuro2a-hAPP cells were transfected with control, ADAM9 siRNA or ADAM9, -10, and -17 siRNAs. Cell lysates were analyzed by Western blot using anti-ADAM9 or anti-ADAM17 Abs. D, Neuro2a-hAPP cells were transfected with control, ADAM9 siRNA or ADAM9, -10, and -17 siRNAs and then stimulated with or without Bz-ATP (1 mm) for 15 min at 37 °C. Supernatants were analyzed by Western blot using anti-APP mAb (22C11). Blots are representative of three independent experiments. E, histograms correspond to densitometric analyses of sAPP release as described under “Experimental Procedures.” Values are the means ± S.E. of three independent experiments.
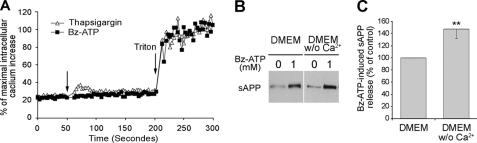
FIGURE 7.
P2X7R-dependent extracellular Ca2+ influx is not involved in sAPP release. A, Neuro2a-hAPP cells were loaded with 2 μm Fura-2 AM for 30 min. Then, these cells were stimulated with 300 μm Bz-ATP (■) or 1 μm thapsigargin (□) in modified Krebs HEPES without Ca2+ at 37 °C and maximal increase was obtained by addition of 0.1% Triton X-100. The Ca2+ increase was determined by dual excitation spectrofluorimetric analysis at 340 and 380 nm (ratio of OD340/OD380). B, supernatants from Neuro2a-hAPP cells stimulated with or without Bz-ATP (1 mm) in DMEM containing or not Ca2+ for 15 min at 37 °C were analyzed by Western blot using anti-APP mAb (22C11). Blots shown are representative of three independent experiments. C, histograms correspond to densitometric analyses of sAPP release as described under “Experimental Procedures.” Values are the means ± S.E. of three independent experiments (Student's t test, **, p < 0.01).
P2X7R-induced sAPPα Release Is Independent of Extracellular Calcium Influx
P2X7R stimulation opens cation-specific ion channels and several enzymatic activities triggered by P2X7R are Ca2+-dependent (33–35). Thus, we investigated the role of P2X7R-mediated extracellular Ca2+ influx upon sAPP release by stimulating Neuro2a cells with Bz-ATP in Ca2+-free medium. We found that sAPP shedding occurs and is significantly increased in the absence of Ca2+ in culture medium (Fig. 7, B and C) (n = 3; p < 0.01, 146.8 ± 15.0%). This phenomenon is probably due to the decrease in divalent cation concentration which increases the concentration of Bz-ATP4−, the fully dissociated tetra-anionic form, considered to be the active ligand of P2X7R. We controlled that we could detect an intracellular Ca2+ influx in the absence of extracellular Ca2+ by treating the cells with thapsigargin. Thapsigargin is a non-competitive inhibitor of a sarco-endoplasmic Ca2+ ATPase which increases cytosolic Ca2+ concentration. As shown in Fig. 7A, a peak of intracellular Ca2+ is observed after thapsigargin treatment and not after Bz-ATP stimulation. Thus, P2X7R stimulation induces no detectable Ca2+ increase in cells incubated in Ca2+-free medium. As metalloproteases are Zn-dependent enzymes, the use of divalent cations chelators such as EGTA may inhibit sAPP release by inactivation of the proteases, independently from P2X7R stimulation. Therefore, Neuro2a-hAPP cells were incubated in calcium-free medium containing low concentrations of EGTA (0.2–1 mm) to chelate residual extracellular Ca2+. We observed no diminution of released sAPP compared with Neuro2a-hAPP in medium containing Ca2+ (data not shown). Altogether, these results strongly suggest that P2X7R-dependent sAPP release is independent of extracellular Ca2+ influx.
Biochemical Pathways Involved in APP Cleavage Mediated by P2X7R
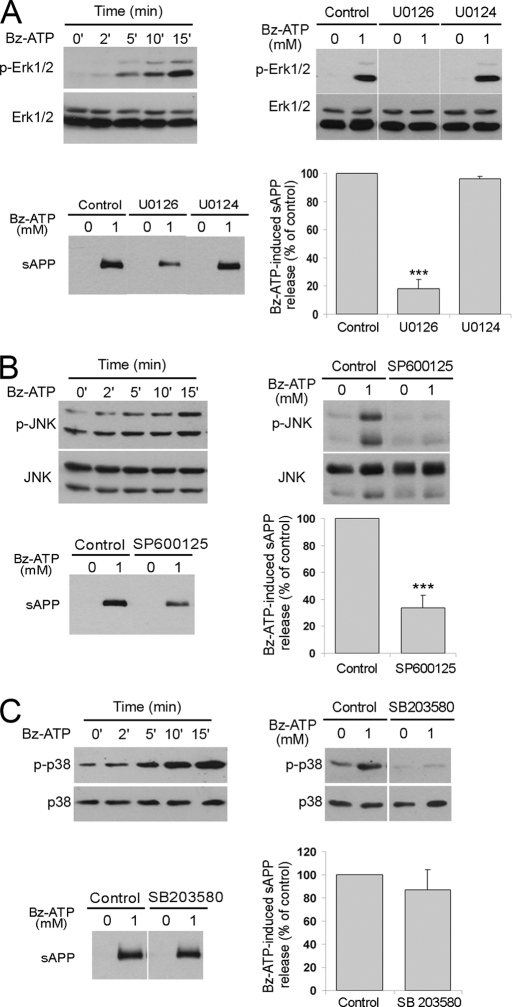
The non-amyloidogenic processing of APP, after stimulation of GPCRs, such as muscarinic receptors or P2Y2 receptors, can be induced by several independent signaling pathways, in particular the phosphorylation of Erk1/2 (7, 8). P2X7R-mediated Erk1/2 and p38 phosphorylation has been reported for different neural cell types (36–38). In Neuro2a-hAPP, we observed Erk1/2 and p38 phosphorylation after 5 min of Bz-ATP treatment (Fig. 8, A and C) and JNK phosphorylation after 15 min (Fig. 8B).
FIGURE 8.
Role of MAP kinase pathways in P2X7R-dependent α-cleavage of APP. A, a kinetic study of Erk1/2 phosphorylation after 1 mm Bz-ATP stimulation is shown in the upper left Western blot. The effect of MEK inhibitor is shown on the upper right and lower left blots. Neuro2a-hAPP cells were stimulated with 1 mm Bz-ATP for 15 min, after 1 h of incubation with inhibitor U0126, control U0124 or vehicle. Phosphorylation of Erk1/2 was analyzed in cell lysates using the anti-phospho-Erk Abs and anti-Erk1/2 Abs (upper right blot) and sAPPα release was identified in the supernatants using anti-APP mAb (22C11) (lower left blot). B, a kinetic study of JNK phosphorylation after Bz-ATP stimulation is shown in the upper left Western blot. The effect of JNK inhibitor on JNK phosphorylation is shown on the upper right and lower left blots. Neuro2a-hAPP cells were stimulated with 1 mm Bz-ATP for 15 min, after 1-h incubation with inhibitor SP600125 or vehicle. Phosphorylation of JNK was analyzed by Western blot of cell lysates using the anti-phospho-JNK antibodies and anti-JNK antibodies. sAPPα release was identified in the supernatants using anti-APP mAb (22C11) (lower left blot). C, effect of the pharmacological inhibitor of p38 (SB203580) on sAPP release by Neuro2a-hAPP cells stimulated by Bz-ATP. The experimental design is identical to those used in panels A and B. The percentage of sAPP released is calculated as described under “Experimental Procedures” and shown in the lower right panel. Values are the means ± S.E. of three independent experiments (Student's t test, ***, p < 0.001).
To determine whether MAP kinases are involved in P2X7R-dependent APP cleavage, we assessed the effect of specific inhibitors of MAP kinases on Bz-ATP-induced sAPPα release from Neuro2a-hAPP cells. Inhibition of sAPPα release was observed after pre-treatment of Neuro2a-hAPP cells with the pharmacologic inhibitors of MAP kinaseErk1/2 module (U0126) and JNK (SP600125). Both inhibitors blocked the phosphorylation of their respective targets (Fig. 8, A and B). Treatment with U0124, a negative control for the MEK1/2 inhibitor U0126, does not block Erk1/2 phosphorylation and sAPPα release (Fig. 8A). In contrast, the specific inhibitor of p38 (SB203580) did not block sAPPα shedding even though it strongly inhibited the phosphorylation of p38 (Fig. 8C).
DISCUSSION
In this report, we have demonstrated that P2X7R stimulation activates enzymatic cascades that trigger α-secretase activity, leading to the proteolytic cleavage of APP and the release of sAPPα. Indeed, after P2X7R stimulation, we observe that sAPPα is released in significant amounts, while, conversely, sAPPβ levels are notably reduced in cell lysates. In parallel, sAPPβ and Aβ peptides are undetected in the supernatants of Bz-ATP-stimulated cells. Thus, P2X7R activation shifts APP processing toward α-cleavage predominantly.
The P2X7R-specific cleavage of APP was observed in mouse and human neuroblastoma cells, in primary murine astrocytes and neural progenitor cells. We also showed that the α-processing of APP is mediated by a metalloprotease different from ADAM9, -10, and -17, which were shown to have α-secretase activity on APP in vivo.
Several lines of evidence demonstrate that ATP- or Bz-ATP-mediated sAPPα release is specifically dependent on P2X7R: (1) three pharmacological inhibitors of P2X7R block the release of sAPPα mediated by Bz-ATP; (2) inhibition of P2X7R synthesis by RNA interference results in 68.5 ± 7.1% reduction of sAPPα shedding (Fig. 3, D and E); and (3) stimulation by Bz-ATP of mouse primary astrocytes and NPCs from P2X7R-deficient mice does not induce sAPPα release while it does in cells derived from C57BL/6 animals (Fig. 3, F and G). Altogether, these experiments establish that ATP or Bz-ATP stimulation of various neural cells triggers P2X7R-dependent α-processing of APP.
Whereas Aβ toxicity is mostly observed on neurons, we have chosen to analyze APP processing on primary astrocytes and neural progenitor cells. Indeed, P2X7R is expressed on a small population of neurons (10) that are difficult to isolate and cultivate whereas P2X7R is expressed at high levels on all glial cells. Astrocytes accumulation has been observed around amyloid deposit on patient brain slices (39) emphazing a role of these cells in the homeostasis of amyloid plaques. Moreover, astrocytes represent a population of interest in AD because of their potential for modulating the neuronal environment (39).
Numerous studies have identified several α-secretases able to cleave membrane APP to generate the non-amyloidogenic sAPPα fragment (reviewed in Refs. 6, 40). ADAM10 and -17 can cut APP at the physiological site between Lys16 and Leu17 of the Aβ peptide, while ADAM9 cleaves it at an alternative site. The first in vivo evidence for a role of ADAM in α-processing of APP came from studies on ADAM10-transgenic mice (41). Transgenic overexpression of ADAM10 inhibits amyloid plaque formation and neurological deficits in an AD mouse model, while overexpression of an inactive form of ADAM10 increases amyloid plaques (41). However, APP processing is normal in mouse embryonic fibroblasts from ADAM10-deficient mice and in neurons of the hippocampus of ADAM9-deficient animals indicating that other proteases replaced them (42, 43). Thus, functional overlap between ADAMs may occur i.e. one substrate can be cleaved by several metalloproteases. Indeed, Le Gall et al. (44) have shown that ADAM10 can cleave ADAM17 substrates in ADAM17-null cells which have been stimulated with phorbol esters, ionomycin or the P2X7R ligand Bz-ATP and vice versa. Furthermore, ADAM9 as well as ADAM15 may be involved in the regulation of ADAM10 by cleaving it and inducing the shedding of its catalytically active ectodomain (45), thus preventing ADAM10 from processing plasma membrane APP (46).For most of the GPCR and NMDA receptors, the evidence for an involvement of ADAMs in APP processing is based mainly on inhibition with pharmacological inhibitors of ADAMs. However, Camden et al. (7) demonstrated, using siRNA, that P2Y2-induced release of sAPPα is due to the activation of ADAM10 and ADAM17. In contrast, our siRNA experiments show that ADAM9, -10, and -17 are not involved in P2X7R-dependent α-cleavage of APP, although the constitutive release of sAPPα was clearly inhibited (64% data not shown). Our results strongly suggest that, in the simultaneous absence of ADAM9, -10, and -17, an alternative sheddase can process APP, most probably another TAPI2-GM6001-sensitive metalloprotease.
Several reports have shown that P2X7R stimulation triggers the proteolytic activity of caspase-1 after NRLP3/ASC inflammasome assembly (reviewed in Ref. 47). In addition, Silverman et al. (48) have shown that P2X7R co-immunoprecipitates with the NRLP1 inflammasome, pannexin-1, ASC and caspase-1 and -11. Thus, using z-VAD, a pan-caspase inhibitor, we show that it does not block sAPPα release, ruling out a role for these caspases in P2X7R-dependent APP processing.
Several enzymatic activities are dependent upon Ca2+ increase consecutive to P2X7R activation (33–35). However, our present experiments indicate that extracellular Ca2+ is not required for sAPPα shedding following P2X7R stimulation. This observation agrees with our previous findings showing that P2XR-dependent thymocyte death, which involves the sequential activation of several enzymes such as Erk1/2, is Ca2+-independent (12). In contrast, the generation of sAPPα after stimulation of P2Y2 (7) or NMDA receptors (49) is dependent on extracellular Ca2+. However, while Ca2+ influx through NMDA receptor is needed for sAPPα cleavage, the P2Y2-stimulated rise in cytosolic Ca2+ is not required for the P2Y2-dependent shedding of sAPPα suggesting that the P2Y2 activated pathway involves an extracellular Ca2+-dependent protein.
Our studies show that P2X7R-induced sAPPα release involves Erk1/2 and JNK phosphorylation. The role of Erk1/2 in α-secretase activation has been observed after stimulation of several GPCR (P2Y2, PAC1, and M1 receptors). In agreement with our results, the involvement of both Erk1/2 and JNK in the α-cleavage of APP has been established in neuroglioma U251 cells (50). Several reports showed that JNK is able to phosphorylate APP at Thr668 (51). However, the physiological role of APP phosphorylation at Thr668 is controversial, some groups claiming that it triggers the amyloidogenic pathway (52) and others that phosphorylation is mainly involved in axonal transport of APP (53). It was also shown that Erk1/2 phosphorylates Thr735 of ADAM17, a step needed for the shedding of TrkA neurotrophin receptor (54). One can hypothesize that P2X7R-dependent Erk1/2 activation is involved in the phosphorylation/activation of the metalloprotease implicated in the release of sAPPα.
It has been shown previously that amyloidogenic processing of APP occurs in rafts in the trans-Golgi network and endosomes, while α-cleavage occurs at the plasma membrane (55), outside raft microdomains (56). In line with this, we found that the vast majority of surface APP is outside of lipid rafts, i.e. in the non-DRM fractions (Fig. 1A). It is worth noting that equal amounts of P2X7R are present inside and outside DRMs, in agreement with several reports (22–24). Garcia-Marcos et al. (23) have shown that the distinct P2X7R pools stimulate different signaling pathways: the pool outside of DRMs seems to be responsible for the nonspecific cation channel activity. Whereas Ca2+ influx is not involved in P2X7R-dependent shedding of APP, both pools of P2X7R may be involved in triggering α-cleavage of APP located outside DRMs via MAP kinases activation.
Several reports have established that some GPCRs are involved in APP processing. The human serotonin 5-HT4 (57), 5-HT2a, 5-HT2c (58), the muscarinic acetylcholine receptors M1 and M3 (59, 60), the metabotropic glutamate receptor (mGluR5) (61), the PAC1 receptor (62), and the purinergic receptor P2Y2 (7) are capable of stimulating the release of sAPPα. Thus, the stimulation of these receptors by pharmacological agonists could improve AD. Indeed, AF267B, a selective M1 muscarinic agonist, was shown to improve the neuropathological lesions and spatial learning in a mouse model of AD (63). However, among the GPCR family, the orphan GPCR-3 was shown to stimulate the amyloidogenic pathway in vivo (64).
Altogether, these findings strongly suggest that identifying receptors capable of modulating the processing of APP is an important goal, which could open new avenues for the treatment of AD. Up to now, inhibitors of β- and γ-secretases for the treatment of AD have been the focus of intense research. Various types of inhibitors have been synthesized, however a decrease in BACE-1 and γ-secretase activities may lead to serious side effects (65). An alternative approach is to stimulate α-secretase. Regulating α-cleavage of APP is pertinent because α-secretase activation prevents the deposition of Aβ peptides and produces the neuroprotective sAPPα fragment. In this respect, sAPPα release can be increased by stimulation of various receptors and by overexpression of proteins such as SNX33, which decreases the rate of APP endocytosis and increase the amount of APP available for α-secretase (66).
Our present results demonstrate that P2X7R triggers a novel non-amyloidogenic pathway, independent of ADAM9, -10, and -17, which open challenging perspectives based on P2X7R being a new therapeutic target in AD.
Acknowledgment
We thank Jocelyne Dujancourt for technical assistance.
This work was supported by the Centre National de la Recherche Scientifique (CNRS), Agence Nationale pour la Recherche (ANR-07-BLAN-0089-02), Association France Alzheimer, and Neuropôle de Recherche francilien (NeRF).
- APP
- amyloid precursor protein
- Aβ
- β-amyloid
- AD
- Alzheimer Disease
- DRM
- detergent-resistant membrane
- NPC
- neural progenitor cells
- Bz-ATP
- benzoylbenzoyl ATP.
REFERENCES
- 1. Gandy S. (2005) J. Clin. Invest. 115, 1121–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilquet V., De Strooper B. (2004) Curr. Opin. Neurobiol. 14, 582–588 [DOI] [PubMed] [Google Scholar]
- 3. Mattson M. P. (1997) Physiol. Rev. 77, 1081–1132 [DOI] [PubMed] [Google Scholar]
- 4. Caillé I., Allinquant B., Dupont E., Bouillot C., Langer A., Müller U., Prochiantz A. (2004) Development 131, 2173–2181 [DOI] [PubMed] [Google Scholar]
- 5. Ohsawa I., Takamura C., Morimoto T., Ishiguro M., Kohsaka S. (1999) Eur. J. Neurosci. 11, 1907–1913 [DOI] [PubMed] [Google Scholar]
- 6. Allinson T. M., Parkin E. T., Turner A. J., Hooper N. M. (2003) J. Neurosci. Res. 74, 342–352 [DOI] [PubMed] [Google Scholar]
- 7. Camden J. M., Schrader A. M., Camden R. E., González F. A., Erb L., Seye C. I., Weisman G. A. (2005) J. Biol. Chem. 280, 18696–18702 [DOI] [PubMed] [Google Scholar]
- 8. Haring R., Fisher A., Marciano D., Pittel Z., Kloog Y., Zuckerman A., Eshhar N., Heldman E. (1998) J. Neurochem. 71, 2094–2103 [DOI] [PubMed] [Google Scholar]
- 9. Surprenant A., North R. A. (2009) Annu. Rev. Physiol. 71, 333–359 [DOI] [PubMed] [Google Scholar]
- 10. Sperlágh B., Vizi E. S., Wirkner K., Illes P. (2006) Prog. Neurobiol. 78, 327–346 [DOI] [PubMed] [Google Scholar]
- 11. Surprenant A., Rassendren F., Kawashima E., North R. A., Buell G. (1996) Science 272, 735–738 [DOI] [PubMed] [Google Scholar]
- 12. Auger R., Motta I., Benihoud K., Ojcius D. M., Kanellopoulos J. M. (2005) J. Biol. Chem. 280, 28142–28151 [DOI] [PubMed] [Google Scholar]
- 13. Delarasse C., Gonnord P., Galante M., Auger R., Daniel H., Motta I., Kanellopoulos J. M. (2009) J. Neurochem. 109, 846–857 [DOI] [PubMed] [Google Scholar]
- 14. Ferrari D., Pizzirani C., Adinolfi E., Lemoli R. M., Curti A., Idzko M., Panther E., Di Virgilio F. (2006) J. Immunol. 176, 3877–3883 [DOI] [PubMed] [Google Scholar]
- 15. Moon H., Na H. Y., Chong K. H., Kim T. J. (2006) Immunol. Lett. 102, 98–105 [DOI] [PubMed] [Google Scholar]
- 16. Gu B., Bendall L. J., Wiley J. S. (1998) Blood 92, 946–951 [PubMed] [Google Scholar]
- 17. Suzuki T., Hide I., Ido K., Kohsaka S., Inoue K., Nakata Y. (2004) J. Neurosci. 24, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu B. J., Wiley J. S. (2006) Blood 107, 4946–4953 [DOI] [PubMed] [Google Scholar]
- 19. Parvathenani L. K., Tertyshnikova S., Greco C. R., Roberts S. B., Robertson B., Posmantur R. (2003) J. Biol. Chem. 278, 13309–13317 [DOI] [PubMed] [Google Scholar]
- 20. McLarnon J. G., Ryu J. K., Walker D. G., Choi H. B. (2006) J. Neuropathol. Exp. Neurol. 65, 1090–1097 [DOI] [PubMed] [Google Scholar]
- 21. Solle M., Labasi J., Perregaux D. G., Stam E., Petrushova N., Koller B. H., Griffiths R. J., Gabel C. A. (2001) J. Biol. Chem. 276, 125–132 [DOI] [PubMed] [Google Scholar]
- 22. Barth K., Weinhold K., Guenther A., Young M. T., Schnittler H., Kasper M. (2007) Febs J. 274, 3021–3033 [DOI] [PubMed] [Google Scholar]
- 23. Garcia-Marcos M., Pérez-Andrés E., Tandel S., Fontanils U., Kumps A., Kabré E., Gómez-Muñoz A., Marino A., Dehaye J. P., Pochet S. (2006) J. Lipid Res. 47, 705–714 [DOI] [PubMed] [Google Scholar]
- 24. Gonnord P., Delarasse C., Auger R., Benihoud K., Prigent M., Cuif M. H., Lamaze C., Kanellopoulos J. M. (2009) Faseb J. 23, 795–805 [DOI] [PubMed] [Google Scholar]
- 25. Carroll W. A., Donnelly-Roberts D., Jarvis M. F. (2009) Purinergic Signal 5, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jarvis M. F., Khakh B. S. (2009) Neuropharmacology 56, 208–215 [DOI] [PubMed] [Google Scholar]
- 27. Duan S., Anderson C. M., Keung E. C., Chen Y., Swanson R. A. (2003) J. Neurosci. 23, 1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suadicani S. O., Brosnan C. F., Scemes E. (2006) J. Neurosci. 26, 1378–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walter L., Dinh T., Stella N. (2004) J. Neurosci. 24, 8068–8074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shemon A. N., Sluyter R., Fernando S. L., Clarke A. L., Dao-Ung L. P., Skarratt K. K., Saunders B. M., Tan K. S., Gu B. J., Fuller S. J., Britton W. J., Petrou S., Wiley J. S. (2006) J. Biol. Chem. 281, 2079–2086 [DOI] [PubMed] [Google Scholar]
- 31. Wiley J. S., Dao-Ung L. P., Gu B. J., Sluyter R., Shemon A. N., Li C., Taper J., Gallo J., Manoharan A. (2002) Lancet 359, 1114–1119 [DOI] [PubMed] [Google Scholar]
- 32. Labasi J. M., Petrushova N., Donovan C., McCurdy S., Lira P., Payette M. M., Brissette W., Wicks J. R., Audoly L., Gabel C. A. (2002) J. Immunol. 168, 6436–6445 [DOI] [PubMed] [Google Scholar]
- 33. Gudipaty L., Munetz J., Verhoef P. A., Dubyak G. R. (2003) Am. J. Physiol. Cell Physiol. 285, C286–C299 [DOI] [PubMed] [Google Scholar]
- 34. Le Stunff H., Auger R., Kanellopoulos J., Raymond M. N. (2004) J. Biol. Chem. 279, 16918–16926 [DOI] [PubMed] [Google Scholar]
- 35. Armstrong S., Pereverzev A., Dixon S. J., Sims S. M. (2009) J. Cell Sci. 122, 136–144 [DOI] [PubMed] [Google Scholar]
- 36. Papp L., Vizi E. S., Sperlágh B. (2007) Biochem. Biophys. Res. Commun. 355, 568–574 [DOI] [PubMed] [Google Scholar]
- 37. Gendron F. P., Neary J. T., Theiss P. M., Sun G. Y., Gonzalez F. A., Weisman G. A. (2003) Am. J. Physiol. Cell Physiol. 284, C571–C581 [DOI] [PubMed] [Google Scholar]
- 38. Panenka W., Jijon H., Herx L. M., Armstrong J. N., Feighan D., Wei T., Yong V. W., Ransohoff R. M., MacVicar B. A. (2001) J. Neurosci. 21, 7135–7142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuller S., Münch G., Steele M. (2009) Expert Rev. Neurother. 9, 1585–1594 [DOI] [PubMed] [Google Scholar]
- 40. Huovila A. P., Turner A. J., Pelto-Huikko M., Kärkkäinen I., Ortiz R. M. (2005) Trends Biochem. Sci. 30, 413–422 [DOI] [PubMed] [Google Scholar]
- 41. Postina R., Schroeder A., Dewachter I., Bohl J., Schmitt U., Kojro E., Prinzen C., Endres K., Hiemke C., Blessing M., Flamez P., Dequenne A., Godaux E., van Leuven F., Fahrenholz F. (2004) J. Clin. Invest. 113, 1456–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., von Figura K., Saftig P. (2002) Hum. Mol. Genet. 11, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 43. Weskamp G., Cai H., Brodie T. A., Higashyama S., Manova K., Ludwig T., Blobel C. P. (2002) Mol. Cell. Biol. 22, 1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Le Gall S. M., Bobé P., Reiss K., Horiuchi K., Niu X. D., Lundell D., Gibb D. R., Conrad D., Saftig P., Blobel C. P. (2009) Mol. Biol. Cell 20, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tousseyn T., Thathiah A., Jorissen E., Raemaekers T., Konietzko U., Reiss K., Maes E., Snellinx A., Serneels L., Nyabi O., Annaert W., Saftig P., Hartmann D., De Strooper B. (2009) J. Biol. Chem. 284, 11738–11747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parkin E., Harris B. (2009) J. Neurochem. 108, 1464–1479 [DOI] [PubMed] [Google Scholar]
- 47. Di Virgilio F. (2007) Trends Pharmacol. Sci. 28, 465–472 [DOI] [PubMed] [Google Scholar]
- 48. Silverman W. R., de Rivero Vaccari J. P., Locovei S., Qiu F., Carlsson S. K., Scemes E., Keane R. W., Dahl G. (2009) J. Biol. Chem. 284, 18143–18151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoey S. E., Williams R. J., Perkinton M. S. (2009) J. Neurosci. 29, 4442–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ma G., Chen S., Wang X., Ba M., Yang H., Lu G. (2005) J. Neurosci. Res. 80, 683–692 [DOI] [PubMed] [Google Scholar]
- 51. Muresan Z., Muresan V. (2005) J. Neurosci. 25, 3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee M. S., Kao S. C., Lemere C. A., Xia W., Tseng H. C., Zhou Y., Neve R., Ahlijanian M. K., Tsai L. H. (2003) J. Cell Biol. 163, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suzuki T., Nakaya T. (2008) J. Biol. Chem. 283, 29633–29637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Díaz-Rodríguez E., Montero J. C., Esparís-Ogando A., Yuste L., Pandiella A. (2002) Mol. Biol. Cell 13, 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thinakaran G., Koo E. H. (2008) J. Biol. Chem. 283, 29615–29619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ehehalt R., Keller P., Haass C., Thiele C., Simons K. (2003) J. Cell Biol. 160, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Robert S. J., Zugaza J. L., Fischmeister R., Gardier A. M., Lezoualc'h F. (2001) J. Biol. Chem. 276, 44881–44888 [DOI] [PubMed] [Google Scholar]
- 58. Nitsch R. M., Deng M., Growdon J. H., Wurtman R. J. (1996) J. Biol. Chem. 271, 4188–4194 [DOI] [PubMed] [Google Scholar]
- 59. Nitsch R. M., Slack B. E., Wurtman R. J., Growdon J. H. (1992) Science 258, 304–307 [DOI] [PubMed] [Google Scholar]
- 60. Buxbaum J. D., Oishi M., Chen H. I., Pinkas-Kramarski R., Jaffe E. A., Gandy S. E., Greengard P. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10075–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee R. K., Wurtman R. J., Cox A. J., Nitsch R. M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8083–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kojro E., Postina R., Buro C., Meiringer C., Gehrig-Burger K., Fahrenholz F. (2006) Faseb J. 20, 512–514 [DOI] [PubMed] [Google Scholar]
- 63. Caccamo A., Oddo S., Billings L. M., Green K. N., Martinez-Coria H., Fisher A., LaFerla F. M. (2006) Neuron 49, 671–682 [DOI] [PubMed] [Google Scholar]
- 64. Thathiah A., Spittaels K., Hoffmann M., Staes M., Cohen A., Horré K., Vanbrabant M., Coun F., Baekelandt V., Delacourte A., Fischer D. F., Pollet D., De Strooper B., Merchiers P. (2009) Science 323, 946–951 [DOI] [PubMed] [Google Scholar]
- 65. Bandyopadhyay S., Goldstein L. E., Lahiri D. K., Rogers J. T. (2007) Curr. Med. Chem. 14, 2848–2864 [DOI] [PubMed] [Google Scholar]
- 66. Schöbel S., Neumann S., Hertweck M., Dislich B., Kuhn P. H., Kremmer E., Seed B., Baumeister R., Haass C., Lichtenthaler S. F. (2008) J. Biol. Chem. 283, 14257–14268 [DOI] [PubMed] [Google Scholar]