Abstract
Ric-8A and Ric-8B are nonreceptor G protein guanine nucleotide exchange factors that collectively bind the four subfamilies of G protein α subunits. Co-expression of Gα subunits with Ric-8A or Ric-8B in HEK293 cells or insect cells greatly promoted Gα protein expression. We exploited these characteristics of Ric-8 proteins to develop a simplified method for recombinant G protein α subunit purification that was applicable to all Gα subunit classes. The method allowed production of the olfactory adenylyl cyclase stimulatory protein Gαolf for the first time and unprecedented yield of Gαq and Gα13. Gα subunits were co-expressed with GST-tagged Ric-8A or Ric-8B in insect cells. GST-Ric-8·Gα complexes were isolated from whole cell detergent lysates with glutathione-Sepharose. Gα subunits were dissociated from GST-Ric-8 with GDP-AlF4− (GTP mimicry) and found to be >80% pure, bind guanosine 5′-[γ-thio]triphosphate (GTPγS), and stimulate appropriate G protein effector enzymes. A primary characterization of Gαolf showed that it binds GTPγS at a rate marginally slower than Gαs short and directly activates adenylyl cyclase isoforms 3, 5, and 6 with less efficacy than Gαs short.
Keywords: Adenylate Cyclase (Adenylyl Cyclase), G Proteins, Phospholipase C, Protein Purification, Signal Transduction, Ric-8, Guanine Nucleotide Exchange Factor, Myristoylation
Introduction
Heterotrimeric G proteins are the foremost signal-transducing molecules used by G protein-coupled-receptors (GPCRs)3 to regulate sensation and cellular physiology. Agonist-stimulated GPCRs are guanine nucleotide exchange factors that stimulate G protein α subunit (Gα) GDP release. Subsequent GTP binding to Gα causes heterotrimer dissociation or rearrangement so that Gα-GTP and Gβγ adopt states for efficient activation of downstream effector enzymes. Purified G protein subunits have been essential reagents used to develop the current understanding of G protein function, structure, and signaling pathways (1, 2). Current knowledge of traditional G protein signaling network complexity is expanding, and G proteins have been assigned new nontraditional signaling roles including regulation of cell division through unique classes of effector and modulatory enzymes (3–5). As cross-disciplinary G protein research proliferates, the need for purified components to elucidate G protein functionality is significant.
G protein heterotrimers are classified by the identity of the guanine nucleotide-binding subunit: Gα. There are four classes of Gα subunits: Gαs, Gαi, Gαq, and Gα12/13. Efficient procedures are in place to produce most Gαi class subunits and Gαs from Escherichia coli (6, 7). Members of the Gαq and Gα12/13 classes can be prepared from an insect cell expression system using a Gβγ co-purification procedure. This method involves tagging the Gγ subunit with a His6 tag, isolating the trimeric G protein by metal chelate chromatography, and eluting the Gα with high specificity using GTP mimicry. This method is tried and true but rather laborious and involves extensive steps of cell membrane preparation, washing, and detergent extraction. The procedure also results in low Gα yields (≤50–200 μg of protein/liter of cell culture) (8–11). To our knowledge, the prime target of the largest class of GPCRs, olfactory-specific Gαolf (a Gαs family member) has not been purified in sufficient, active quantity to permit its characterization.
While characterizing the G protein guanine nucleotide exchange factor activity of Ric-8 (resistance to inhibitors of cholinesterase 8), a series of observations were made that led us to hypothesize that Ric-8 proteins could be used as molecular tools to prepare recombinant Gα subunits: 1) Ric-8A and Ric-8B collectively bound all four Gα subunit classes (12, 13); 2) the Ric-8A·Gα interaction could be manipulated with guanine nucleotides. Gαi1 formed a stable complex with GST-Ric-8A in the presence of GDP but was dissociated by GTP(γS) (13, 14); 3) reduction of Ric-8 expression through genetic interventions reduced plasma membrane localization of different Gα subunits (15–19), implying that Ric-8 proteins may positively affect G protein expression; and 4) Ric-8B transfection in mammalian cells promoted Gαs/Gαolf expression (18, 20).
Here we introduce a method for Gα subunit purification that substantially improves upon established methods in its simplicity, uniformity of application toward all Gα subunit classes, and yield and purity of G protein obtained. Co-expression of GST-tagged Ric-8A or B and Gα subunits in insect cells permitted the isolation of GST-Ric-8·Gα complexes from whole cell detergent lysates with glutathione-Sepharose. Gα subunits were recovered specifically from this matrix by elution with AlF4− and desalted. This procedure allowed the first production of active Gαolf, an olfactory/brain-specific stimulator of adenylyl cyclase. Using in vitro effector enzyme reconstitution assays, we show that the Gα subunits produced by these means are functional proteins and demonstrate that Gαolf is a less potent and efficacious activator of adenylyl cyclase isoforms than equivalently produced Gαs short.
EXPERIMENTAL PROCEDURES
Quantitative Gα-YFP Expression Assays
HEK293 cells were co-transfected as described (21) with pcDNA3.1-Gαi1-YFP (22) or pcDNAI/Amp-Gαs-YFP (a gift from Dr. Catherine H. Berlot, Geisinger Health System, Danville, PA) (23) and pcDNA3.1 constructs that expressed Ric-8A (13), Ric-8BFL, or Ric-8BΔ9. Fluorescence measurements were performed as described previously (21). Forty-eight hours after transfection, the cells were harvested with Tyrode's solution (140 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 0.37 mm NaH2PO4, 24 mm NaHCO3, 10 mm HEPES-KOH, pH 7.4, and 0.1% glucose (m/v)) and distributed in triplicate at 1 × 105 cells/well into gray 96-well plates. Total fluorescence (excitation, 485 nm; emission, 535 nm) was measured to quantify Gαi1-YFP or Gαs-YFP expression using a TriStar LB 941 plate reader (Berthold Technologies, Oak Ridge, TN). The data are plotted in relative fluorescent units and are the averages of three independent transfection experiments.
Insect Cell Culture and Protein Expression
GST-tagged Ric-8A and untagged G protein α subunit baculoviruses were described previously (8–11, 13, 24). A GST-Ric-8B baculovirus-targeting construct was created using linker-based PCR to amplify full-length mouse Ric-8B from a purchased clone (Invitrogen LLAM collection clone 6490136 in pCMV-Sport6). The amplified product was digested and ligated into the EcoRI and SalI restriction sites of pFASTBac GST-tobacco etch virus (13). The resultant amino acid sequences of the tagged Ric-8 proteins were N′-GST-tobacco etch virus site -Glu(E)-Phe(F)-Ric-8-C′. If cleaved by tobacco etch virus protease digestion, the sequences become N′-Gly(G)-Glu(E)-Phe(F)-Ric-8-C′. Recombinant baculoviruses were produced after transfection of adherent Sf9 cells per the manufacturer's instructions (Bac-to-Bac system; Invitrogen). The transfection viral medium supernatants were harvested after 9 days, and culture volumes were amplified twice for 5 days in log phase Sf9 suspension cells grown in shake flasks at 2.0 × 106 cells/ml. Suspension Sf9 cells were grown in IPL41 medium containing 10% (v/v) heat-inactivated FBS. Secondarily amplified viruses (5–10 ml of GST-Ric-8 and 5–15 ml of Gα) were used to co-infect 1-liter High Five insect cell cultures growing at 2.0 × 106 cells/ml in Sf900II medium (Invitrogen). After 48 h expression, the cells were harvested by centrifugation at 2000 × g and stored as a cell paste at −80 °C until use. The optimal amounts and ratios of secondary amplified viruses used were determined empirically in smaller sized culture (50–200 ml) prior to conducting large scale (1 liter) preparations (supplemental Fig. 1S).
HiTrap Q Anion Exchange Chromatography
High Five insect cells (200 ml) grown in suspension to a density of 2.0 × 106 cells/ml were infected with volumes of twice amplified GST-Ric-8A and/or Gαq baculovirus stocks for 48 h. The cell pellets were collected by centrifugation at 1500 × g and lysed in 200 ml of Buffer N (20 mm HEPES-KOH, pH 8.0, 2 mm MgCl2, 1 mm EDTA, 1 mm DTT, 11 mm CHAPS, and protease inhibitor mixture) by Parr bomb nitrogen cavitation. The detergent whole cell lysates were clarified by centrifugation at 100,000 × g for 45 min, passaged through a 0.22-μm filter and loaded onto a 5-ml Hi trap Q column at 1 ml/min using a Bio-Rad Duoflow system. The column was washed with Buffer N and eluted with a linear gradient to 500 mm NaCl in Buffer N. Fractions of the eluate were collected as the gradient developed. Fractions containing Gαq were analyzed by Western blot with anti-Gαq/11 antibody, C-19 (Santa Cruz, Inc. SC-392), Coomassie-stained SDS-polyacrylamide gel analysis, and the GTPγS nitrocellulose filter binding assay.
Glutathione-Sepharose Chromatography
Cell pastes were suspended in 300 ml of detergent lysis buffer (20 mm HEPES-KOH, pH 8.0, 150 mm NaCl, 1 mm DTT, 1 mm EDTA, 0.05% (m/v) Genapol C100 detergent (Calbiochem), containing protease inhibitor mixture; (23 μg/ml phenylmethylsulfonyl fluoride, 21 μg/ml Nα-p-tosyl-l-lysine-chloromethyl ketone, 21 μg/ml l-1-p-tosylamino-2-phenylethyl-chloromethyl ketone, 3.3 μg/ml leupeptin, and 3.3 μg/ml lima bean trypsin inhibitor)) and stirred at 4 °C for 30 min. The detergent lysates were homogenized/disrupted by nitrogen cavitation using a Parr Bomb (Parr Instrument, Moline, IL), or by tight pestle Dounce homogenization (Kontes, Vineland, NJ). The lysates were centrifuged sequentially at 3000 × g for 10 min and 100,000 × g for 45 min. The clarified detergent supernatants were loaded onto packed 5-ml bed volume glutathione-Sepharose 4B columns driven by gravity. The column flow through was reapplied to this matrix one time. The columns were washed with 20 column volumes of CHAPS buffer (20 mm HEPES-KOH, pH 8.0, 100 mm NaCl, 1 mm DTT, 11 mm CHAPS, and protease inhibitor mixture) and then warmed to 22 °C. To elute Gα subunits, 50 ml of 30 °C AMF buffer (20 mm HEPES-KOH, pH 8.0, 100 mm NaCl, 50 mm MgCl2, 1 mm DTT, 10 mm NaF, 30 μm AlCl3, 11 mm CHAPS, and 100 μm GTP) was applied to the columns and allowed to flow through slowly. Gα subunits were typically eluted in the first 10–15 ml with this elution buffer. Ric-8 proteins were then eluted with CHAPS buffer containing 20 mm reduced glutathione. Gα yield was measured by Bradford assay, and purity was estimated by Image J (version 10.2) analysis of full Coomassie-stained SDS-polyacrylamide gel lanes.
PD-10-desalting Gel Filtration
AlF4− and excess MgCl2 removal could be accomplished by passaging Gα subunits through PD-10 desalting columns (GE Healthcare). Gα subunits in AMF buffer were concentrated in Vivaspin-20 30,000 molecular weight cut-off ultrafiltration centrifugal concentrators (Sartorius Stedim Biotech, Goettingen, Germany) to a final volume of 2.5 ml (no more than 5 mg/ml protein) and passaged onto a PD-10 column pre-equilibrated with CHAPS storage buffer (20 mm HEPES-KOH, pH 8.0, 1 mm DTT, 0.5 mm EDTA, 1 μm GDP, and 11 mm CHAPS). Gα subunits were eluted by gravity in 3.5 ml of storage buffer and concentrated by ultrafiltration. Aliquoted Gα concentrated proteins were snap frozen in liquid N2 and stored at −80 °C.
Superdex Gel Filtration
The preferred method of AlF4− and MgCl2 removal was gel filtration of concentrated Gα subunits through Superdex 75 and 200 10/300 GL columns arranged in tandem (GE Healthcare). Superdex chromatography thoroughly removed the chemical and some minor protein impurities. Gα subunits (2.5 mg) eluted from the glutathione-Sepharose columns with Mg·GDP·AlF4 were concentrated to 550 μl in CHAPS storage buffer by ultrafiltration. The Superdex columns were equilibrated with CHAPS storage buffer and precalibrated with gel filtration sizing standards (Bio-Rad). Gα subunits were pumped through the columns at 0.3 ml/min using a Bio-Rad Duoflow System, and fractions of the column eluate were collected using a fraction collector. Fractions containing monomeric Gα subunits were pooled, concentrated by ultrafiltration, snap frozen in liquid N2, and stored at −80 °C.
Subcellular Fractionation
High Five insect cells (25 ml of suspension culture) were grown to 2.0 × 106 cells/ml in Sf900 II medium (Invitrogen) and infected with 250 μl of twice amplified Gαi1, and GST or GST-Ric-8A baculovirus stocks. The cells were collected by centrifugation and lysed in 12.5 ml of detergent-free buffer (20 mm HEPES-KOH, pH 8.0, 150 mm NaCl, 1 mm DTT, 1 mm EDTA, and protease inhibitor mixture) by nitrogen cavitation using a Parr bomb. The nuclei were removed by centrifugation at 500 × g, and the membranes were then separated from soluble proteins by centrifugation of the 500 × g supernatant at 100,000 × g for 1 h. Reducing Laemmli sample buffer was added to the supernatant (soluble) and membrane fractions, and the samples were boiled and resolved on 10% SDS-polyacrylamide gels containing 4 m urea in the resolving gel. The gels were Western blotted with anti-Gαi1/2 antiserum (BO84) to detect myristoylated and unmodified Gαi1 (25).
Trypsin Protection Assays
Gα trypsin protection assays were performed as described with minor modifications (10, 26–29). Gα subunits (2.5 μm each) were incubated with 100 μm GDP in HEDL buffer (20 mm HEPES-KOH, pH 8.0, 1 mm EDTA, 0.05% m/v deionized polyoxyethylene 10 lauryl ether (C12E10)) alone or in HEDL buffer containing 30 μm AlCl3, 50 mm MgCl2, 10 mm NaF on ice for 30 min. Gα subunits were then incubated for 10 or 30 min with the following concentrations of trypsin that had been pretreated with 25 ng/ml l-1-p-tosylamino-2-phenylethyl-chloromethyl ketone; Gαq, 0.1% (m/v) (22 °C); Gα13, 0.25% (m/v) (22 °C); Gαi1, 0.25% (m/v) (30 °C); and Gαs short and Gαolf, 0.5% (m/v) (30 °C). The reactions were quenched by the addition of 40 μg/ml lima bean trypsin inhibitor and reducing SDS-PAGE Laemmli sample buffer. The samples were boiled and resolved by SDS-PAGE, and the protein fragments were visualized by Coomassie Blue staining.
GTPγS Binding Assays
Intrinsic and Ric-8-assisted GTPγS binding assays were performed as reported previously (13, 30). Purified untagged Ric-8A or Ric-8BFL (200 nm) were mixed with Gα (100 nm) in 20 mm HEPES-KOH, pH 8.0, 100 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA, 10 mm MgCl2, 0.05% (m/v) deionized polyoxyethylene (10) lauryl ether, C12E10 (Gαi1, Gαs short, and Gα13) or 0.05% (m/v) Genapol C-100 (Calbiochem) (Gαq and Gαolf), and 10 μm [35S]GTPγS (specific activity, 20,000 cpm/pmol). Duplicate aliquots were taken from the reactions at specific time points, quenched in 20 mm Tris, pH 7.7, 100 mm NaCl, 10 mm MgCl2, 1 mm GTP, and 0.08% (m/v) deionized polyoxyethylene 10 lauryl ether C12E10, and filtered onto BA-85 nitrocellulose filters (GE Healthcare). The filters were washed with 20 mm Tris, pH 7.7, 100 mm NaCl, 2 mm MgCl2, dried, and subjected to scintillation counting. To quantify the amount of GTPγS-binding proteins present in the HiTrap Q Gαq column eluate fractions, 400 nm purified Ric-8A was mixed with each fraction, and the assay was performed for 30 min at 30 °C.
Phospholipase Cβ Assays
Phospholipid vesicles were prepared as described previously so that the final reaction (60 μl) contained 200 μm phosphatidylethanolamine and 50 μm [inositol-2-3H(N)]-phosphatidylinositol 4,5-bisphosphate (PIP2) at 6–8000 cpm/assay (31). PLCβ2 or PLCβ3 were added at 10 ng/assay. Gαq was diluted in buffer containing 20 mm HEPES-KOH, pH 7.2, 100 mm NaCl, 1 mm DTT, 2 mm MgCl2, 0.5 mm EDTA, 1 μm GDP, and 0.15% (m/v) β-octylglucoside (final assay concentration). To activate Gαq, Gαq was diluted in the same buffer but with 10 mm NaF and 30 μm AlCl3. The PLC reactions were initiated by the addition of 2.8 mm CaCl2 (1 μm free Ca2+), and the samples were incubated at 30 °C. The reactions were terminated by the addition of 200 μl of 10% (m/v) trichloroacetic acid, followed by the addition of 100 μl of 10 mg/ml BSA. Precipitated proteins and lipids were centrifuged, and 300 μl of the inositol trisphosphate-containing supernatant was analyzed by liquid scintillation counting. In all assays, blank solutions corresponding to the storage buffers for each of the proteins were included such that all of the reactions had exactly the same solution components.
Adenylyl Cyclase Assays
Sf9 cells were infected with recombinant adenylyl cyclase (AC) 3, 5, or 6 baculoviruses. The cells were collected 48 h after infection, suspended in lysis buffer (20 mm HEPES-KOH, pH 8.0, 150 mm NaCl, 5 mm EDTA, 1 mm EGTA, 2 mm DTT, protease inhibitor mixture), and lysed by nitrogen cavitation using a Parr bomb. The cell lysate was centrifuged at 500 × g. The supernatant was centrifuged at 70,000 × g for 30 min to isolate total cell membranes. The membranes were washed and homogenized into membrane storage buffer (20 mm HEPES-KOH, pH 8.0, 20% (m/v) sucrose, 1 mm DTT + protease inhibitor mixture) using a Dounce homogenizer with tight pestle. Membrane homogenates were frozen and stored at −80 °C until use. G proteins were loaded with [35S]GTPγS and isolated by gel filtration chromatography as described previously (14). Precisely determined Gα-GTPγS concentrations were measured by scintillation counting of a fixed volume of each gel-filtered, monomeric Gα pool. Forskolin and/or G proteins in ATP regeneration buffer (50 mm HEPES-KOH, pH 8.0, 10 mm MgCl2, 10 mm phosphocreatine, 10 units/ml creatine phosphokinase, 10 μm GTP, 200 μm ATP, 100 μm 3-isobutyl-1-methylxanthine, 100 μm rolipram) were added to 625 ng of membrane homogenate in stimulation buffer (50 mm HEPES-KOH, pH 8.0, 0.05% (m/v) BSA, 100 μm 3-isobutyl-1-methylxanthine, 100 μm rolipram) in 96-well format and incubated for 5 min at 22 °C. Produced cAMP was detected using a PerkinElmer Life Sciences LANCE cAMP detection kit according to the manufacturer's instructions and measured in a Victor 3V (PerkinElmer Life Sciences) plate reader.
RESULTS
Ric-8 Proteins Promote Recombinant Gα Subunit Expression in Cells
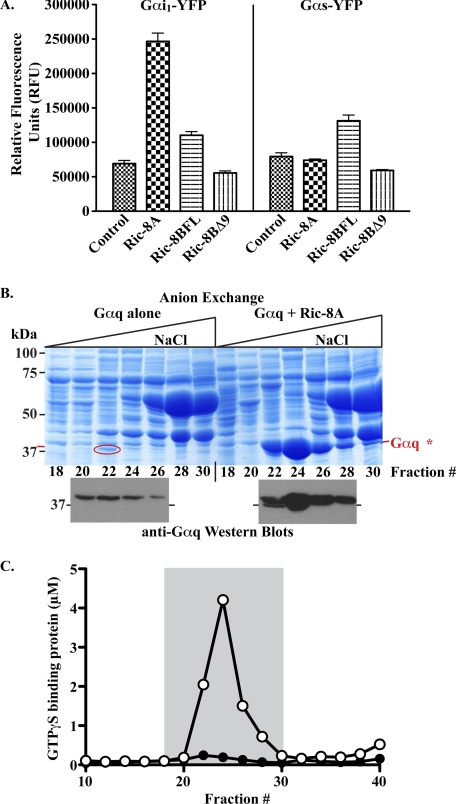
Genetic ablation of Ric-8 genes in various organisms leads to defects in efficient Gα subunit expression (15–19). Ric-8A binds all Gα subunits in vitro except the Gαs class, and Ric-8B preferentially binds Gαs and Gαq (12, 13). We tested whether the expression of Gαs short or Gαi1 was up-regulated by co-overexpression of Ric-8 homologs. Ric-8A or two Ric-8B isoforms were co-transfected in HEK293 cells with YFP-tagged Gαs short or Gαi1 subunits. Fluorescence intensity measurements of intact cells were used (excitation, 485 nm; emission, 535 nm) to quantify the relative amounts of expressed YFP-Gα in each condition of Ric-8 expression. Ric-8BFL specifically potentiated YFP-Gαs expression, whereas Ric-8A and, to a lesser degree, Ric-8BFL potentiated YFP-Gαi expression (Fig. 1A). These results are consistent with the observed Ric-8 binding specificities to Gα subunits, with the exception that Ric-8BΔ9 binds Gαs but did not enhance its expression.
FIGURE 1.
Ric-8 proteins promote recombinant Gα subunit expression. A, HEK293 cells were transfected with 100 ng of pcDNA3.1-Gαi1-YFP or pcDNAI/Amp-Gαs-YFP and 500 ng of pcDNA3.1-Ric8A, Ric8BFL, Ric8BΔ9, or control empty pcDNA3.1. Forty-eight hours post-transfection, relative fluorescence intensity (excitation, 485 nm; emission, 535 nm) was measured and quantified as described under “Experimental Procedures.” B, High Five insect cells were infected with a recombinant Gαq baculovirus or GST-Ric-8A and Gαq baculoviruses. The cells were lysed in CHAPS buffer, and the clarified lysates were chromatographed over a HiTrap Q anion exchange column. The column was eluted with a linear NaCl gradient, and the eluates were fractionated. Equal portions of the Gαq-containing fractions were resolved by SDS-PAGE in duplicate. One gel was stained with Coomassie Brilliant Blue, and the other was Western blotted with a Gαq/11-specific antiserum. The position of Gαq on the Coomassie gel is indicated by a red line and is circled in the non-Ric-8A experiment. C, HiTrap Q column fractions (1 ml each) from the Gαq plus GST-Ric-8A (○) or Gαq alone (●) expression experiments were assayed to determine the concentration of protein present capable of binding GTPγS using the nitrocellulose filter binding assay. Purified, untagged Ric-8A (400 nm) was supplemented in the assayed aliquots of each fraction to promote evaluation of stoichiometric Gαq GTPγS binding. The portion of the graph with a gray background denotes the same range of fractions analyzed by SDS-PAGE in B.
The insect cell protein expression system is the method of choice for purification of G protein subunits resistant to expression in E. coli (8–11). Purification of many insect cell-expressed Gα subunits (Gαq and Gα12/13 families) is laborious and results in low yield of final product. We know of no example in which Gαolf has been purified by this method successfully. Because Ric-8 proteins promoted Gα subunit expression in mammalian cells, we tested whether they could also potentiate recombinant Gα subunit expression in insect cells for the eventual purpose of using this system to develop an enhanced method of Gα subunit purification. High Five insect cells were infected with untagged Gαq, or Gαq and GST-Ric-8A recombinant baculoviruses. Whole detergent lysates of pelleted cells were prepared, clarified, and chromatographed over HiTrap Q anion exchange columns. The columns were washed and eluted with a linear NaCl gradient. Consecutive fractions of the column eluates that contained Gαq were resolved by SDS-PAGE and Coomassie stained or Western blotted with an anti-Gαq/11 antibody. Co-expression of GST-Ric-8A with Gαq dramatically potentiated the amount of Gαq recovered from the column ∼25-fold in comparison with the condition where GST-Ric-8A was not expressed (Fig. 1B). The Gαq obtained from this one-step purification procedure was ≥50% pure and was tested functionally in respect to its capacity to bind GTPγS in a Ric-8A-dependent manner. An equal portion of each HiTrap Q column eluate fraction was supplemented with purified Ric-8A (400 nm) and allowed to bind radiolabeled GTPγS for 30 min at 25 °C. The amount of active G protein present in each fraction was determined by quantifying the amount of protein-bound nucleotide using a nitrocellulose filter binding method. The peak Gαq-containing fractions (1 ml each) from the GST-Ric-8A and Gαq or Gαq alone experiments as judged by the Coomassie gels and Western blots also contained the highest levels of protein-bound GTPγS (4.2 and 0.15 μm active G protein, respectively) (Fig. 1C). The peak fraction from the GST-Ric-8A and Gαq experiment did not bind appreciable GTPγS without purified Ric-8A supplementation, further showing that the GTP-binding protein is recombinant Gαq, because Gαq does not bind appreciable GTPγS in solution in the absence of Ric-8A (data not shown).
Surprisingly, endogenous insect cell G protein expression (Gαi, Gαq, and Gβ) was actually reduced by GST-Ric-8A or GST-Ric-8BFL but not GST expression (supplemental Fig. 3S). The mechanism of this reduction is not understood but could be a consequence of the supersaturating levels of GST-Ric-8 overexpression (in comparison with endogenous G protein expression) achieved from baculovirus vectors. Nonetheless, reduction of endogenous G protein subunit expression was a positive attribute to the system for the recombinant Gα subunit purification scheme.
GST-Ric-8 Purification of Gα Subunits
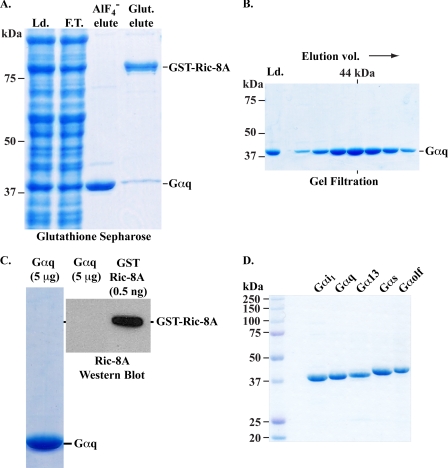
To determine whether co-expression and co-purification of GST-Ric-8 proteins with Gα subunits could be used as a method to isolate highly pure Gα subunits, High Five insect cells were co-infected with Gαq and GST-tagged Ric-8A baculoviruses, and GST-Ric-8A·Gαq complexes were isolated from detergent whole cell lysates over a gravity-driven glutathione-Sepharose 4B resin column (GE Healthcare). The column was washed and treated with a CHAPS-detergent buffer that contained GDP, AlF4−, and MgCl2 to elute Gαq. Mg-GDP-AlF4 mimics the Gα transition state during GTP hydrolysis and induces an activated conformational state of Gα that has greatly reduced affinity for either Gβγ or Ric-8A (13, 32). GST-Ric-8A was then eluted from the resin with reduced glutathione. Fig. 2A shows that the majority of the Gαq eluted with the Mg-GDP-AlF4 buffer, whereas GST-Ric-8A and some residual Gαq eluted with reduced glutathione. The Gαq was estimated by Image J (version 10.2) line profile analysis of the Coomassie-stained SDS gel to be ∼86% pure (Fig. 2A and Table 1). Similar results were obtained from High Five cell purification experiments when GST-Ric-8A was co-expressed with Gαi1 or Gα13, and GST-Ric-8B was co-expressed with Gαs or Gαolf (Table 1). When a native (detergent-free) GST-Ric-8A purification of Gαq was performed from the soluble fraction of insect cells co-expressing GST-Ric-8A and Gαq using identical lysis buffer that lacked detergent, approximately one-half the yield of Gαq was obtained in comparison with the whole cell detergent extraction procedure presented in Fig. 2 (not shown). This indicates that a substantial portion of functional Gα resides in the cytosol of insect cells co-expressing GST-Ric-8 proteins.
FIGURE 2.
Purification of Gα subunits by GST-Ric-8 association. A, GST-Ric-8A and Gαq were co-expressed in High Five insect cells from recombinant baculoviruses. A cell lysate (column load, Ld.) was prepared and adsorbed to a glutathione-Sepharose column (column flow through, F.T.). The column was washed, and Gαq was eluted with buffer that contained AlF4− (AlF4− elute). GST-Ric-8A was eluted with reduced glutathione (Glut. elute). The proteins (∼5 μg of each sample) were resolved by SDS-PAGE and visualized with Coomassie Blue. B, the Gαq (AlF4− eluate) was gel-filtered over Superdex 75 and 200 columns arranged in tandem. Proteins (Gαq) present in the fractionated Superdex eluate were visualized by Coomassie Blue-stained SDS-PAGE. The volume at which a 44-kDa size standard eluted in a calibration run is indicated. C, gel-filtered Gαq (5 μg) was resolved in duplicate SDS gel lanes alongside purified GST-Ric-8A (0.5 ng). One Gαq gel lane was stained with Coomassie Blue, and the other, plus the GST-Ric-8A standard were Western blotted with an anti-Ric-8A antiserum. D, Gαi1, Gαq, and Gα13 were isolated using the Ric-8A co-purification method, and Gαs short and Gαolf were isolated using the Ric-8B co-purification method. Gα subunits were desalted and purified using Superdex gel filtration chromatography, and ∼2.0 μg were resolved by SDS-PAGE. The gel was stained with Coomassie Blue. The positions of molecular mass markers are indicated.
TABLE 1.
Purified G protein α subunits
| Gα | Ric-8 co-purification |
Gel filtration |
βγ-Co-purification (Yield/liter) | GTPγS binding |
|||
|---|---|---|---|---|---|---|---|
| Yield/liter | Estimated purity | Yield/liter | Estimated Purity | Stoichiometry | Rate | ||
| mg | % | mg | % | μg | mol GTPγS/mol Gα | min−1 | |
| Gαq | 8.1 | 86.0 | 2.5 | 96.6 | 125a | 0.75d | 0.068d |
| Gα13 | 4.3 | 81.6 | 2.5 | 84.8 | 100a | 0.35d | 0.077d |
| Gαi1 | 12.0 | 89.6 | 4.8 | 95.2 | 500–750a | 0.51 | 0.055 |
| Gαs short | 25.1 | 93.1 | 6.0 | 97.4 | 525b | 0.68 | 0.094 |
| Gαolf | 8.5 | 85.8 | 4.2 | 87.1 | NAc | 0.63 | 0.078 |
In two eukaryotic systems (insect and HEK cells), Ric-8 proteins potentiated Gα subunit expression. Combined evidence from the experiments in Figs. 1 and 2 indicate that Ric-8 proteins work predominantly in a stoichiometric fashion to promote Gα overexpression, although a small portion of overexpressed Gα seemed to be free from GST-Ric-8. Yields close to a 1:1 molar ratio of GST-Ric-8A (∼12–18 mg) to Gαq (∼8 mg) were recovered from glutathione-Sepharose, as typified by the experiment shown in Fig. 2A. After anion exchange chromatographic resolution of the GST-Ric-8A- and Gαq-expressing insect whole cell lysate, a peak of ∼700 μg of active Gαq eluted at ∼75 mm NaCl as determined by GTPγS binding (Fig. 1C). The Gαq in this peak did not contain (bound) GST-Ric-8A, because no Coomassie-stained GST-Ric-8A band (97 kDa) could be discerned that approached the level of Gαq (Fig. 1B), and the peak fraction did not bind GTPγS without Ric-8A supplementation (not shown). When pure GST-Ric-8A and Gαq were mixed and chromatographed over the anion exchange column, a formed GST-Ric-8A·Gαq complex remained intact and eluted at a distinctly higher ionic strength (∼220–240 mm NaCl) than free Gαq (∼60–80 mm NaCl) (supplemental Fig. 2S). This shows that the GST-Ric-8A·Gαq complex remains intact when bound and eluted from the anion exchange column and demonstrates that the portion of Gαq isolated from the whole cell lysate was free from GST-Ric-8A in the cell, or conceivably, could have dissociated from GST-Ric-8A during lysis and/or chromatography.
To use the purified Gα subunits in downstream applications, it was necessary to remove the AlF4− and high concentration of MgCl2 from each preparation. If purity ≥80% was sufficient, each Gα preparation could be processed most simply by passage through gravity driven PD-10 (GE Healthcare) desalting columns (not shown, but described under “Experimental Procedures”). Gα subunits could be enriched further with concomitant removal of MgCl2/AlF4− by concentration in centrifugal ultrafiltration devices and gel filtration over precalibrated Superdex 75 and 200 10/300 GL columns hooked in tandem (GE Healthcare). In Fig. 2B, Gαq was gel-filtered, and the eluate from the Superdex columns was fractionated. Proteins present in the fractions were visualized by Coomassie-stained SDS gel. The Gαq eluted from the Superdex columns at a volume coincident with a 44-kDa sizing standard, indicating that the preparation was mono-disperse. Superdex gel filtration increased the purity of each Gα preparation (Table 1). To test whether GST-Ric-8A was a contaminant in the Gαq preparation, 5 μg of Gαq was resolved by SDS-PAGE alongside 0.5 ng of purified GST-Ric-8A and Western blotted with a Ric-8A polyclonal antiserum (33). No GST-Ric-8A was detected in the Gαq preparation (Fig. 2C). No GST-Ric-8B was detected in the Gαs short preparation, and <0.5% mol/mol GST-Ric-8 proteins were detected by quantitative Western blotting of the purified Gαi1, Gαolf, and Gα13 (not shown).
Final Gα subunit purity after GST-Ric-8 co-purification and Superdex chromatography was shown by resolving ∼2.0 μg of each preparation on a Coomassie-stained SDS gel (Fig. 2D). Purity was quantified by performing an Image J line profile analysis (Table 1). In each instance, Gα subunit purity was found to be as good or better than that obtained using the Gβγ co-purification method (8–11). The Gαq and Gαs short preparations appeared nearly homogenous, whereas minor contaminants were present in the Gαi1, Gα13, and Gαolf preparations. Each Gα preparation and prepared High Five insect cell membranes (50 μg) were then analyzed by Western blot analyses to determine whether contaminating endogenous insect cell G proteins were present. Endogenous insect cell Gαi and a potential Gαs-like protein were detected with P960 antiserum (34). Insect Gβ was detected with B600 antiserum (34), and a Gαq-like protein was detected with the Gαq/11 antibody, C19 (Santa Cruz). In supplemental Fig. 4S, no insect cell Gαi-, Gβ-, or Gαs-like proteins were detected when 100 ng of the GST-Ric-8-purified Gαi1, Gαq, Gα13, Gαs short, or Gαolf were analyzed by Western blot. The C-19 Gαq/11 antibody (Santa Cruz) was highly selective for Gαq but weakly cross-reacted with Gαs short and Gαolf. The presence of a doublet band in C-19 Western blots of the Gαs short and Gαolf preparations raised the possibility that insect cell Gαq (lower band of the doublet) was a trace contaminant in these preparations (supplemental Fig. 4S). No doublet was detected in the Gαs short and Gαolf preparations when blotted with P960 (supplemental Fig. 4S).
The most significant result from the GST-Ric-8 Gα co-purification procedure was that the yield of Gα obtained from each preparation was unprecedented. When 1 liter of High Five insect culture expressing GST-Ric-8A and Gαq was processed, ∼2.5 mg of Gαq was purified. Table 1 compares the final yields of each Gα prepared by GST-Ric-8 co-purification versus yields reportedly obtained using Gβγ co-purification methods (compare columns 4–6) (8–11). The yields were increased ∼20-fold for Gαq, ∼25-fold for Gα13, ∼8-fold for Gαi1, and ∼11-fold for Gαs short. Gαolf has not been purified successfully by any method, in significant quantity, yet here the yield was quite high (4.2 mg).
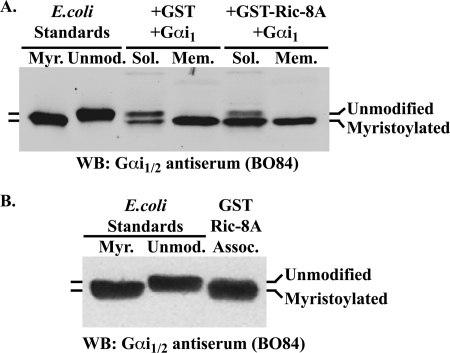
Post-translational covalent lipid attachment to Gα subunits influences Gα functional interactions with binding protein partners and the membrane. Gαi class members are modified permanently by myristoylation, and all Gα classes are modified by reversible palmitoylation (35, 36). The influence of Ric-8A on expressed Gαi1 myristoylation and subcellular fractionation was assessed by SDS-PAGE and Western blot analysis. Myristate attachment increases the apparent mobility of Gαi1 through SDS-PAGE (7, 36). High Five cells were infected with Gαi1 and GST or with GST-Ric-8A baculoviruses. The infected cells were lysed in a native buffer and subjected to crude subcellular fractionation by centrifugation of the post-nuclear supernatant at 100,000 × g. Soluble and membrane attached proteins were processed in reducing SDS Laemmli sample buffer, resolved by 4 m urea SDS-PAGE alongside myristoylated and unmodified Gαi1 standards prepared in E. coli, and Western blotted with anti-Gαi1/2 specific antiserum (BO84) (6, 7, 25, 34). Fig. 3A shows that approximately equal portions of membrane-attached, myristoylated Gαi1 were expressed in insect cells co-expressing GST or GST-Ric-8A. However, the amount of soluble, myristoylated Gαi1 was greatly enhanced by GST-Ric-8A co-expression. The Gαi1 prepared by the GST-Ric-8A co-purification method co-migrated exclusively with the E. coli myristoylated Gαi1 standard, indicating that the preparation is likely myristoylated completely (Fig. 3B). Measurements of G protein palmitoylation status are not as forthcoming and cannot be assessed by simple SDS-PAGE analysis. Assessment of the palmitoylation status of Gα subunits purified by Ric-8 co-purification will answer a significant question about the functionality of the prepared Gα subunits and may also provide important insight regarding the stage(s) at which Ric-8 proteins regulate Gα overexpression in cells.
FIGURE 3.
Ric-8A enhances soluble myristoylated Gαi1 levels, and Gαi1 purified by GST-Ric-8A association appears to be myristoylated fully. A, High Five insect cells were infected with recombinant Gαi1 and GST or GST-Ric-8A baculoviruses. The cells were lysed in detergent-free lysis buffer and sequentially centrifuged at 500 × g to remove nuclei, and at 100,000 × g to separate soluble (Sol.) from crude membrane fractions (Mem.). Subcellular fractions were processed in reducing SDS Laemmli sample buffer, and the protein concentrations of each were quantified by Amido Black protein assay. Subcellular fractions (10 μg each) and myristoylated Gαi1 and unmodified Gαi1 purified standards (7.5 ng each) produced in E. coli were resolved on a 10% SDS-polyacrylamide gel containing 4 m urea. Gαi1 proteins were detected by Western blot (WB) using Gαi1/2 antiserum (BO84). B, myristoylated (Myr.) and unmodified (Unmod.) Gαi1 E. coli standards (10 ng each) were resolved by SDS-PAGE alongside 10 ng of GST-Ric-8A-co-purified Gαi1. The proteins were transferred to nitrocellulose and Western blotted with BO84 antiserum. Myristoylated, E. coli-produced Gαi1, and GST-Ric-8A-co-purified Gαi1 had faster apparent mobilities through SDS-PAGE than unmodified Gαi1.
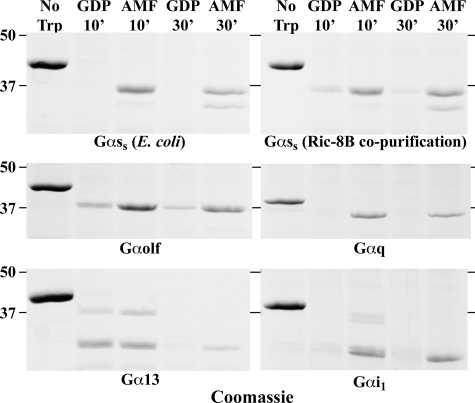
One test of G protein functionality is the ability to become resistant to limited trypsinolysis after adopting the active GTP- or Mg-GDP-AlF4-bound conformations (26, 28). Preparations of Gα subunits not capable of achieving trypsin resistance when activated are generally considered inactive. Each Gα prepared by GST-Ric-8 co-purification or a Gαs short standard prepared from E. coli were treated with GDP or GDP and AlF4− and incubated with trypsin for 10 and 30 min. The trypsinization reactions were quenched with trypsin inhibitor, and the denatured G proteins and G protein fragments were resolved by SDS-PAGE and visualized by Coomassie staining. All of the Gα subunit preparations displayed AlF4−-specific trypsin resistance (Fig. 4). The Gα13 preparation was only partially resistant under the conditions used. These data show that the Gα subunits prepared by GST-Ric-8 co-purification are active and capable of switching between the active and inactive conformations.
FIGURE 4.
Purified Gα subunits are resistant to trypsin digestion when activated with Mg·GDP·AlF4. GST-Ric-8 co-purified Gα subunits and Gαs purified from E. coli (2.5 μm each) were incubated with GDP or Mg·GDP·AlF4 and subjected to limited trypsin digestion for 10 or 30 min. The trypsinization reactions were stopped by the addition of trypsin inhibitor and Gα proteins, and tryptic fragments were resolved by reducing SDS-PAGE. The gels were stained with Coomassie Blue to visualize proteins (shown as gray scale).
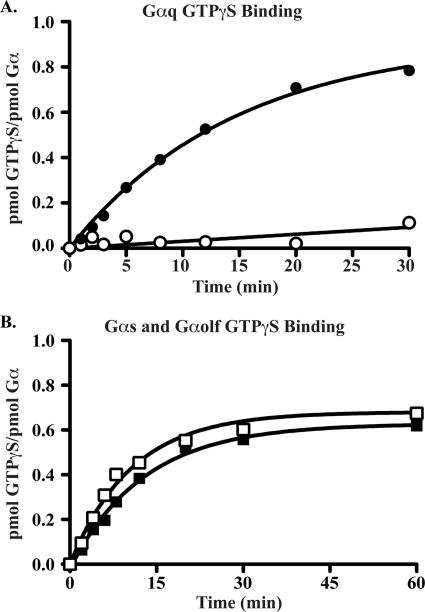
Functional G proteins bind GTP with high stoichiometry and at defined rates. The kinetics and end point stoichiometries of GTPγS binding of each Gα preparation were determined. Gα subunits (100 nm) were incubated in timed reactions containing 10 μm radiolabeled GTPγS. The amount of GTPγS that was bound to each Gα over time was quantified using a nitrocellulose filter binding method (13, 30). Gαq and Gα13 do not bind GTPγS readily in solution, so these rates were measured in the presence of Ric-8A catalyst (Ric-8A-assisted GTPγS binding). Gαq achieved a final GTPγS binding stoichiometry of 0.75 mol/mol in the presence of Ric-8A but did not bind GTPγS appreciably in its absence (Fig. 5A and Table 1). These results are in-line with reported results of Ric-8A-assisted Gαq GTPγS binding using Gαq purified with the Gβγ affinity method (13). Ric-8A-co-purified Gαi1 and Gα13 GTPγS binding rates were also consistent with those reported (Table 1) (11, 13, 37). The GTPγS binding rate for Gαolf has never been reported and was found to be slightly slower (0.078 min−1) than the GTPγS binding rate of Ric-8B-co-purified Gαs short (0.094 min−1) (Fig. 5B and Table 1). Nitrocellulose filter binding assay measurements of final (Ymax) GTPγS binding stoichiometries of Ric-8-co-purified Gα subunits were within a range (0.51–0.75 mol/mol) typically observed for Gα subunits prepared from E. coli or from insect cells by Gβγ affinity purification (Table 1). The only exception was that the Gα13 preparation bound a lower amount of GTPγS (0.35 mol/mol) when assisted by Ric-8A.
FIGURE 5.
Purified Gαq, Gαolf, and Gαs bind GTPγS. The kinetics of GTPγS binding to Gαq (100 nm) in the absence (○) or presence (●) of Ric-8A (A, 200 nm) and Gαs (□) or Gαolf (■) (B, 100 nm each) were measured at 25 °C. The reactions were initiated by the addition of G protein to 10 μm [35S]GTPγS (specific activity, 20,000 cpm/pmol). Aliquots were withdrawn from the reactions at the indicated times, quenched, and filtered onto nitrocellulose. The filters were washed, dried, and subjected to scintillation counting to quantify the amount of G protein-bound GTPγS. The reactions were performed in duplicate and are representative of three independent experiments.
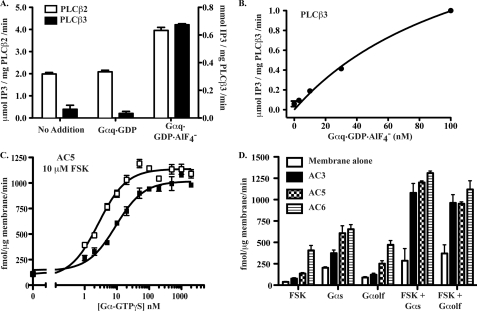
Ric-8-co-purified Gαq, Gαs, and Gαolf were tested for the abilities to stimulate the effector enzymes phospholipase Cβ and adenylyl cyclase isoforms, respectively, using in vitro reconstitution assays. Gαq was treated with GDP or Mg-GDP-AlF4, and stimulation of purified PLCβ2 and PLCβ3 inositol trisphosphate production from prepared lipid vesicles was measured using an established assay (31). The activated form of Gαq (Mg-GDP-AlF4-bound) but not Gαq-GDP stimulated PLCβ2 and PLCβ3 activities 2- and 22-fold, respectively (Fig. 6A). Dose-dependent activation of PLCβ3 by activated-Gαq was also determined (Fig. 6B). The potency of the Gαq preparation was found to be consistent with previously established values (38).
FIGURE 6.
Ric-8-co-purified Gαq, Gαolf, and Gαs stimulate effector enzymes. A, PLCβ2 (□) and PLCβ3 (■) inositol trisphosphate (IP3) release from PIP2-containing lipid vesicles was measured in response to 0 (no addition) or 50 nm Gαq·GDP or Gαq·Mg·GDP·AlF4. B, PLCβ3 PIP2 hydrolysis activity (●) was measured in response to increasing doses of Gαq·Mg·GDP·AlF4 (0–100 nm). All of the PLCβ assay results are representative of at least two independent experiments that contained two or three replicates/assay. C, forskolin-enhanced (10 μm) adenylyl cyclase 5 activity was measured in response to increasing doses of Gαs short-GTPγS (□) or Gαolf-GTPγS (■) (0–2 μm). All of the adenylyl cyclase assays were conducted in triplicate. cAMP levels were quantified using the PerkinElmer Life Sciences Lance cAMP detection system and Victor3 plate reader. The data were fit to sigmoidal dose response functions using Graph Pad prism 5.0. D, adenylyl cyclase activity assays of control and AC3-, AC5-, and AC6-expressing insect cell membranes were conducted in the presence of Gαs-GTPγS or Gαolf-GTPγS (1 μm each) and/or 10 μm forskolin in 5-min reactions at 22 °C.
A direct comparison of Gαs short and Gαolf stimulation of adenylyl cyclase using purified proteins has not been possible because of the inability to purify active Gαolf. To compare these activities, it was necessary to prepare with precision known active concentrations of both G proteins bound to GTPγS. Purified Gαs short and Gαolf were loaded to completion with [35S]GTPγS and gel-filtered through Superdex columns to remove unbound nucleotide. The concentrations of active monomeric Gα-GTPγS were quantified by scintillation counting. AC isoforms 3, 5, and 6 were expressed from recombinant baculoviruses in Sf9 cells. Membranes were prepared from these and control cells. G proteins and/or forskolin were incubated with the membrane preparations for 5 min, and the amount of cAMP produced was measured using a LANCE cAMP detection kit (PerkinElmer Life Sciences). A titration of Gαs short-GTPγS and Gαolf-GTPγS activation of AC5 was first performed in the presence of a low concentration of forskolin (10 μm). Gαs short was more potent than Gαolf at activating AC5 (EC50 values of ∼1.9 nm versus ∼4.7 nm) (Fig. 6C). Gαs short also activated AC5 with higher efficacy than Gαolf. Saturating concentrations of Gαolf-GTPγS stimulated AC5 production of cAMP to a maximum value that was only 88% that of the Gαs short-GTPγS stimulated value (∼1017 versus ∼1149 fmol·μg−1·min−1).
Next, we explored the possibility that Gαs short and Gαolf might have preferences for activation of different adenylyl cyclase isoforms. Gαolf and AC3 are required for olfaction (39, 40). Gαolf may preferentially activate AC3 (in comparison with Gαs short), or more simply, Gαolf and AC3 work together because they are co-expressed in olfactory tissues. Insect cell membranes that expressed AC3 had the lowest measurable forskolin-stimulated cAMP production in comparison with AC5 or AC6 membranes, but the AC3 activity level was more than double that of control, non-AC-expressing membranes. In assays with each AC isoform, inclusion of a saturating amount of Gαs short-GTPγS (1 μm) consistently resulted in greater cAMP accumulation when compared with the levels achieved by the addition of saturating Gαolf-GTPγS (1 μm) (Fig. 6D). Very little AC isoform activity was observed when GDP-bound G proteins were used in equivalent assays (data not shown). Gαs short and Gαolf also displayed the characteristic synergism with forskolin, when a low concentration (10 μm) of this second site allosteric AC activator was included in the assays. With each AC isoform, more forskolin-stimulated activity was observed in the presence of Gαs short-GTPγS versus Gαolf-GTPγS. Because precisely controlled concentrations of active G proteins were included in these assays, we conclude that Gαs short indiscriminately activates AC isoforms with higher efficacy than Gαolf in vitro.
DISCUSSION
We introduce a method of G protein α subunit preparation that substantially improves upon established methods both in its ease, yield of G protein obtained, and applicability to all four Gα subunit classes. The method involves the co-expression in insect cells and subsequent co-purification of GST-tagged Ric-8 nonreceptor guanine nucleotide exchange factors and the Gα subunit of interest. The GST-Ric-8·Gα complex is isolated from whole cell detergent lysates with glutathione-Sepharose resin. Gα is dissociated from GST-Ric-8 with GDP-AlF4− and thereby eluted from the resin with high purity (>85%). The Gα subunits are then gel-filtered to remove the excess AlF4− with modest enrichment. The produced Gα subunits were functional and bound GTPγS and stimulated effector enzymes appropriately. We anticipate that this will become the method of choice for Gα subunit production because the procedure can be conducted easily in any facility with the capability to culture insect cells.
The tried and true method of Gα subunit purification involved co-expression of Gα, Gβ, and His6-tagged Gγ in insect cells and isolation and extraction of cell membranes with detergent. G protein trimers were isolated from the detergent extract with nickel chelate resin, and Gα was eluted specifically from the His6-Gβγ resin with GDP-AlF4− and polished by subsequent ion exchange chromatographies (8–11). The yields of Gα subunits obtained using the present GST-Ric-8-based method exceeded Gβγ co-purification 8–25-fold depending on Gα species and allowed the first production of active Gαolf. Increased Gα yield was likely a contribution of at least two parameters of the GST-Ric-8-insect cell expression/purification system: 1) Ric-8 and Gα protein co-expression increased overall Gα subunit expression levels by an unknown mechanism as shown in both insect and HEK293 cells (Fig. 1); and 2) the material loaded on the glutathione-Sepharose column was whole insect cell detergent extracts, so this included Gα present in both the soluble and membrane fractions.
What could be the mechanism by which Ric-8 proteins potentiate Gα subunit expression? Genetic ablation or RNAi reduction of Ric-8 homologs in worms, flies, and mammalian cells resulted in reduction of Gα expression (15–19). These data combined with our Ric-8A and Ric-8B overexpression data and that of others suggest a positive role for Ric-8 proteins in mediating some aspect of G protein biosynthesis, defined as the complete process from Gα translation to stable plasma membrane residence (18, 20). Because intracellular G protein trimer assembly is a requirement for efficient Gα and Gβγ expression and trafficking to the plasma membrane (41, 42), we sought to test whether Ric-8 potentiated recombinant Gβγ expression in insect cells. In supplemental Fig. 5S, GST-Ric-8A or GST were co-expressed with hexahistidine-tagged-Gαi1, Gβ1, and Gγ2. Gβ1γ2 dimers were then purified from insect cell membranes using the established method (9). Similar degrees of Gβ1γ2 purity were obtained from both preparations. Equivalent yields were also obtained (0.6 and 0.61 mg of Gβ1γ2/liter of culture), but slightly more product was obtained when GST-Ric-8A was co-expressed (16% more than GST expression) when calculated as a function of membrane input into the purification procedure. This slight enhancement of Gβ1γ2 yield per mass of membrane input did not mark the same degree to which GST-Ric-8 potentiated Gα expression and recovery (up 25-fold). Because Gαi myristoylation precedes intracellular Gi trimer formation, it is enticing to speculate that the modest increase in Gβ1γ2 recovery resulted from the presence of more myristoylated Gαi1 subunit in the GST-Ric-8A-expressing cells. It will be tempting to explore the possibility that GST-Ric-8 isoforms may selectively aid Gβγ dimer combination co-purification with Gα subunits that are otherwise difficult to express in insect cells when compared with Gαi (i.e. Gαq, Gα13, and Gαolf).
Approximately 50% of the functional overproduced Gαq was present in the cytosol of insect cells co-overexpressing GST-Ric-8A. GST-Ric-8 proteins are mostly cytosolic, whereas functional endogenously expressed G proteins are viewed to reside predominantly on the plasma membrane with only a small subfraction present in the cytosol. Studies have shown that a population of some G proteins becomes released from the plasma membrane into a soluble fraction upon GPCR agonist treatment (42–45). Perhaps a function of Ric-8 is to bind the Gα released into the cytosol and recycle it back to the plasma membrane. Systems with intentionally perturbed Ric-8 expression would lack this Ric-8-dependent plasma membrane Gα replacement activity and result in a slow leach of Gα from the plasma membrane, eventually leading to the observed dramatic reductions in steady-state Gα expression. In this capacity, it is somewhat difficult to imagine how Ric-8 overexpression could promote the magnitude of Gα overexpression observed in the insect cells, if it were not for the large cytosolic Gα pool. Membrane binding sites for Gα could become saturated at the levels of Gα overexpression observed.
This study prompts further investigation into the mechanism of Ric-8-induced Gα subunit overexpression. The combined chromatography experiments of Figs. 1 and 2 and supplemental Fig. 2S indicate that the bulk of overexpressed GST-Ric-8A and Gαq are likely bound to each other in the cell and that ∼15% of the overexpressed Gαq was free from GST-Ric-8A. It will be interesting to determine precisely whether endogenously expressed (levels of) Ric-8 work to promote Gα subunit expression processively or in a stoichiometric manner. This knowledge would provide insight to discriminate whether Ric-8 proteins exert a positive role upon Gα biosynthesis or a protective role in Gα subunit degradation.
Purification of Gαolf with GST-Ric-8B allowed primary characterization of this G protein. Seifert and co-workers (46) previously compared Gαolf and Gαs short biochemical properties using Gα-βAR fusion proteins expressed in isolated insect cell membranes. The Gαolf-βAR fusion had reduced GDP affinity and correspondingly faster basal and hormone-stimulated GTPγS binding rates when compared with the equivalent Gαs short-βAR fusion protein. In the detergent solution-based GTPγS binding kinetic assays that we conducted, purified Gαolf was found to bind GTPγS at a rate slightly slower than the Gαs short produced with GST-Ric-8B (Fig. 5). We speculate that the discrepancy between these observations may be attributed to either the membrane environment in which the βAR-Gα fusion protein assays were conducted, including the presence of Gβγ, or were due to different ways that the fused βAR allosterically influenced Gαolf versus Gαs.
Gαolf has also been analyzed for its capacity to stimulate adenylyl cyclases by assay of membranes deficient in Gαs (Cyc-) made to express Gαolf or with membranes that expressed the βAR-Gαolf fusion protein (46, 47). Both studies concluded that Gαolf activated adenylyl cyclases with less efficacy than Gαs. However, it could not be ascertained whether one contribution to the reduced efficacy was a consequence of the nature of the direct interaction between Gαolf-GTP and AC enzyme or was due to a combination of differences in Gα expression, Gα guanine nucleotide occupancy or turnover, and/or efficiency of G protein/receptor coupling. By using purified activated Gαs short and Gαolf of known concentration, we were able to demonstrate that at least one factor that explains in part the observed decreased efficacy of Gαolf for AC activation is due to the way that Gαolf-GTP interacts with AC enzymes. Gαolf is a less efficacious and potent direct activator of adenylyl cyclase when compared with equivalently produced Gαs short-GTP. The degree to which Gαolf is less efficacious likely does not account fully for the magnitude of observed differences in the agonist stimulation assays.
One proposal for the existence of an olfactory specific AC stimulatory protein is that unique signaling requirements for olfaction pressured evolution of Gαolf and that Gαolf may regulate a unique adenylyl cyclase isoform in olfactory tissues. AC3 is highly expressed in olfactory tissues and is a target of odorant receptor/Golf signaling. We show here that Gαolf is actually less efficacious at activating AC3 than Gαs short, because saturating concentrations of Gαs short activated AC3 better than Gαolf did. This shows directly that Gαolf is not a better or preferential activator of AC3 in comparison with Gαs short and suggests that features of the olfactory signaling system (such as controlled detection of odorant threshold) require a less potent or efficacious AC activator to facilitate the physiology of olfaction.
Supplementary Material
Acknowledgments
We thank Dr. Catherine H. Berlot (Geisinger Health System, Danville, PA) for the pcDNAI-Gαs-YFP expression construct and Carmen Dessauer (University of Texas Health Science Center at Houston Medical School, Houston, TX) for adenylyl cyclases 3, 5, and 6 isoform baculoviruses.
This work was supported, in whole or in part, by National Institutes of Health Grants GM088242 (to G. G. T.), GM053536 (to A. V. S.), GM086510 (to J. B. B.), and NS24821 and DA025896 (to S. M. L.). This work was also supported by New York State Stem Cell Science Grant C024307 (to G. G. T.) and National Institute on Drug Abuse Grant T32 DA07232 (to P. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1S–5S.
- GPCR
- G protein-coupled receptor
- AC
- adenylyl cyclase
- βAR
- β-adrenergic receptor
- Gαs short
- G protein αs short isoform
- GTPγS
- guanosine 5′-[γ-thio]triphosphate
- PLCβ
- phospholipase Cβ
- YFP
- yellow fluorescent protein
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- PIP2
- [inositol-2-3H(N)]-phosphatidylinositol 4,5-bisphosphate
- PIP2
- phosphatidylinositol 4,5-bisphosphate.
REFERENCES
- 1. Gilman A. G. (1987) Annu. Rev. Biochem. 56, 615–649 [DOI] [PubMed] [Google Scholar]
- 2. Sprang S. R. (1997) Annu. Rev. Biochem. 66, 639–678 [DOI] [PubMed] [Google Scholar]
- 3. Blumer J. B., Cismowski M. J., Sato M., Lanier S. M. (2005) Trends Pharmacol. Sci. 26, 470–476 [DOI] [PubMed] [Google Scholar]
- 4. Siderovski D. P., Willard F. S. (2005) Int. J. Biol. Sci. 1, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilkie T. M., Kinch L. (2005) Curr. Biol. 15, R843–R954 [DOI] [PubMed] [Google Scholar]
- 6. Lee E., Linder M. E., Gilman A. G. (1994) Methods Enzymol. 237, 146–164 [DOI] [PubMed] [Google Scholar]
- 7. Linder M. E., Ewald D. A., Miller R. J., Gilman A. G. (1990) J. Biol. Chem. 265, 8243–8251 [PubMed] [Google Scholar]
- 8. Hepler J. R., Kozasa T., Gilman A. G. (1994) Methods Enzymol. 237, 191–212 [DOI] [PubMed] [Google Scholar]
- 9. Kozasa T. (1999) Purification of Recombinant G Protein α and βγ Subunits from Sf9 Cells, pp. 23–38, CRC Press, Baco Raton, FL [Google Scholar]
- 10. Kozasa T., Gilman A. G. (1995) J. Biol. Chem. 270, 1734–1741 [DOI] [PubMed] [Google Scholar]
- 11. Singer W. D., Miller R. T., Sternweis P. C. (1994) J. Biol. Chem. 269, 19796–19802 [PubMed] [Google Scholar]
- 12. Klattenhoff C., Montecino M., Soto X., Guzmán L., Romo X., García M. A., Mellstrom B., Naranjo J. R., Hinrichs M. V., Olate J. (2003) J. Cell. Physiol. 195, 151–157 [DOI] [PubMed] [Google Scholar]
- 13. Tall G. G., Krumins A. M., Gilman A. G. (2003) J. Biol. Chem. 278, 8356–8362 [DOI] [PubMed] [Google Scholar]
- 14. Tall G. G., Gilman A. G. (2004) Purification and Functional Analysis of Ric-8A: A Guanine Nucleotide Exchange Factor for G-Protein α Subunits Methods in Enzymology (Siderovski D. P. ed) pp. 377–388, Volume 390, Academic Press; [DOI] [PubMed] [Google Scholar]
- 15. Afshar K., Willard F. S., Colombo K., Siderovski D. P., Gönczy P. (2005) Development 132, 4449–4459 [DOI] [PubMed] [Google Scholar]
- 16. David N. B., Martin C. A., Segalen M., Rosenfeld F., Schweisguth F., Bellaiche Y. (2005) Nat. Cell Biol. 7, 1083–1090 [DOI] [PubMed] [Google Scholar]
- 17. Hampoelz B., Hoeller O., Bowman S. K., Dunican D., Knoblich J. A. (2005) Nat. Cell Biol. 7, 1099–1105 [DOI] [PubMed] [Google Scholar]
- 18. Nagai Y., Nishimura A., Tago K., Mizuno N., Itoh H. (2010) J. Biol. Chem. 285, 11114–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H., Ng K. H., Qian H., Siderovski D. P., Chia W., Yu F. (2005) Nat. Cell Biol. 7, 1091–1098 [DOI] [PubMed] [Google Scholar]
- 20. Kerr D. S., Von Dannecker L. E., Davalos M., Michaloski J. S., Malnic B. (2008) Mol. Cell. Neurosci. 38, 341–348 [DOI] [PubMed] [Google Scholar]
- 21. Oner S. S., Maher E. M., Breton B., Bouvier M., Blumer J. B. (2010) J. Biol. Chem. 285, 20588–20594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibson S. K., Gilman A. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hein P., Rochais F., Hoffmann C., Dorsch S., Nikolaev V. O., Engelhardt S., Berlot C. H., Lohse M. J., Bünemann M. (2006) J. Biol. Chem. 281, 33345–33351 [DOI] [PubMed] [Google Scholar]
- 24. Biddlecome G. H., Berstein G., Ross E. M. (1996) J. Biol. Chem. 271, 7888–8007; Correction (1996) J. Biol. Chem.271, 33705 [DOI] [PubMed] [Google Scholar]
- 25. Mumby S. M., Kahn R. A., Manning D. R., Gilman A. G. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fung B. K., Nash C. R. (1983) J. Biol. Chem. 258, 10503–10510 [PubMed] [Google Scholar]
- 27. Halliday K. R., Stein P. J., Chernoff N., Wheeler G. L., Bitensky M. W. (1984) J. Biol. Chem. 259, 516–525 [PubMed] [Google Scholar]
- 28. Hudson T. H., Roeber J. F., Johnson G. L. (1981) J. Biol. Chem. 256, 1459–1465 [PubMed] [Google Scholar]
- 29. Winslow J. W., Van Amsterdam J. R., Neer E. J. (1986) J. Biol. Chem. 261, 7571–7579 [PubMed] [Google Scholar]
- 30. Sternweis P. C., Robishaw J. D. (1984) J. Biol. Chem. 259, 13806–13813 [PubMed] [Google Scholar]
- 31. Lehmann D. M., Yuan C., Smrcka A. V. (2007) Methods Enzymol. 434, 29–48 [DOI] [PubMed] [Google Scholar]
- 32. Coleman D. E., Berghuis A. M., Lee E., Linder M. E., Gilman A. G., Sprang S. R. (1994) Science 265, 1405–1412 [DOI] [PubMed] [Google Scholar]
- 33. Malik S., Ghosh M., Bonacci T. M., Tall G. G., Smrcka A. V. (2005) Mol. Pharmacol. 68, 129–136 [DOI] [PubMed] [Google Scholar]
- 34. Mumby S. M., Gilman A. G. (1991) Methods Enzymol. 195, 215–233 [DOI] [PubMed] [Google Scholar]
- 35. Linder M. E., Middleton P., Hepler J. R., Taussig R., Gilman A. G., Mumby S. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3675–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mumby S. M., Heukeroth R. O., Gordon J. I., Gilman A. G. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tall G. G., Gilman A. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16584–16589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smrcka A. V., Sternweis P. C. (1993) J. Biol. Chem. 268, 9667–9674 [PubMed] [Google Scholar]
- 39. Belluscio L., Gold G. H., Nemes A., Axel R. (1998) Neuron 20, 69–81 [DOI] [PubMed] [Google Scholar]
- 40. Wong S. T., Trinh K., Hacker B., Chan G. C., Lowe G., Gaggar A., Xia Z., Gold G. H., Storm D. R. (2000) Neuron 27, 487–497 [DOI] [PubMed] [Google Scholar]
- 41. Dupré D. J., Robitaille M., Rebois R. V., Hébert T. E. (2009) Annu. Rev. Pharmacol. Toxicol. 49, 31–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marrari Y., Crouthamel M., Irannejad R., Wedegaertner P. B. (2007) Biochemistry 46, 7665–7677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Allen J. A., Yu J. Z., Donati R. J., Rasenick M. M. (2005) Mol. Pharmacol. 67, 1493–1504 [DOI] [PubMed] [Google Scholar]
- 44. Svoboda P., Kvapil P., Insel P. A., Ransnäs L. A. (1992) Eur. J. Biochem. 208, 693–698 [DOI] [PubMed] [Google Scholar]
- 45. Svoboda P., Milligan G. (1994) Eur. J. Biochem. 224, 455–462 [DOI] [PubMed] [Google Scholar]
- 46. Liu H. Y., Wenzel-Seifert K., Seifert R. (2001) J. Neurochem. 78, 325–338 [DOI] [PubMed] [Google Scholar]
- 47. Jones D. T., Masters S. B., Bourne H. R., Reed R. R. (1990) J. Biol. Chem. 265, 2671–2676 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.