Abstract
HES1 (hairy and enhancer of split) is a transcription factor that regulates osteoblastogenesis in vitro. The skeletal effects of HES1 misexpression were studied. Transgenic mice where a 3.6-kilobase fragment of the collagen type 1 α1 promoter directs HES1 overexpression were created. Transgenics were osteopenic due to decreased osteoblast function in female and increased bone resorption in male mice. HES1 impaired osteoblastogenesis in vitro, and transgenic osteoblasts enhanced the resorptive activity of co-cultured osteoclast precursors. Mice homozygous for a Hes1 loxP-targeted allele were bred to transgenics, where the paired-related homeobox gene enhancer or the osteocalcin promoter direct Cre recombinase expression to inactivate Hes1 in the limb bud or in osteoblasts. To avoid genetic compensation, Hes1 was inactivated in the context of the global deletion of Hes3 and Hes5. Hes3 and Hes5 null mice had no skeletal phenotype. Hes1 inactivation in the limb bud increased femoral length and trabecular number. Hes1 inactivation in osteoblasts increased trabecular bone volume, number, and connectivity due to increased mineral apposition rate and suppressed bone resorption. Hes1 inactivation in vitro increased alkaline phosphatase expression and suppressed the resorptive activity of co-cultured osteoclast precursors. In conclusion, by inhibiting osteoblast function and inducing bone resorption, HES1 is an intracellular determinant of bone mass and structure.
Keywords: Bone, Cell Differentiation, Gene Knock-out, Helix-Loop-Helix Transcription Factors, Notch Pathway, Bone Formation, Bone Resorption, Osteoblast, Osteoclast, Trabecular Microarchitecture
Introduction
HES is a family of evolutionary conserved basic helix-loop-helix transcription factors that comprises seven members, termed HES1 through HES7 (1–4). HES proteins are homologues of Drosophila Hes and were first identified as targets of canonical Notch signaling. Notch signaling is activated following cell to cell contact interactions and is critical for developmental processes and the regulation of tissue renewal and maintenance (5–9). HES proteins generally act as repressors of transcription (3). HES1, -3, and -5 have partially overlapping functions, are involved in binary cell fate decisions, and maintain precursor multipotent cells in an undifferentiated state in several tissues during development and adult life (3, 10–13). HES1 is necessary for neural development, and its global inactivation results in early embryonic or perinatal lethality (14).
Bone remodeling is a process carried out in discrete multicellular units, where osteoclasts resorb bone and osteoblasts form new bone in a coordinated fashion to renew skeletal tissue (15). Osteoblasts are derived from multipotent bone marrow mesenchymal stem cells, whereas osteoclasts are derived from multipotent hematopoietic cells (16). Bone marrow mesenchymal stem cells can differentiate toward the osteoblastic, chondrocytic, and adipocytic lineages (17). The fate of mesenchymal cells and their differentiation into osteoblasts is controlled by a signaling network that includes bone morphogenetic proteins, Wnt and Notch (18–23). Notch signaling inhibits osteoblast differentiation and causes osteopenia in vivo, but even though HES1 is a target of Notch, it does not recapitulate all of the effects of Notch (24). HES1 participates in cell fate determination of mesenchymal multipotent cells and blocks the initial differentiation of preadipocytes, although it is necessary for their terminal differentiation (25, 26). HES1 suppresses the expression of osteocalcin and induces the transcription of osteopontin (27, 28). In addition, HES1 interacts with RUNX2 (runt-related transcription factor 2) prolonging its half-life, and, as a consequence, enhances the differentiation of murine osteoblastic cells (28–31). However, the function of HES1 in skeletal tissue has not been established.
Osteoclasts are multinucleated cells that form through the aggregation of bone marrow mononuclear cell precursors. Osteoclast formation requires RANKL (receptor activator of NF-κB ligand), a membrane-bound ligand of RANK, and its activity is opposed by the soluble decoy receptor osteoprotegerin (32, 33). RANKL and osteoprotegerin are expressed by bone marrow stromal cells and osteoblasts, and their ratio regulates osteoclastogenesis. Notch signaling inhibits the maturation of bone marrow osteoclast precursors by inducing the expression of osteoprotegerin (21, 34, 35). However, the role of HES1 in osteoclastogenesis is not known.
The intent of this study was to define the function of HES1 on skeletal development and postnatal bone remodeling. For this purpose, transgenic mice overexpressing HES1 under the control of a 3.6-kb rat collagen type 1 α1 (Col1a1) promoter fragment were created. In addition, a conditional deletion approach to ablate Hes1 in the skeleton and avoid the lethality of the global Hes1 inactivation was employed (14). Mice, where Hes1 sequence coding exons were flanked by loxP sequences (Hes1loxP/loxP), were bred to transgenics expressing the Cre recombinase under the control of the paired-related homeobox 1 (Prx1) enhancer (Prx1-Cre) or the osteocalcin promoter (Oc-Cre), to inactivate Hes1 in the limb bud or in mature osteoblasts, respectively. Hes1 conditional null mice were created in a Hes3 and Hes5 null background (Hes3−/−Hes5−/−), to avoid a possible phenotypic compensation following the inactivation of Hes1. The skeletal phenotype of mice misexpressing HES1 was studied by histomorphometric and structural analysis of the femur. To understand the mechanisms of HES1 action in bone, the differentiation and function of skeletal cells misexpressing HES1 were examined in vitro.
EXPERIMENTAL PROCEDURES
Hes1 Transgenic Mice
After introduction of a Kozak consensus sequence upstream of the translation initiation codon, a 1.5-kb cDNA fragment coding for murine HES1 was cloned downstream of the 3.6-kb fragment of the rat Col1a1 promoter and upstream of the bovine growth hormone polyadenylation signal (36). Microinjection of linearized DNA into pronuclei of fertilized oocytes from Friend leukemia virus strain B (FVB) inbred mice (Charles River Laboratories, Wilmington, MA), and transfer of microinjected embryos into pseudopregnant mice were carried out at the Gene Targeting and Transgenic Facility of the University of Connecticut Health Center (Farmington, CT). Positive founders were identified by Southern blot analysis of tail DNA and bred to wild type FVB mice to create transgenic lines (37). Heterozygous transgenic mice were mated to wild type FVB mice to generate heterozygous HES1 transgenic mice and wild type littermate controls for the experiments described. Presence of the Hes1 transgene was documented by PCR in tail DNA (Table 4).
TABLE 4.
Femoral histomorphometry and bone microarchitecture of 1-, 3-, and 6-month-old Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− (Hes1/3/5 null) male mice and Hes1loxP/loxP;Hes3−/−;Hes5−/− (Control) controls
Femoral histomorphometry and microcomputed tomography were performed on femurs from 1-, 3-, and 6-month-old Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− male mice and Hes1loxP/loxP;Hes3−/−;Hes5−/− male littermate controls. Values are means ± S.E.; n = 5–12.
| Males | 1-month-old |
3-month-old |
6-month-old |
|||
|---|---|---|---|---|---|---|
| Control | Hes1/3/5 null | Control | Hes1/3/5 null | Control | Hes1/3/5 null | |
| Femoral histomorphometry | ||||||
| Bone volume/tissue volume (%) | 9.5 ± 2.0 | 16.7 ± 2.2a | 18.2 ± 1.3 | 25.1 ± 0.9a | 16.4 ± 1.2 | 27.9 ± 4.0a |
| Trabecular separation (μm) | 282 ± 98 | 142 ± 26 | 135 ± 8 | 97 ± 5a | 175 ± 9 | 115 ± 18a |
| Trabecular no. (mm−1) | 4.1 ± 0.8 | 6.5 ± 0.9 | 6.1 ± 0.3 | 7.8 ± 0.3a | 4.9 ± 0.2 | 6.8 ± 0.6a |
| Trabecular thickness (μm) | 22.9 ± 0.7 | 25.8 ± 0.6a | 29.9 ± 1.1 | 32.5 ± 1.0 | 33.5 ± 1.8 | 40.1 ± 4.0 |
| Osteoblast surface/bone surface (%) | 14.8 ± 1.7 | 18.7 ± 4.0 | 6.5 ± 0.4 | 6.3 ± 0.8 | 9.2 ± 0.6 | 8.3 ± 1.4 |
| No. of osteoblasts/bone perimeter (mm−1) | 13.8 ± 1.5 | 17.0 ± 3.9 | 6.7 ± 0.4 | 6.5 ± 0.7 | 9.7 ± 0.7 | 8.9 ± 1.4 |
| Osteoid surface/bone surface (%) | 1.88 ± 0.40 | 1.08 ± 0.33 | 0.17 ± 0.07 | 0.20 ± 0.07 | 0.16 ± 0.05 | 0.07 ± 0.03 |
| Osteoclast surface/bone surface (%) | 12.0 ± 1.0 | 10.0 ± 0.6 | 7.5 ± 0.6 | 7.2 ± 0.5 | 9.4 ± 0.5 | 8.5 ± 0.6 |
| No. of osteoclasts/bone perimeter (mm−1) | 5.8 ± 0.5 | 4.6 ± 0.3a | 3.4 ± 0.2 | 3.2 ± 0.2 | 4.0 ± 0.2 | 3.7 ± 0.2 |
| Eroded surface/bone surface (%) | 25.0 ± 1.9 | 20.6 ± 1.4 | 14.6 ± 1.1 | 13.6 ± 0.8 | 16.3 ± 0.9 | 15.3 ± 0.9 |
| Mineral apposition rate (μm day−1) | 1.44 ± 0.09 | 1.95 ± 0.10a | 0.62 ± 0.04 | 0.50 ± 0.04a | 0.50 ± 0.06 | 0.49 ± 0.03 |
| Mineralizing surface/bone surface (%) | 3.1 ± 0.2 | 2.4 ± 0.5 | 4.6 ± 1.0 | 2.8 ± 0.6 | 5.0 ± 1.5 | 5.1 ± 1.3 |
| Bone formation rate (μm2 μm−3 day−1) | 0.045 ± 0.006 | 0.043 ± 0.006 | 0.031 ± 0.007 | 0.014 ± 0.004a | 0.030 ± 0.010 | 0.026 ± 0.007 |
| Microcomputed tomography | ||||||
| Bone volume fraction (%) | 10.8 ± 1.9 | 16.7 ± 1.8a | 16.5 ± 0.9 | 26.0 ± 1.5a | 14.6 ± 2.8 | 26.9 ± 1.8a |
| Trabecular separation (μm) | 341 ± 67 | 195 ± 16 | 176 ± 8 | 137 ± 6a | 222 ± 17 | 155 ± 7a |
| Trabecular no. (mm−1) | 3.6 ± 0.6 | 5.3 ± 0.4a | 5.4 ± 0.2 | 6.5 ± 0.2a | 4.4 ± 0.4 | 5.8 ± 0.2a |
| Trabecular thickness (μm) | 40 ± 1 | 41 ± 1 | 43 ± 1 | 46 ± 2 | 50 ± 2 | 53 ± 3 |
| Connectivity density (mm−3) | 200 ± 49 | 361 ± 45a | 247 ± 22 | 385 ± 30a | 149 ± 25 | 273 ± 14a |
| Structure model index | 1.87 ± 0.05 | 1.73 ± 0.14 | 1.7 ± 0.1 | 0.9 ± 0.2a | 1.97 ± 0.18 | 0.80 ± 0.29a |
| Cortical thickness (μm) | 118 ± 6 | 115 ± 4 | 223 ± 11 | 212 ± 4 | 229 ± 13 | 188 ± 11a |
a Significantly different from wild type controls, p < 0.05.
Hes1 Conditional Null Mice
Mice where the Hes1 sequence comprised between exons 2 and 4 was flanked by loxP sequences and deletion mutant mice for Hes3 and Hes5, created in a CD1 genetic background, were obtained from R. Kageyama (Kyoto University, Kyoto, Japan) (38–40). Hes1loxP/loxP mice were bred to Hes3−/−;Hes5−/− to create Hes1loxP/loxP;Hes3−/−;Hes5−/− mice. To study the consequences of the inactivation of Hes1 during early limb development, Prx1-Cre mice created in a C57BL/6 genetic background, were obtained from The Jackson Laboratory (Bar Harbor, ME) (41). To study the inactivation of Hes1 in mature osteoblasts, transgenic mice expressing the Cre recombinase under the control of a 3.9 kb human osteocalcin promoter, created in an FVB genetic background, were obtained from T. Clemens (Baltimore, MD) (42). Transgenics expressing Cre were bred to Hes1loxP/loxP;Hes3−/−;Hes5−/− mice to create heterozygous Prx1-Cre/+;Hes1loxP/+;Hes3+/−;Hes5+/− or Oc-Cre/+;Hes1loxP/+;Hes3+/−;Hes5+/− mice. These were mated with Hes1loxP/loxP;Hes3−/−;Hes5−/− to create Prx1-Cre/+;Hes1loxP/loxP;Hes3−/−;Hes5−/− and Oc-Cre/+;Hes1loxP/loxP;Hes3−/−;Hes5−/−. These mice were mated with Hes1loxP/loxP;Hes3−/−;Hes5−/− to generate an experimental cohort, in which Cre excises the loxP-flanked sequences from the Hes1loxP allele (Hes1Δ/Δ;Hes3−/−;Hes5−/−) and a control littermate group (Hes1loxP/loxP;Hes3−/−;Hes5−/−). Male and female conditional null mice were compared with littermate controls of the same sex at 15 and 18 days post coitum, at birth, and at 1, 3, and 6 months of age, and their skeletal phenotype was analyzed. To ensure the validity of Hes1loxP/loxP;Hes3−/−;Hes5−/− mice as controls, the skeletal phenotype of Hes3−/−;Hes5−/− mice was compared with that of wild type littermates, and the phenotype of Hes1loxP/loxP;Hes3−/−;Hes5−/− mice was compared with that of Hes3−/−;Hes5−/− littermates at 1 month of age. Detection of Hes1loxP, Hes3−, and Hes5− alleles was carried out by PCR in tail DNA extracts in newborns and adult mice (Table 1). Excision of loxP-flanked sequences by Cre recombinase was documented by PCR in DNA extracted from calvariae of 1-, 3-, and 6-month-old mice, using specific primers (Table 1). All animal experiments were approved by the Animal Care and Use Committee of Saint Francis Hospital and Medical Center.
TABLE 1.
Primers used for genotyping by PCR and for mRNA level determination by real-time RT-PCR
[FAM] indicates the position of the guanine labeled with the fluorophore.
X-ray Analysis, BMD, and Femoral Length
X-Rays were performed on eviscerated mice at an intensity of 30 kW for 20 s on a Faxitron X-Ray system (model MX 20, Faxitron X-Ray Corp., Wheeling, IL). Total bone mineral density (BMD2; g cm−2) was measured on anesthetized mice using the PIXImus small animal DEXA system (GE Medical System/LUNAR, Madison, WI) (43). Femoral images were used to determine femoral length (mm). Calibrations were performed with a phantom of defined value, and quality assurance measurements were performed before each use. The coefficient of variation for total BMD is <1%.
Bone Histomorphometric Analysis
Static and dynamic histomorphometry of femurs was carried out after injection with 20 mg/kg calcein and 50 mg/kg demeclocycline, at an interval of 2 days for 1-month-old mice and 7 days for 3- and 6-month-old mice. Animals were sacrificed by CO2 inhalation 2 days after the demeclocycline injection. Femoral longitudinal sections, 5-μm thick, were cut on a microtome (Microm, Richards-Allan Scientific, Kalamazoo, MI) and stained with 0.1% toluidine blue or von Kossa. Static parameters of bone formation and resorption were measured in a defined area between 360 and 2160 μm from the growth plate, using an OsteoMeasure morphometry system (Osteometrics, Atlanta, GA) (44). For dynamic histomorphometry, mineralizing surface per bone surface and mineral apposition rate were measured on unstained sections under ultraviolet light, using a triple diamino-2-phenylindole fluorescein set long pass filter, and bone formation rate was calculated. The terminology and units used are those recommended by the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research (45).
Microcomputed Tomography
Femurs were scanned using a Microcomputed Tomographic Instrument (μCT 40, Scanco Medical AG, Bassersdorf, Switzerland), calibrated weekly using a phantom provided by the manufacturer. Femurs were scanned in 70% ethanol at an energy level of 55 peak kilo voltage, intensity of 145 μA, and integration time of 200 ms. Trabecular bone volume and microarchitecture were evaluated starting ∼1.2 mm proximal to the femoral condyles. A total of 160 consecutive slices acquired at an isotropic voxel size of 6 μm3 and a slice thickness of 6 μm, were chosen for analysis. Contours were manually drawn every 10 slices a few voxels away from the endocortical boundary to define the region of interest for analysis. The contours of the remaining slices were iterated automatically. Trabecular regions were assessed for bone volume fraction, trabecular thickness, number and separation, connectivity density, and structural model index, using a Gaussian filter (σ = 0.8, support = 1) and a user-defined threshold (46). A total of 100 slices for the cortical region were measured at the mid-diaphysis of each femur with an isotropic voxel size of 6 μm3 and a slice thickness of 6 μm. For mid-diaphysis analysis, contours were iterated across the 100 slices along the cortical shell, excluding the bone marrow cavity. Analysis for cortical thickness was performed using a Gaussian filter (σ = 0.8, support = 1) and a user-defined threshold (46).
Osteoblastic Cell Cultures
Primary osteoblasts were isolated from parietal bones of 3- to 5-day-old mice by sequential collagenase digestion, as described (47). Cells were cultured in DMEM (Invitrogen) supplemented with nonessential amino acids, 20 mm HEPES, 100 μg/ml ascorbic acid, and 10% FBS (Atlanta Biologicals, Norcross, GA) at 37 °C in a humidified 5% CO2 incubator. Primary bone marrow stromal cells were recovered by centrifugation of femurs that were aseptically removed from 4-week-old HES1 transgenic and littermate control mice, as described (44). Cells were cultured in α-minimum essential medium (Invitrogen) containing 15% FBS at 37 °C in a humidified 5% CO2 incubator.
Osteoblast-Splenocyte Co-cultures and Pit Formation Assay
Primary osteoblasts were seeded on BioCoat discs (BD Biosciences), and after reaching confluence, cultured in the presence of 100 μg/ml ascorbic acid and 5 mm β-glycerophosphate. Primary splenocytes were harvested from spleens aseptically removed from 5- to 8-week-old wild type FVB mice, and 1 × 106 cells/cm2 were seeded on the layer of primary osteoblasts in the presence of 10 nm 1,25-dihydroxyvitamin D3 (BIOMOL Intl., Plymouth Meeting, PA) or PBS, as control (48–50). Splenocytes and primary osteoblasts were cultured for 7 days; the cells were removed with bleach for 5 min, and BioCoat discs were stained with von Kossa. Discs were photographed on a white light background, and the digital image was analyzed with Adobe Photoshop (Adobe Systems, Inc., San Jose, CA). Brightness and contrast were maximized, the file was inverted to yield the negative image, and the area of resorption was calculated as the percent of pixels contained in half of the grayscale, determined by using the histogram function of Adobe Photoshop.
Real-time RT-PCR
Total RNA was extracted from cultures of primary osteoblastic cells, and mRNA levels were determined by real-time RT-PCR (51, 52). For this purpose, 1–3 μg of total RNA were reverse-transcribed using the SuperScript III Platinum Two-step Quantitative RT-PCR kit (Invitrogen), according to the manufacturer's instructions and amplified in the presence of specific primers (Table 1) and Platinum Quantitative PCR SuperMix-UDG (Invitrogen) at 60 °C for 30 cycles. The copy number was estimated by comparison with a standard curve constructed using alkaline phosphatase and HES1 (both from ATCC, Manassas, VA) and osteocalcin (from J. B. Lian, University of Massachusetts, Worcester, MA) cDNAs and corrected for ribosomal protein L38 (Rpl38; ATCC) expression (53–55). Reactions were conducted in a 96-well spectrofluorometric thermal iCycler (Bio-Rad), and fluorescence was monitored during every PCR cycle at the annealing step. Data are expressed as the copy number corrected for Rpl38.
Statistical Analysis
Data are expressed as means ± S.E. Statistical differences were determined by analysis of variance or Student's t test.
RESULTS
HES1 Overexpression Causes Osteopenia
Two transgenic lines overexpressing HES1 under the control of the 3.6-kb rat Col1a1 promoter were established. Transgenic mice from both lines were osteopenic, and one line was studied in detail. Following heterozygous intermatings, only heterozygous transgenics were born, suggesting intrauterine lethality of homozygous transgenics. Therefore, heterozygous HES1 transgenic mice were used for subsequent analysis. These mice developed normally but did not live beyond 3 months of age; consequently, the analysis of their skeletal phenotype was performed in 1- and 3-month-old animals.
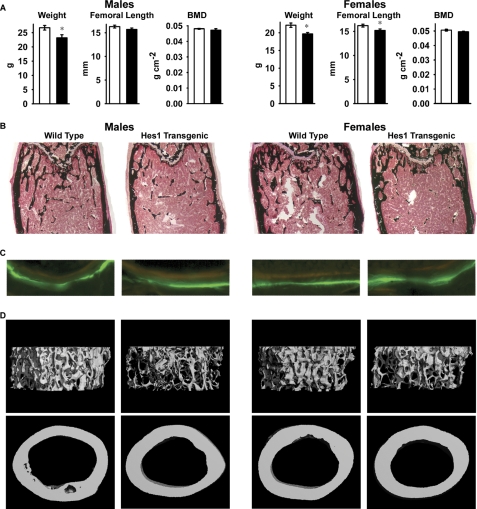
At 1 month of age, HES1 transgenics appeared normal, and no skeletal abnormalities were detected by contact radiography. Similarly, bone histomorphometric and microarchitectural analysis of 1-month-old transgenics did not reveal differences from littermate controls. HES1 transgenic mice at 3 months of age did not exhibit skeletal abnormalities by contact radiography and had normal BMD. However, transgenics of both sexes had significantly lower weight, and female transgenics displayed shortened femoral length (Fig. 1A). Bone histomorphometric analysis revealed a 20–25% decrease in trabecular bone volume, secondary to a reduced number of trabeculae, demonstrating that HES1 overexpression causes osteopenia (Table 2 and Fig. 1B). Although transgenics from both sexes were osteopenic, the mechanisms responsible appeared to be different. In male transgenics, there was a modest increase in the number of osteoclasts, leading to an increase in eroded surface, whereas in female transgenics, there was a decrease in osteoblast number associated with reduced osteoid surface. Bone formation and mineral apposition rate were not affected in either sex (Table 2 and Fig. 1C). Microcomputed tomography confirmed the decrease in trabecular bone volume, number, and increased separation observed by bone histomorphometry and revealed decreased connectivity density in HES1 transgenics of both sexes. Measurement of the structural model index revealed a higher ratio of rod-shaped versus plate-like trabeculae in HES1 transgenics. There were no differences in cortical bone thickness between transgenics and controls (Table 2 and Fig. 1D). These results indicate that overexpression of HES1 directed by a 3.6-kb Col1a1 promoter fragment has a negative effect on trabecular microarchitecture without affecting cortical bone.
FIGURE 1.
Skeletal phenotype of 3-month-old male (left) and female (right) heterozygous HES1 transgenic mice (black bars) and wild type littermate controls (white bars). In A, the weight (g), femoral length (mm), and total BMD (g cm−2) are shown. Values are means ± S.E., n = 6–8. *, Significantly different from control mice, p < 0.05. Shown in B and C are von Kossa staining (B; final magnification, 40×) and calcein/demeclocycline labeling (C; final magnification, 100×) of representative femoral sections. D, microcomputed tomography of representative femurs.
TABLE 2.
Femoral histomorphometry and bone microarchitecture of 3-month-old male and female HES1 transgenic mice and wild type controls
Femoral histomorphometry and microcomputed tomography were performed on femurs from 3-month-old male and female heterozygous HES1 transgenic mice and littermate wild type controls. Values are means ± S.E.; n = 3–8.
| Males |
Females |
|||
|---|---|---|---|---|
| Wild type | HES1 | Wild type | HES1 | |
| Histomorphometry | ||||
| Bone volume/tissue volume (%) | 7.1 ± 0.4 | 5.5 ± 0.6a | 7.9 ± 0.6 | 5.8 ± 0.4a |
| Trabecular separation (μm) | 268 ± 6 | 362 ± 41a | 254 ± 14 | 337 ± 16a |
| Trabecular no. (mm−1) | 3.5 ± 0.1 | 2.8 ± 0.3a | 3.7 ± 0.2 | 2.8 ± 0.1a |
| Trabecular thickness (μm) | 20.3 ± 0.9 | 19.7 ± 0.4 | 21.4 ± 0.9 | 20.2 ± 0.7 |
| Osteoblast surface/bone surface (%) | 11.0 ± 0.8 | 13.6 ± 1.7 | 13.7 ± 1.0 | 8.9 ± 1.3a |
| No. of osteoblasts/bone perimeter (mm−1) | 12.5 ± 0.9 | 15.7 ± 2.0 | 14.8 ± 1.0 | 10.5 ± 1.2a |
| Osteoid surface/bone surface (%) | 0.8 ± 0.3 | 0.7 ± 0.3 | 1.7 ± 0.3 | 0.5 ± 0.2a |
| Osteoclast surface/bone surface (%) | 10.4 ± 0.3 | 11.6 ± 0.3a | 13.3 ± 0.5 | 13.6 ± 0.6 |
| No. of osteoclasts/bone perimeter (mm−1) | 5.3 ± 0.1 | 6.0 ± 0.2a | 6.9 ± 0.3 | 6.9 ± 0.3 |
| Eroded surface/bone surface (%) | 18.5 ± 0.6 | 20.8 ± 0.9a | 23.9 ± 0.9 | 23.5 ± 1.2 |
| Mineral apposition rate (μm day−1) | 0.63 ± 0.03 | 0.61 ± 0.03 | 0.82 ± 0.04 | 0.85 ± 0.06 |
| Mineralizing surface/bone surface (%) | 11.2 ± 0.8 | 12.8 ± 1.0 | 8.9 ± 1.0 | 9.0 ± 1.3 |
| Bone formation rate (μm2 μm−3 day−1) | 0.071 ± 0.006 | 0.077 ± 0.005 | 0.073 ± 0.008 | 0.077 ± 0.013 |
| Microcomputed tomography | ||||
| Bone volume fraction (%) | 11.8 ± 0.7 | 7.6 ± 0.6a | 13.9 ± 1.0 | 10.4 ± 0.8a |
| Trabecular separation (μm) | 199 ± 3 | 242 ± 8a | 215 ± 5 | 241 ± 8a |
| Trabecular no. (mm−1) | 4.9 ± 0.1 | 4.1 ± 0.1a | 4.7 ± 0.1 | 4.2 ± 0.1a |
| Trabecular thickness (μm) | 37 ± 1 | 37 ± 1 | 42 ± 1 | 41 ± 1 |
| Connectivity density (mm−3) | 280 ± 16 | 138 ± 17a | 267 ± 15 | 181 ± 17a |
| Structure model Index | 2.0 ± 0.1 | 2.4 ± 0.1a | 1.6 ± 0.1 | 2.0 ± 0.1a |
| Cortical thickness (μm) | 179 ± 2 | 186 ± 4 | 199 ± 3 | 200 ± 2 |
a Significantly different from wild type controls, p < 0.05.
HES1 Overexpression Impairs Osteoblastogenesis and Induces Osteoclastogenesis in Vitro
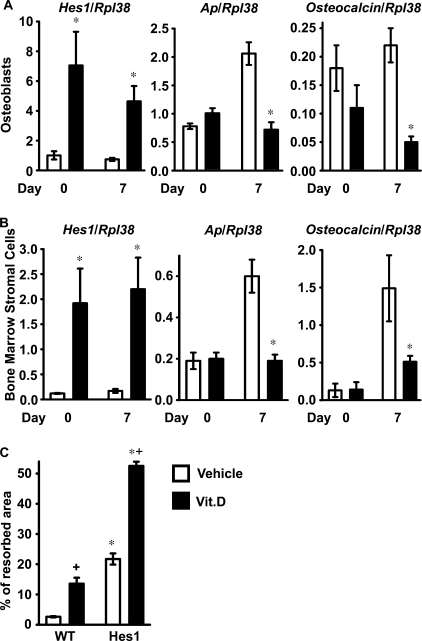
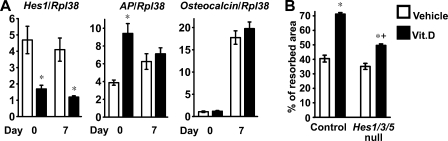
To understand the mechanisms responsible for the effects of HES1 overexpression, calvarial osteoblasts and bone marrow stromal cells were harvested from HES1 transgenics and from wild type littermate controls of both sexes. HES1 transcripts were increased in primary osteoblastic cells from HES1 transgenics throughout the culture period, and calvarial osteoblasts expressed decreased levels of alkaline phosphatase and osteocalcin mRNA after 7 days of culture (Fig. 2A). In accordance with the results obtained in osteoblasts, alkaline phosphatase and osteocalcin mRNA levels were suppressed in transgenic bone marrow stromal cells after 7 days of culture (Fig. 2B). These findings confirm that HES1 overexpression impairs osteoblast differentiation and could explain the decreased number of osteoblasts observed in female HES1 transgenics.
FIGURE 2.
Effects of HES1 overexpression on osteoblastic function in vitro. In A and B, calvarial osteoblasts (A) and bone marrow stromal cells (B) were harvested from HES1 transgenics (black bars) and wild type littermate controls (white bars). Total RNA was extracted at confluence and after 7 days of culture in conditions favoring osteoblastogenesis; mRNA was reverse-transcribed and amplified by real time RT-PCR in the presence of specific primers. Data are expressed as Hes1, alkaline phosphatase (Ap), and osteocalcin copy number, determined by real time RT-PCR, corrected for Rpl38 expression. Values are means ± S.E., n = 4. In C, calvarial osteoblasts harvested from HES1 transgenics (HES1) and wild type littermate controls (WT) were co-cultured with splenocytes harvested from wild type mice, in the presence of 10 nm 1,25-dihydroxyvitamin D3 (Vit.D; black bars) or control vehicle (white bars). Data are expressed as % of resorbed area. Values are means ± S.E., n = 4–6. *, significantly different from wild type cells, p < 0.05. + significantly different from cells treated with vehicle, p < 0.05.
To ascertain whether the overexpression of HES1 in osteoblasts increased osteoclast differentiation or function, calvarial osteoblasts from HES1 transgenics or wild type littermate controls of both sexes were co-cultured with splenocytes from wild type FVB mice, as a source of mononuclear osteoclast precursors. Osteoclast activity was determined by the pit formation assay in a BD BioCoat cell culture system. Treatment with 1,25-dihydroxyvitamin D3 enhanced the resorptive activity of wild type splenocytes (48). In accordance with the increased number of osteoclasts and eroded surface observed in male HES1 transgenics, osteoblasts from HES1 transgenics enhanced resorption in comparison to control osteoblasts. The effect was present whether the cultures were treated or not with 1,25-dihydroxyvitamin D3 (Fig. 2C). These results indicate that HES1 overexpression in osteoblasts increases bone resorption and offer an explanation for the resorptive phenotype observed in male HES1 transgenics.
Controls for the Hes1 Conditional Null Mouse Model
The conditional inactivation of Hes1 was studied in the context of the global ablation of Hes3 and Hes5 to prevent possible compensatory effects from HES3 and HES5 because they are expressed by osteoblasts, and their function partially overlaps with that of HES1 (3, 10, 12). One-month-old Hes3−/−;Hes5−/− mice appeared normal and bone histomorphometry did not reveal differences between Hes3−/−;Hes5−/− mice and wild type sex-matched littermates. To assess whether the presence of loxP sequences caused a skeletal phenotype, 1-month-old Hes1loxP/loxP;Hes3−/−;Hes5−/− mice were compared with Hes3−/−;Hes5−/− sex-matched littermates. Absence of spontaneous recombination of the Hes1loxP allele was documented by PCR in calvarial extracts, and Hes1loxP/loxP;Hes3−/−;Hes5−/− mice were not different from Hes3−/−;Hes5−/− mice when assessed by bone histomorphometry. These results indicate that Hes1loxP/loxP;Hes3−/−;Hes5−/− mice are valid controls for Hes1Δ/Δ;Hes3−/−;Hes5−/− experimental mice.
Inactivation of Hes1 in Limb Bud Mesenchyme Increases Bone Volume
To induce the conditional deletion of Hes1 in the limb bud starting at day 10.5 of embryonic life, Prx1-Cre/+;Hes1loxP/loxP;Hes3−/−;Hes5−/− mice were mated with Hes1loxP/loxP;Hes3−/−;Hes5−/− mice to create Prx1-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− conditional null and Hes1loxP/loxP;Hes3−/−;Hes5−/− littermate controls (41). The skeletal phenotype was investigated in 15- and 18-day-old embryos, newborns, and 1-month-old mice. Skeletal staining of embryos and newborn mice with alcian blue and alizarin red did not reveal obvious differences between Hes1 conditional null mice and controls in cartilage and mineralized tissue (data not shown).
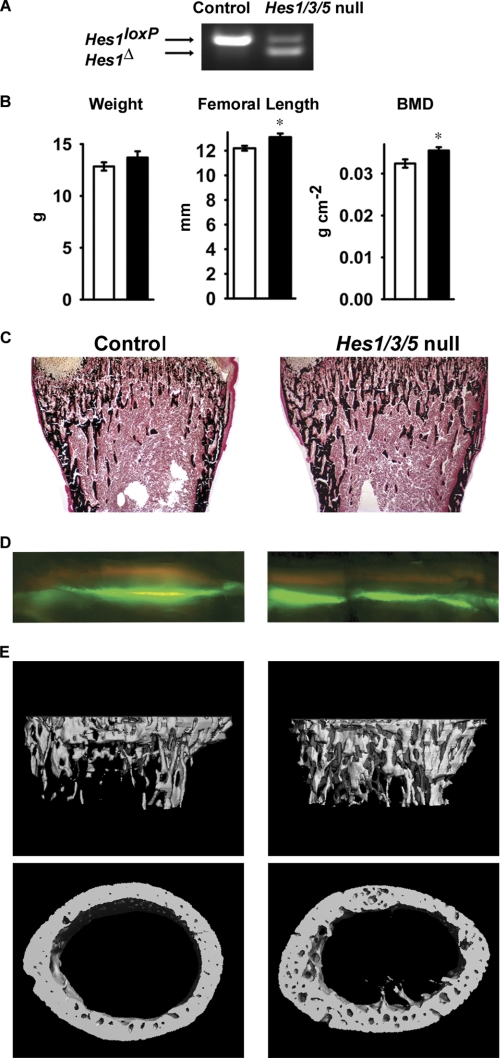
Recombination of Hes1loxP was documented in calvarial DNA extracts from 1-month-old Prx1-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− mice (Fig. 3A). One-month-old conditional null mice appeared normal and contact radiography did not reveal differences when compared with littermate controls (data not shown). Hes1 conditional null male mice exhibited increased femoral length and total BMD, indicating that Hes1 inhibits longitudinal bone growth (Fig. 3B). Bone histomorphometric analysis of 1-month-old male Hes1 conditional null mice revealed a 70% increase in trabecular bone volume that was secondary to an increased number of trabeculae (Table 3 and Fig. 3C). The osteoblast and osteoclast numbers were not changed, and no differences were observed in parameters of osteoblast or osteoclast function, suggesting that Hes1 inactivation in the limb bud does not affect postnatal bone remodeling (Table 3 and Fig. 3D). The results from histomorphometric analysis were confirmed by microcomputed tomography. Prx1-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− male mice exhibited a tendency toward increased bone volume fraction (p < 0.065) and a significant increase in connectivity density and trabecular number coupled to a decrease in trabecular separation, whereas cortical thickness was not affected (Table 3 and Fig. 3E). The phenotype was restricted to male mice, and Hes1Δ/Δ;Hes3−/−;Hes5−/− female mice were not different from sex-matched littermate controls.
FIGURE 3.
Skeletal phenotype of 1-month-old male Prx1-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− conditional null mice (Hes1/3/5 null) and Hes1loxP/loxP;Hes3−/−;Hes5−/− littermate controls (Control) of the same sex. In A, a representative PCR demonstrates the recombination of the Hes1loxP allele in calvarial DNA extracts. In B, the weight (g), femoral length (mm), and total BMD (g cm−2) of Prx1-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− mice (black bars) and controls (white bars) are shown. Values are means ± S.E., n = 5–6. *, significantly different from control mice, p < 0.05. C and D, von Kossa staining (C; final magnification, 40×) and calcein/demeclocycline labeling (D; final magnification, 100×) of representative femoral sections. Shown in E are microcomputed tomography of representative femurs.
TABLE 3.
Femoral histomorphometry and bone microarchitecture of 1-month-old Prx1-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− (Hes1/3/5 null) male mice and Hes1loxP/loxP;Hes3−/−;Hes5−/− controls
Femoral histomorphometry and microcomputed tomography were performed on femurs from 1-month-old Prx1-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− male mice and Hes1loxP/loxP;Hes3−/−;Hes5−/− male littermate controls. Values are means ± S.E.; n = 4–6.
| Males | Control | Hes1/3/5 null |
|---|---|---|
| Histomorphometry | ||
| Bone volume/tissue volume (%) | 11.0 ± 1.3 | 18.8 ± 2.5a |
| Trabecular separation (μm) | 199 ± 19 | 132 ± 25 |
| Trabecular no. (mm−1) | 4.6 ± 0.4 | 6.6 ± 0.7a |
| Trabecular thickness (μm) | 23.5 ± 1.0 | 27.3 ± 1.5 |
| Osteoblast surface/bone surface (%) | 27.7 ± 1.9 | 30.8 ± 2.3 |
| No. of osteoblasts/bone perimeter (mm−1) | 27.1 ± 1.3 | 30.8 ± 2.6 |
| Osteoid surface/bone surface (%) | 4.6 ± 0.7 | 3.5 ± 0.6 |
| Osteoclast surface/bone surface (%) | 9.4 ± 1.1 | 9.6 ± 0.6 |
| No. of osteoclasts/bone perimeter (mm−1) | 4.5 ± 0.6 | 4.4 ± 0.3 |
| Eroded surface/bone surface (%) | 20.3 ± 2.6 | 20.4 ± 1.2 |
| Mineral apposition rate (μm day−1) | 3.00 ± 0.23 | 2.67 ± 0.36 |
| Mineralizing surface/bone surface (%) | 1.2 ± 0.3 | 1.2 ± 0.3 |
| Bone formation rate (μm2 μm−3 day−1) | 0.038 ± 0.011 | 0.030 ± 0.001 |
| Microcomputed tomography | ||
| Bone volume fraction (%) | 9.7 ± 1.7 | 17.0 ± 2.5 |
| Trabecular separation (μm) | 286 ± 12 | 179 ± 26a |
| Trabecular no. (mm−1) | 3.6 ± 0.2 | 5.8 ± 0.7a |
| Trabecular thickness (μm) | 40 ± 1 | 39 ± 1 |
| Connectivity density (mm−3) | 148.6 ± 34.5 | 355.4 ± 65.5a |
| Structure model index | 2.09 ± 0.17 | 1.81 ± 0.14 |
| Cortical thickness (μm) | 107 ± 5 | 97 ± 6 |
a Significantly different from controls, p < 0.05.
Hes1 Inactivation in Mature Osteoblasts Increases Bone Volume
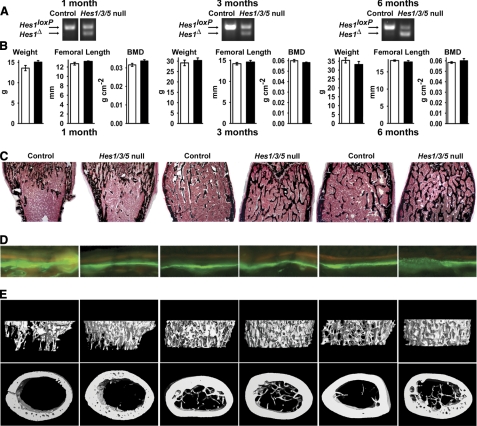
For the conditional deletion of Hes1 in mature osteoblasts, Oc-Cre/+;Hes1loxP/loxP;Hes3−/−;Hes5−/− mice were crossed with Hes1loxP/loxP;Hes3−/−;Hes5−/− mice to create Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− conditional null and Hes1loxP/loxP;Hes3−/−;Hes5−/− littermate controls. Recombination of Hes1loxP was documented by PCR analysis in calvarial DNA extracts from Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− mice (Fig. 4A). Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− mice appeared normal, contact radiography did not reveal skeletal abnormalities (data not shown), and there were no changes in weight, BMD, or femoral length when compared with littermate controls (Fig. 4B).
FIGURE 4.
Skeletal phenotype of 1 month (left), 3 month (middle) and 6 month (right) old male Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/−conditional null mice (Hes1/3/5 null) and Hes1loxP/loxP;Hes3−/−;Hes5−/− littermate controls (Control) of the same sex. A, representative PCR demonstrates the recombination of the Hes1loxP allele in calvarial DNA extracts. B, the weight (g), femoral length (mm), and total BMD (g cm−2) of Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− mice (black bars) and controls (white bars). C and D, von Kossa staining (C; final magnification, 40×) and calcein/demeclocycline labeling (D; final magnification, 100×) of representative femoral sections. E, microcomputed tomography of representative femurs.
The deletion of Hes1 in mature osteoblasts caused an increase in trabecular bone volume in male mice, due to an increase in trabecular number that was observed from 1 to 6 months of age (Table 4 and Fig. 4C). In accordance with the results obtained from the conditional deletion of Hes1 in the limb bud, female mice did not exhibit a skeletal phenotype (data not shown). Osteoblast number was not changed and Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− male mice displayed a transient increase in mineral apposition rate at 1 month of age, followed by a transient decrease at 3 months of age (Table 4 and Fig. 4D). In accordance with the results observed in HES1 male transgenics, the number of osteoclasts was decreased in Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− mice at 1 month of age. This indicates that the deletion of Hes1 in osteoblasts affects osteoclast number and possibly function, because there was a tendency toward decreased eroded surface (Table 4). Confirming the histomorphometric findings, microcomputed tomography revealed increased trabecular bone volume, number, and connectivity density in Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− mice. The structural model index was closer to zero, demonstrating a preponderance of plate-like trabecular structures (Table 4; Fig. 4E). Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− mice exhibited a modest decrease in cortical thickness at 6 months of age (Table 4 and Fig. 4E). These results indicate that inactivation of Hes1 affects the cortical bone compartment by inducing an age-related loss in cortical thickness.
Hes1 Regulates Osteoblastogenesis and Osteoclastogenesis in Vitro
To study the mechanisms responsible for the consequences of Hes1 inactivation, the differentiation of calvarial osteoblasts from Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− conditional null or Hes1loxP/loxP;Hes3−/−;Hes5−/− littermate controls of both sexes was analyzed. Hes1 mRNA levels were decreased in confluent cultures and in cells maintained in differentiation medium for 7 days, and alkaline phosphatase transcripts levels were induced upon down-regulation of Hes1 in confluent cultures (Fig. 5A). However, osteocalcin transcripts (Fig. 5A) and levels of alkaline phosphatase activity and mineralization of the culture were not increased (data not shown).
FIGURE 5.
Consequences of Hes1 inactivation on osteoblastic function in vitro. In panel A, Calvarial osteoblasts were harvested from Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− conditional null mice (black bars) and Hes1loxP/loxP;Hes3−/−;Hes5−/− littermate controls (white bars). Total RNA was extracted at confluence and after 7 days of culture in conditions favoring osteoblastogenesis; mRNA was reverse-transcribed and amplified by real time RT-PCR in the presence of specific primers. Data are expressed as Hes1, alkaline phosphatase (Ap), and osteocalcin copy number, determined by real time RT-PCR, corrected for Rpl38 expression. Values are means ± S.E., n = 4. *, significantly different from control cells, p < 0.05. In B, calvarial osteoblasts harvested from Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− conditional null mice (Hes1/3/5 null) and Hes1loxP/loxP;Hes3−/−;Hes5−/− littermate controls (Control) were co-cultured with splenocytes harvested from wild type mice, in the presence of 10 nm 1,25-dihydroxyvitamin D3 (Vit.D, black bars) or control vehicle (white bars). Data are expressed as % of resorbed area. Values are means ± S.E., n = 4–6. *, significantly different from control cells, p < 0.05. +, significantly different from cells treated with vehicle, p < 0.05.
To determine whether inactivation of Hes1 in osteoblasts impaired osteoclast formation and function, calvarial osteoblasts from Oc-Cre/+;Hes1Δ/Δ;Hes3−/−;Hes5−/− conditional null or Hes1loxP/loxP;Hes3−/−;Hes5−/− littermate controls of both sexes were co-cultured with splenocytes from wild type FVB mice. Under basal conditions, resorption was not different between experimental and control cultures. Treatment with 1,25-dihydroxyvitamin D3 enhanced resorption in control cultures, and this effect was blunted in co-cultures of Hes1 conditional null osteoblasts (Fig. 5B). This is in agreement with the suppressed number of osteoclasts and eroded surface observed in male Hes1 conditional null mice.
DISCUSSION
In the present study, we demonstrate that Hes1 modulates bone remodeling, as perturbing its expression in osteoblastic cells results in significant alterations in cancellous bone volume and microarchitecture. Transgenic mice overexpressing HES1 under the control of the 3.6-kb Col1a1 promoter display impaired growth and shortened life span. This could be due to excessive dosage of HES1 in extraskeletal tissues, because expression of the 3.6-kb Col1a1 promoter fragment is not entirely specific to skeletal cells (36). HES1 transgenic mice were moderately osteopenic and whereas males exhibited increased bone resorption, females displayed a reduced number of osteoblasts. These differences could be explained by different degrees of penetrance of the Hes1 transgene in the two sexes or by a sexually dimorphic mechanism of HES1 action in the skeleton. In vitro studies of osteoblastic differentiation of transgenic cells confirmed that HES1 suppressed osteoblastogenesis, and by acting on osteoblastic cells, it increased bone resorption. These results are in accordance with the inhibitory effects of HES1 on osteocalcin expression in osteoblastic cells and explain the osteopenic phenotype observed in HES1 transgenics (27).
The consequences of the conditional inactivation of Hes1 were analyzed in the context of the global deletion of Hes3 and Hes5. Although HES3 and HES5 are expressed in osteoblasts, Hes3−/−;Hes5−/− mice had no skeletal phenotype, suggesting that these genes are dispensable for normal skeletal development and bone remodeling, and their functions may be carried out by HES1. Inactivation of Hes1 in the limb bud or in mature osteoblasts caused a skeletal phenotype only in male mice, and this may be explained by earlier studies demonstrating sexual dimorphism in murine skeletal maturation and trabecular microarchitecture (56, 57). We could not detect differences in the recombination of the Hes1loxP allele between male and female Hes1 conditional null mice (data not shown); therefore, it is unlikely that Cre recombination was more efficient in skeletal cells from male than from female mice.
Even though inactivation of Hes1 in the limb bud increased the number and connectivity of trabeculae, osteoblast number and function were not changed. These observations are consistent with an inhibitory effect of HES1 on chondrogenesis and suggest that Hes1 impairs endochondral bone formation (58). This observation also is in accordance with the suppression of collagen type 2 α1 transcription by HES1 (58). Studies on the conditional inactivation of Hes1 in a Hes5 null background have suggested that HES1 and HES5 are dispensable for cartilage development (59). This discrepancy with our results could be explained by the fact that the phenotypic analysis was conducted in mice with a functional Hes3 allele, which could have compensated for the loss of Hes1 and Hes5.
Inactivation of Hes1 in mature osteoblasts caused a dramatic increase in trabecular bone volume and in the structural properties of cancellous bone, favoring the formation of plate-like trabecular structures and increasing connectivity. Although the phenotype of the Hes1 inactivation in the skeleton was striking, the mechanisms responsible were less evident. The increased bone volume observed seemed to be secondary to a transient increase in osteoblast function and a decrease in osteoclast function. It is of interest that despite the fact that functional changes were short-lived, the consequences were long lasting and observed up to 6 months of age. Cellular experiments confirmed an increase in osteoblast function and a decrease in the resorptive capacity of osteoclast precursors exposed to Hes1 null osteoblasts. Although the effects were relatively modest, they provide an explanation for the phenotype observed in vivo.
Although Hes1 is a target of Notch signaling in osteoblasts, the phenotype observed in these studies indicates that the misexpression of HES1 does not phenocopy that of Notch. HES1 transgenics exhibit a less pronounced skeletal phenotype than mice overexpressing the Notch1 intracellular domain under the control of the 3.6-kb Col1a1 promoter (23). Conditional inactivation of Hes1 in the limb bud caused a modest phenotype, whereas inactivation of Notch1 and Notch2 in the limb bud resulted in severe skeletal malformations due to abnormal accumulation of hypertrophic chondrocytes (22). An analogous phenotype to the Notch1 and Notch2 inactivation in the limb bud was observed following the conditional inactivation of Epstein-Barr virus latency C promoter binding factor 1, suppressor of Hairless and Lag-1 (Csl) in chondrocytes (60). CSL forms a complex with the Notch intracellular domain to induce transcription of Notch target genes in the canonical Notch signaling pathway (9). These observations would confirm a distinct function of Notch canonical signaling in skeletal development that cannot be fully accounted for by its induction of HES1 expression. This would suggest that other Notch target genes, such as Hey1, Hey2, and HeyL may be responsible, at least in part, for the effects of Notch in the skeleton. The distinct function of Hes1 is further substantiated by studies demonstrating that the conditional inactivation of Notch1 and Notch2 in mature osteoblasts causes no alteration in osteoblastic function, whereas the inactivation of Hes1 causes a distinct phenotype (22). In vivo and in vitro studies have revealed that in contrast to the stimulatory effects of Hes1 on osteoclast function, Notch suppresses osteoclastogenesis (21, 34, 35). These observations demonstrate that the skeletal effects of HES1 in osteoblasts are largely independent from those of Notch signaling (21, 34, 35). It is also important to note that the expression of Hes1 in skeletal cells is regulated by alternative signals, such as connective tissue growth factor, which induces Hes1 by mechanisms independent from Notch transactivation (24).
Osteoporosis, a disease characterized by reduction in bone mass, is a major health care problem due to the large number of people affected and the morbidity and mortality associated with osteoporotic fractures (15, 61). We have demonstrated that inactivation of Hes1, Hes3, and Hes5 not only increases bone mass but also enhances the microarchitectural properties of the skeleton. Although inhibition of intracellular protein function is difficult to achieve, targeted down-regulation of Hes1 expression or activity in osteoblasts could be considered as a possible strategy in the development of novel therapies for osteoporosis. Intracellular and extracellular inhibition of Notch signaling has been proposed for the treatment of a variety of disorders, such as Alzheimer disease and malignancies, and similar approaches could be considered for the inactivation of Hes1 (62). In conclusion, HES1 suppresses osteoblast function and enhances bone resorption and, as a consequence, regulates bone mass and bone microarchitecture.
Acknowledgments
We thank Drs. R. Kageyama for the Hes1loxP/loxP, Hes3−/−, and Hes5−/− mice, T. Clemens for osteocalcin-Cre transgenic mice, J. B. Lian for osteocalcin cDNA, and J. A. Lorenzo for helpful discussions. The authors thank A. Kent, K. Parker, and M. Monarca for technical help and M. Yurczak for secretarial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant DK045227 from the NIDDK.
- BMD
- bone mineral density
- Col1a1
- collagen type 1 α1
- FVB
- Friend leukemia virus strain B
- Prx
- paired-related homeobox gene
- Rpl38
- ribosomal protein L38.
REFERENCES
- 1. Iso T., Kedes L., Hamamori Y. (2003) J. Cell. Physiol. 194, 237–255 [DOI] [PubMed] [Google Scholar]
- 2. Katoh M., Katoh M. (2004) Int. J. Oncol. 25, 529–534 [PubMed] [Google Scholar]
- 3. Kageyama R., Ohtsuka T., Kobayashi T. (2007) Development 134, 1243–1251 [DOI] [PubMed] [Google Scholar]
- 4. Fischer A., Gessler M. (2007) Nucleic Acids Res. 35, 4583–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cornell R. A., Eisen J. S. (2005) Semin. Cell Dev. Biol. 16, 663–672 [DOI] [PubMed] [Google Scholar]
- 6. Androutsellis-Theotokis A., Leker R. R., Soldner F., Hoeppner D. J., Ravin R., Poser S. W., Rueger M. A., Bae S. K., Kittappa R., McKay R. D. (2006) Nature 442, 823–826 [DOI] [PubMed] [Google Scholar]
- 7. Gridley T. (2007) Development 134, 2709–2718 [DOI] [PubMed] [Google Scholar]
- 8. Lee J., Basak J. M., Demehri S., Kopan R. (2007) Development 134, 2795–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zanotti S., Canalis E. (2010) Mol. Cell. Biol. 30, 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murtaugh L. C., Stanger B. Z., Kwan K. M., Melton D. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kodama Y., Hijikata M., Kageyama R., Shimotohno K., Chiba T. (2004) Gastroenterology 127, 1775–1786 [DOI] [PubMed] [Google Scholar]
- 12. Hatakeyama J., Bessho Y., Katoh K., Ookawara S., Fujioka M., Guillemot F., Kageyama R. (2004) Development 131, 5539–5550 [DOI] [PubMed] [Google Scholar]
- 13. van Es J. H., van Gijn M. E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D. J., Radtke F., Clevers H. (2005) Nature 435, 959–963 [DOI] [PubMed] [Google Scholar]
- 14. Ishibashi M., Ang S. L., Shiota K., Nakanishi S., Kageyama R., Guillemot F. (1995) Genes Dev. 9, 3136–3148 [DOI] [PubMed] [Google Scholar]
- 15. Canalis E., Giustina A., Bilezikian J. P. (2007) N. Engl. J. Med. 357, 905–916 [DOI] [PubMed] [Google Scholar]
- 16. Canalis E. (2005) N. Engl. J. Med. 352, 2014–2016 [DOI] [PubMed] [Google Scholar]
- 17. Bianco P., Gehron Robey P. (2000) J. Clin. Invest. 105, 1663–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Canalis E., Economides A. N., Gazzerro E. (2003) Endocr. Rev. 24, 218–235 [DOI] [PubMed] [Google Scholar]
- 19. Westendorf J. J., Kahler R. A., Schroeder T. M. (2004) Gene 341, 19–39 [DOI] [PubMed] [Google Scholar]
- 20. Canalis E. (2008) Sci. Signal. 1, pe17. [DOI] [PubMed] [Google Scholar]
- 21. Engin F., Yao Z., Yang T., Zhou G., Bertin T., Jiang Chen Y., Wang L., Zheng H., Sutton R. E., Boyce B. F., Lee B. (2008) Nat. Med. 14, 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hilton M. J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H. M., Teitelbaum S. L., Ross F. P., Kopan R., Long F. (2008) Nat. Med. 14, 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zanotti S., Smerdel-Ramoya A., Stadmeyer L., Durant D., Radtke F., Canalis E. (2008) Endocrinology 149, 3890–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smerdel-Ramoya A., Zanotti S., Deregowski V., Canalis E. (2008) J. Biol. Chem. 283, 22690–22699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ross D. A., Rao P. K., Kadesch T. (2004) Mol. Cell. Biol. 24, 3505–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ross D. A., Hannenhalli S., Tobias J. W., Cooch N., Shiekhattar R., Kadesch T. (2006) Mol. Endocrinol. 20, 698–705 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2009) J. Cell. Biochem. 108, 651–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shen Q., Christakos S. (2005) J. Biol. Chem. 280, 40589–40598 [DOI] [PubMed] [Google Scholar]
- 29. McLarren K. W., Lo R., Grbavec D., Thirunavukkarasu K., Karsenty G., Stifani S. (2000) J. Biol. Chem. 275, 530–538 [DOI] [PubMed] [Google Scholar]
- 30. Lee J. S., Thomas D. M., Gutierrez G., Carty S. A., Yanagawa S., Hinds P. W. (2006) J. Bone Miner Res. 21, 921–933 [DOI] [PubMed] [Google Scholar]
- 31. Suh J. H., Lee H. W., Lee J. W., Kim J. B. (2008) Biochem. Biophys. Res. Commun. 367, 97–102 [DOI] [PubMed] [Google Scholar]
- 32. Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 33. Teitelbaum S. L. (2007) Am. J. Pathol. 170, 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamada T., Yamazaki H., Yamane T., Yoshino M., Okuyama H., Tsuneto M., Kurino T., Hayashi S., Sakano S. (2003) Blood 101, 2227–2234 [DOI] [PubMed] [Google Scholar]
- 35. Bai S., Kopan R., Zou W., Hilton M. J., Ong C. T., Long F., Ross F. P., Teitelbaum S. L. (2008) J. Biol. Chem. 283, 6509–6518 [DOI] [PubMed] [Google Scholar]
- 36. Kalajzic I., Kalajzic Z., Kaliterna M., Gronowicz G., Clark S. H., Lichtler A. C., Rowe D. (2002) J. Bone Miner. Res. 17, 15–25 [DOI] [PubMed] [Google Scholar]
- 37. Irwin N. (1989) in Sambrook (Fritsch E. F., Maniatis T. eds) pp. 9.32–9.36, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 38. Imayoshi I., Shimogori T., Ohtsuka T., Kageyama R. (2008) Development 135, 2531–2541 [DOI] [PubMed] [Google Scholar]
- 39. Hirata H., Tomita K., Bessho Y., Kageyama R. (2001) EMBO J. 20, 4454–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cau E., Gradwohl G., Casarosa S., Kageyama R., Guillemot F. (2000) Development 127, 2323–2332 [DOI] [PubMed] [Google Scholar]
- 41. Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N., Tabin C. J. (2002) Genesis. 33, 77–80 [DOI] [PubMed] [Google Scholar]
- 42. Zhang M., Xuan S., Bouxsein M. L., von Stechow D., Akeno N., Faugere M. C., Malluche H., Zhao G., Rosen C. J., Efstratiadis A., Clemens T. L. (2002) J. Biol. Chem. 277, 44005–44012 [DOI] [PubMed] [Google Scholar]
- 43. Nagy T. R., Prince C. W., Li J. (2001) J. Bone Miner. Res. 16, 1682–1687 [DOI] [PubMed] [Google Scholar]
- 44. Gazzerro E., Pereira R. C., Jorgetti V., Olson S., Economides A. N., Canalis E. (2005) Endocrinology 146, 655–665 [DOI] [PubMed] [Google Scholar]
- 45. Parfitt A. M., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., Recker R. R. (1987) J. Bone Miner. Res. 2, 595–610 [DOI] [PubMed] [Google Scholar]
- 46. Bouxsein M. L., Boyd S. K., Christiansen B. A., Guldberg R. E., Jepsen K. J., Müller R. (2010) J Bone Miner Res. 25, 1468–1486 [DOI] [PubMed] [Google Scholar]
- 47. McCarthy T. L., Centrella M., Canalis E. (1990) J. Biol. Chem. 265, 15353–15356 [PubMed] [Google Scholar]
- 48. Takahashi N., Akatsu T., Udagawa N., Sasaki T., Yamaguchi A., Moseley J. M., Martin T. J., Suda T. (1988) Endocrinology 123, 2600–2602 [DOI] [PubMed] [Google Scholar]
- 49. Wyzga N., Varghese S., Wikel S., Canalis E., Sylvester F. A. (2004) Bone 35, 614–620 [DOI] [PubMed] [Google Scholar]
- 50. Lee S. K., Kalinowski J., Jastrzebski S., Lorenzo J. A. (2002) J. Immunol. 169, 2374–2380 [DOI] [PubMed] [Google Scholar]
- 51. Nazarenko I., Pires R., Lowe B., Obaidy M., Rashtchian A. (2002) Nucleic Acids Res. 30, 2089–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nazarenko I., Lowe B., Darfler M., Ikonomi P., Schuster D., Rashtchian A. (2002) Nucleic Acids Res. 30, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Akazawa C., Sasai Y., Nakanishi S., Kageyama R. (1992) J. Biol. Chem. 267, 21879–21885 [PubMed] [Google Scholar]
- 54. Lian J., Stewart C., Puchacz E., Mackowiak S., Shalhoub V., Collart D., Zambetti G., Stein G. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kouadjo K. E., Nishida Y., Cadrin-Girard J. F., Yoshioka M., St-Amand J. (2007) BMC Genomics 8, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Glatt V., Canalis E., Stadmeyer L., Bouxsein M. L. (2007) J. Bone Miner. Res. 22, 1197–1207 [DOI] [PubMed] [Google Scholar]
- 57. DeMambro V. E., Clemmons D. R., Horton L. G., Bouxsein M. L., Wood T. L., Beamer W. G., Canalis E., Rosen C. J. (2008) Endocrinology 149, 2051–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Grogan S. P., Olee T., Hiraoka K., Lotz M. K. (2008) Arthritis Rheum. 58, 2754–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Karlsson C., Brantsing C., Kageyama R., Lindahl A. (2010) Cells Tissues Organs 192, 17–27 [DOI] [PubMed] [Google Scholar]
- 60. Mead T. J., Yutzey K. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14420–14425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Looker A. C., Orwoll E. S., Johnston C. C., Jr., Lindsay R. L., Wahner H. W., Dunn W. L., Calvo M. S., Harris T. B., Heyse S. P. (1997) J. Bone Miner. Res. 12, 1761–1768 [DOI] [PubMed] [Google Scholar]
- 62. Moellering R. E., Cornejo M., Davis T. N., Del Bianco C., Aster J. C., Blacklow S. C., Kung A. L., Gilliland D. G., Verdine G. L., Bradner J. E. (2009) Nature 462, 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]