Abstract
Mesencephalic astrocyte-derived neurotrophic factor (MANF) protects neurons and repairs the Parkinson disease-like symptoms in a rat 6-hydroxydopamine model. We show a three-dimensional solution structure of human MANF that differs drastically from other neurotrophic factors. Remarkably, the C-terminal domain of MANF (C-MANF) is homologous to the SAP domain of Ku70, a well known inhibitor of proapoptotic Bax (Bcl-2-associated X protein). Cellular studies confirm that MANF and C-MANF protect neurons intracellularly as efficiently as Ku70.
Keywords: Apoptosis, Neurodegeneration, Neurotrophic Factor, NMR, Parkinson Disease, Bax, Ku70
Introduction
Parkinson disease is a chronic, progressive neurodegenerative disease where dopaminergic cells die most prominently in the area of substantia nigra (1). Neurotrophic factors are secreted proteins, which, upon binding to their target receptors, trigger survival pathways to prevent neuronal loss. Three of four major families of the neurotrophic growth factors are as follows: the neurotrophins, the glial cell line-derived neurotrophic factor (GDNF)4 family of ligands (GFL), and neuropoietic cytokines. The neurotrophins nerve growth factor (NGF), brain-derived neurotrophic factor, neurotrophin 3, and neurotrophin 4/5, share homologous classic cysteine knot growth factor structure, which form a head-to-head noncovalently assembled dimer (2). Neurotrophins act through two distinct receptors types: tropomyosin-related kinase and low affinity p75 neurotrophin receptor (p75NTR) with a cysteine-rich extracellular region (3). GDNF and other GFLs, neurturin artemin and persephin, are also cysteine knot proteins, forming homodimers covalently linked by a disulfide bond in head-to-toe arrangement (2). Upon activation, the dimer of GDNF forms a complex with the co-receptor GDNF family receptor α1. The heterotetrameric complex induces dimerization and activation of rearranged during transfection (receptor tyrosine kinase) receptor (4). Unlike neurotrophins and GFLs, which both include three pairs of antiparallel β-strands, neuropoietic cytokine ciliary neurotrophic factor is a four-helix bundle protein, which binds to heterotrimeric receptor formed by a specific ciliary neurotrophic factor α, the β-subunit of the leukemia inhibitory factor (LIFRβ), and glycoprotein gp130 to activate the signals for neuronal survival (5).
Mesencephalic astrocyte-derived neurotrophic factor (MANF) (also recognized as ARMET or arginine-rich mutated in early stages of tumor) (6) and cerebral dopamine neurotrophic factor (CDNF) (7) belong to fourth family of neurotrophic factors. Mammals have CDNF and MANF, whereas invertebrate animals have one gene homologous to MANF/CDNF (8). MANF and CDNF are the most potent factors protecting and repairing the dopaminergic neurons in the rat 6-hydroxydopamine model of Parkinson disease (7, 9). MANF also efficiently rescued cortical neurons in the rat stroke model (10). In non-neuronal cells, MANF is also identified as a protein up-regulated by unfolded protein response and protecting against various forms of endoplasmic reticulum stress (11). Although the mode of action of MANF is unclear, it has been suggested to act both as a secreted neurotrophic factor and as an intracellular protein. The crystal structure of MANF revealed a well defined N-terminal domain belonging to the saposin family and a mostly disordered C-terminal domain, supporting a bifunctional role of MANF (12). Although the results regarding neuronal protection and repair are highly interesting and promising, the cellular and neuroprotective mechanisms of action of MANF and CDNF have remained entirely unclear.
Here, we report a three-dimensional solution structure of full-length human MANF, determined by multidimensional NMR spectroscopy. The structure confirms earlier suggested two-domain architecture (6), additionally revealing novel structural features, which differ drastically from all neurotrophic factors known so far. Unlike the crystal structure of MANF that showed poorly defined, highly disordered C-terminal domain (12), the solution structure reveals a well defined globular structural module. Remarkably, the C-terminal domain is homologous to the SAP (SAF-A/B, Acinus, and PIAS) domain of Ku70 (C-Ku70), a well known inhibitor of proapoptotic Bax (13). During preparation of this article, Hoseki and co-workers (14) have independently determined the NMR structure of MANF (Protein Data Bank code 2RQY). Although the three-dimensional structure is highly similar to our solution structure with an r.m.s.d. of 2.77 Å (residues 13–88) and 1.08 Å (residues 113–123 and 133–147) for N- and C-terminal domains, no structure-function relationships to neuroprotective and cytoprotective features of MANF were found.
EXPERIMENTAL PROCEDURES
Protein Production
Conventional molecular biology protocols have been utilized for the preparation of proteins for NMR and cell cultivation studies. Detailed strategies are introduced in the supplemental “Experimental Procedures.” 15N- and/or 13C-labeled full-length MANF and C-MANF were expressed in OrigamiB(DE3) cells to obtain native protein with disulfide bonds. Cells were grown at 37 °C until the OD reached 0.5, the temperature was decreased to 16 °C, and cells were induced when OD reached 0.6 with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested after 16 h of induction. 15NH4Cl and 13C-labeled d-glucose were used as sole sources of nitrogen and carbon, respectively.
Structural studies were carried out using 1 mm uniformly 13C/15N-labeled sample of full-length MANF in 10 mm Bis-Tris, pH 6.8, 50 mm NaCl, supplemented with 7% (v/v) D2O. NMR buffer for C-MANF was 20 mm Na-PO4, pH 6.5, 50 mm NaCl, supplemented with 7% (v/v) D2O with protein concentration of 1 mm. A standard set of NMR spectra was recorded to complete the assignments and described in detail elsewhere (15).
NMR Spectroscopy
All NMR spectra were recorded at 35 °C on a Varian Unity INOVA 800 NMR spectrometer, operating at 800 MHz of 1H frequency. Interproton distance restraints were determined from the three-dimensional 15N-separated NOESY-HSQC, and a 13C-separated NOESY-HSQC spectrum modified to simultaneously excite aliphatic and aromatic carbon resonances. The NOESY-15N-HSQC spectrum was recorded with two transients, using 150, 80, and 1408 complex points in 1H(ω1), 15N(ω2), and 1H(ω3) dimensions, corresponding to acquisition times of 13.6, 36.4, and 128 ms in t1, t2, and t3, respectively. The NOESY-13C-HSQC was recorded with eight transients, using 90, 72, and 1408 complex points in 1H(ω1), 13C(ω2), and 1H(ω3) dimensions, corresponding to acquisition times of 8.2, 4.8, and 128 ms in t1, t2, and t3, respectively.
The heteronuclear steady-state NOE spectra were acquired with 200 and 1024 complex points in ω1 (15N) and ω2 (1H) dimensions. The corresponding spectral widths for 15N and 1H were 2000 and 8000 Hz, respectively. The spectra were accumulated using 32 transients per spectrum.
Structure Calculations
For protein-structure calculations, structural information derived from multidimensional solution state NMR spectroscopy was utilized. Structure calculations, based on automated NOE peak assignments, were performed by using Cyana structure calculation package (version 2.0) (16, 17), and structures were subsequently refined by AMBER (version 8.0) (18). A more detailed description of structure calculations is given in the supplemental “Experimental Procedures.”
Neuronal Cultures
Cell cultivation and microinjection of the sympathetic superior cervical ganglion (SCG) neurons was performed as described earlier (19). The details are shown in the supplemental “Experimental Procedures.” Plasmids for Ku70 and Bcl-xL were described earlier (19).
Experiments with Iodinated Proteins
MANF (20) and GDNF (PeproTech, Ltd.) were iodinated using the lactoperoxidase method, as described (9). For the binding assay, cultured SCG neurons were treated with 125I-labeled MANF or GDNF at 5 ng/ml in PBS containing 1% BSA, in the presence or absence of 200-fold excess of respective unlabeled factor. After incubation with the factors for 2 h on ice, the cultures were washed three times with PBS to remove unbound factors and solubilized with 1 n NaOH, and the radioactivity was counted using a 1480 WIZARD 3 scintillation counter (Wallac). For the internalization assay, the neurons were treated with the factors in the same way on ice for 5 h, followed by incubation for 30 min at 37 °C. The neurons were then washed once with 0.2 m acetic acid/0.5 m NaCl, pH 4.5, washed three times with PBS/1% BSA, washed once with 0.2 m acetic acid/0.5 m NaCl, pH 2.8, washed once with PBS/1% BSA, and then solubilized with 1 n NaOH, and the radioactivity was counted.
RESULTS AND DISCUSSION
We used NMR spectroscopy to solve a three-dimensional solution structure of full-length MANF (FL-MANF) protein as well as its isolated C-terminal domain (C-MANF) comprising residues 96–158 (numbered according to mature protein). The primary sequence of MANF contains eight conserved cysteines, corresponding to residues Cys6, Cys9, Cys40, Cys51, Cys82, Cys93, Cys127, and Cys130, which according to electrospray-ionization mass spectrometry and crystal structure analysis, all form disulfide bonds in maturated protein (7, 12, 20, 21).
The solution structure of MANF reveals that it is composed of two domains, which are connected with a short linker (Fig. 1 and supplemental Fig. S1). Individual N- and C-terminal domains are very well defined, and statistics of structure calculations describe the quality of the structure of FL-MANF and that of C-MANF summarized in Table 1 and Table 2, respectively. No NOEs were observed between the two domains, resulting in a relatively high fluctuation between orientations of two domains due to lack of NOE-mediated interdomain restraints. The heteronuclear {1H}-15N NOEs of the backbone amides are sensitive to molecular motions taking place in ps-ns timescale. {1H}-15N NOE data recorded in FL-MANF (Fig. 1C) indicate that the linker region (residues 96–103) as well as the N- and C termini (residues 1–4 and 148–158, respectively) are highly flexible as manifested by low heteronuclear {1H}-15N NOE values (< 0.6) and by the lack of long and medium range NOE distance restraints (see also supplemental Fig. S1). Based on these data, we concluded that domains are not tightly packed to each other, but instead tumble as independent structural modules separated by the flexible linker between them. The N-terminal domain of MANF is a globular, highly α-helical structure comprising five α-helices and one 310 helix, and is stabilized by three disulfide bridges (Fig. 1A and supplemental text). Comparison of the existing crystal structure (PDB code 2W51) indicates that its overall fold is highly similar to solution structure of the N-terminal domain of MANF with an r.m.s.d. of 1.78 Å. The N-terminal domain of MANF is homologous to the lipid and membrane binding protein superfamily, saposins. They share a characteristic “closed leaf” fold, comprised of five helices and three conserved disulfide bridges (22, 23).
FIGURE 1.
Schematic ribbon presentation of overall fold of MANF and plot of heteronuclear {1H}-15N NOE values. A, structure of FL-MANF is color-coded from N to C terminus starting from blue via green to red. N- and C-terminal domains are connected with flexible linker region (see also supplemental Fig. S1). B, the structure of isolated C-MANF is shown. Color-coding of C-MANF is similar to FL-MANF. The structure of C-MANF is composed of three helices, the first helix (α6) is loosely formed and the two consecutive helices (α7 and α8) are found in parallel orientation forming a helix-loop-helix arrangement. Disulfide bridges are shown in magenta. The three-dimensional structure of isolated C-MANF is highly similar to the C-terminal domain of intact full-length MANF. C, shown is a plot of heteronuclear steady-state {1H}-15N NOE values as a function of primary structure. The y axis represents the {1H}-15N NOE values obtained from the ratios of peak intensities in the saturated spectrum and in the unsaturated spectrum. Ratios of <0.6 were ranked as flexible regions. These were found between two domains and at both termini.
TABLE 1.
NMR and refinement statistics for the solution structure of FL-MANF
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 3090 |
| Intra-residue | 772 |
| Inter-residue | |
| Sequential (|i − j| = 1) | 761 |
| Medium range (|i − j| ≤ 4) | 950 |
| Long range (|i − j| ≥ 5) | 607 |
| Total dihedral angle restraints (used for the program Cyana) | 172 |
| φ | 86 |
| ψ | 86 |
| Structure statistics | |
| Violations (mean and S.D.) | |
| Distance constraints (Å) | 0.009 ± 0.0003 |
| Maximum distance constraint violation (Å) | 0.29 |
| Dihedral angle constraints (°) | |
| Maximum dihedral angle violation (°) | |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0104 ± 0.0001 |
| Bond angles | 2.1999 ± 0.0170° |
| Average pairwise r.m.s.d. (Å)a | |
| Backbone | 0.472 (0.346) |
| Heavy | 0.819 (0.762) |
a Pairwise r.m.s.d. was calculated among 15 refined structures for residues 7–91 (112–147).
TABLE 2.
NMR and refinement statistics for the solution structure of C-MANF
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 845 |
| Intra-residue | 272 |
| Inter-residue | |
| Sequential (|i − j| = 1) | 215 |
| Medium range (|i − j| ≤ 4) | 217 |
| Long range (|i − j| ≥ 5) | 141 |
| Total dihedral angle restraints | 46 |
| φ | 23 |
| ψ | 23 |
| Structure statistics | |
| Violations (mean and S.D.) | |
| Distance constraints (Å) | 0.0024 ± 0.0008 |
| Maximum distance constraint violation (Å) | 0.289 |
| Dihedral angle constraints | 0.03 ± 0.11° |
| Max. dihedral angle violation | 0.836° |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0108 ± 0.0001 |
| Bond angles | 1.9171 ± 0.0311° |
| Average pairwise r.m.s.d. (Å)a | |
| Backbone | 0.54 |
| Heavy | 1.06 |
a Pairwise r.m.s.d. was calculated among 15 refined structures for residues 112–147.
The structure of C-MANF is composed of three helices, where the first helix (α6) is loosely formed and the two consecutive helices (α7 and α8) are found in parallel orientation forming a helix-loop-helix arrangement. Two of eight cysteine residues found in MANF are located in C-MANF. Cys127 and Cys130 form a CXXC motif residing in the loop region, which connects two helices (helix α7 and helix α8) running in the same direction.
To study stability, structural integrity, as well as a plausible independent functional role of the isolated small C-terminal domain of MANF, we designed the C-terminal construct comprising residues 96–158. This construct includes a linker region, i.e. starts directly after the last 310 helix in the N-terminal saposin domain of mature full-length MANF. Amide proton-nitrogen correlation experiment (15N-HSQC) exhibited a well dispersed correlation map with similar cross-peak intensities indicative of independently folding globular domain (15). The resulting three-dimensional structure of the isolated C-terminal domain is identical to the structure of the corresponding C-terminal domain in full-length MANF (Fig. 1).
Although the C-terminal domain of MANF is poorly defined in the crystal structure, it is unlikely that this reflects conditions used for crystallization (100 mm sodium cacodylate buffer, pH 6.5, 0.2 m magnesium acetate, and 12–18% (w/v) PEG 8000) (8). According to our data, C-MANF is not an intrinsically unfolded protein, and the lack of electron density in the crystal structure merely stems from the dynamic linker, which increases the number of different orientations for the C-terminal domain to adopt in crystals, yielding poorly defined crystal coordinates for C-MANF.
We carried out the structural alignment using the program Dali (24), which showed that C-MANF is structurally similar to members of the SAP protein superfamily. They are putative DNA binding domains found in diverse nuclear proteins involved in chromosomal organization, but the exact role of the small helical bundle is still unknown (25). The highest structural similarity were found to the C-terminal SAP domain of Ku70 (26) (PDB code 1JEQ), residues 559–609 (Z-score, 4.3; r.m.s.d., 2.0 Å), SAP domain of human E1B-55 kDa associated protein 5 isoform C (PDB code 1ZRJ; Z-score, 4.2; r.m.s.d., 4.9), FYVE RING finger protein Sakura (27) (PDB code 1Y02; Z-score, 4.1; r.m.s.d., 2.8 Å), SAP domain of human nuclear protein HCC-1 (PDB code 2DO1; Z-score, 4.1; r.m.s.d., 4.7), and recombination endonuclease VII of bacteriophage T4 (28) (PDB code 1E7D; Z-score, 3.9; r.m.s.d., 4.9 Å). All proteins showing the highest Z-scores are members of the SAP protein superfamily (Fig. 2).
FIGURE 2.
Structural comparison of C-MANF with its homologue proteins with Z-score values >4 and r.m.s.d. value <3. A, ribbon presentation of C-MANF in green (PDB code 2KVE). B, C-Ku70 in cyan (PDB code 1JEQ) (25). C, SAP domain of FYVE RING finger protein Sakura in purple (PDB code 1Y02) (26). D, sequences aligned according to structural comparison. Secondary structure elements (helices) of the proteins are labeled with sequences as yellow boxes. Sequences are numbered according to maturated full-length proteins. VPMLKE motif of Ku70 and corresponding region of C-MANF and SAP domain of FYVE RING finger protein Sakura are marked with blue boxes, and the regions aligned to the CXXC motif of C-MANF are shown as green boxes. According to the structural alignment, the first helix in the C-Ku70 and FYVE RING finger protein Sakura is better defined than the corresponding helix α6 in C-MANF. Disulfide bond of C-MANF formed by CXXC motif marked in yellow on the structure. The first cysteine of SAP domain of FYVE RING finger protein Sakura is replaced by threonine. The loop connecting helices α7 and α8 in C-MANF is two residues shorter in C-Ku70, but the disulfide bond can be aligned against the backbone of Ku70. The VPMLKE motif (residues 578–583) of Ku70 is marked in blue ribbons, and the corresponding regions of C-MANF and FYVE RING finger protein Sakura are marked in red and pink ribbons, respectively. The side chains of conserved lysines are marked as red, and conserved hydrophobic amino acids are localized into the α-helical regions (gray side chains and boxes) as a part of the hydrophobic core.
The C-terminal SAP domain of Ku70 exhibited the lowest r.m.s.d. and yielded the highest Z-score with C-MANF (Fig. 2). Ku70 is a relatively large, multifunctional protein. Together with Ku80, it forms a heterodimeric Ku protein, which is essential for nonhomologous DNA double-strand break repair (26). Importantly, Ku70 has also cytoplasmic antiapoptotic function where it, via its C-terminal SAP domain, binds the proapoptotic protein Bax, thereby keeping it in the inactive conformation. In the apoptotic cells, Ku70 dissociates from Bax, thereby allowing its activation and triggering of the mitochondrial cell death pathway (13, 29, 30). Notably, Bax is the main proapoptotic effector in neurons. Cytoprotective activity of Ku70 has been localized within residues 536–609, which form the most C-terminal part of the protein, or more specifically to VPMLKE motif (residues 578–583). Ku70-derived pentapeptide (VPMLK) is also able to arrest Bax-mediated apoptosis (31). Strikingly, the C-terminal domain of MANF and C-terminal domain of Ku70 (C-Ku70) share a similar epitope, located at the beginning of the helix α7 (Fig. 2).
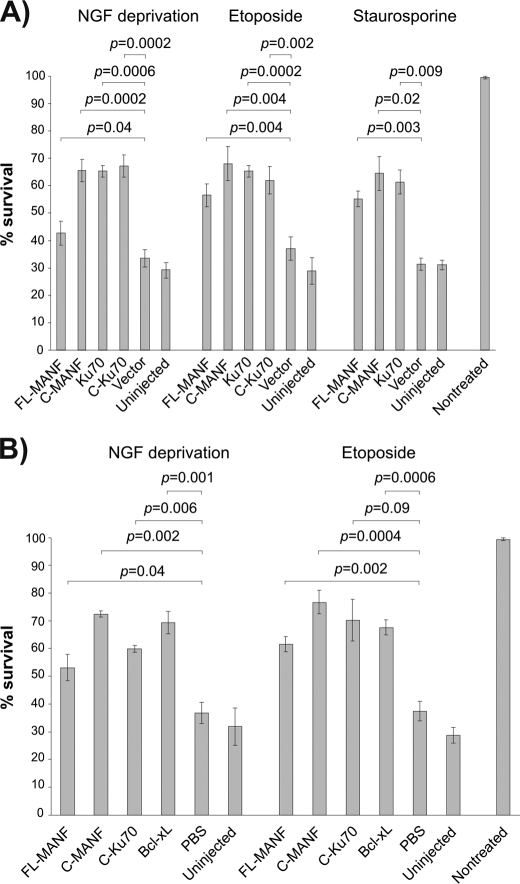
Owing to the highest structural similarity with the SAP domain of Ku70, we investigated plausible functional similarity between C-Ku70 and MANF, particularly whether C-MANF is able to intracellularly block the Bax-dependent cell death of the neurons. To that end, we overexpressed the plasmids encoding for FL-MANF, C-MANF, Ku70, and C-Ku70 in the sympathetic neurons from SCG and induced their death by three apoptotic stimuli, all killing the neurons in the Bax-dependent manner: i) deprivation of NGF, a physiological apoptotic stimulus (18, 32); ii) treatment with etoposide, a topoisomerase II inhibitor that causes double-strand DNA breaks (33); or iii) staurosporine, a broad-spectrum protein kinase inhibitor (13).
FL-MANF and, more prominently, C-MANF protected the neurons in all three conditions analogously to Ku70 and C-Ku70 (Fig. 3). Recombinant MANF or C-MANF proteins also rescued the NGF-deprived or etoposide-treated neurons as efficiently as the well established antiapoptotic protein Bcl-xL, when microinjected directly into the cytoplasm (Fig. 3). Thus, similarly to Ku70, MANF, and, more efficiently, C-MANF are able to protect the neurons intracellularly against the Bax-dependent apoptosis.
FIGURE 3.
Overexpressed FL-MANF and C-MANF protect apoptotic neurons. Newborn mouse SCG neurons were cultured for 5–6 days with NGF, and then microinjected with the indicated expression plasmids (A) or recombinant proteins (B) and either deprived of NGF or treated with etoposide (30 μm) or staurosporine (200 nm) for 3 days. Live injected neurons were then counted and expressed as percent of initially injected neurons. The mean ± S.E. of eight (A, NGF deprivation, etoposide), three (staurosporine), or four (B) independent experiments is shown. Data of each experimental group were compared with control plasmid pcDNA3 (vector) in A, or control PBS in B, by one-way analysis of variance and post hoc Dunnett's t test. The null hypothesis was rejected at p < 0.05.
In this work, we have shown that the three-dimensional structure of MANF differs drastically from other known neurotrophic factors. MANF has a two-domain architecture, and both domains may carry out very distinct roles during neuroprotection. The N-terminal domain is homologous to the lipid and membrane binding saposin protein family and the C-terminal domain to the SAP domain of Ku70, which has been shown to inhibit the proapoptotic Bax and prevent mitochondrial cell death signaling. Accordingly, MANF and yet more efficiently C-MANF are also able to arrest mitochondrial cell death pathway, induced by etoposide, staurosporine or NGF deprivation in SCG neurons with efficiency comparable with that of C-Ku70.
MANF has been localized to endoplasmic reticulum and Golgi and is also a secreted protein (6, 11, 20). Accordingly, two mechanisms of action have been shown for MANF. On one hand, intracranially (extracellularly) injected MANF or CDNF efficiently protected dopaminergic neurons in a rat 6-hydroxydopamine induced Parkinson disease model and the model of ischemia (10), thereby suggesting them to function as secreted neurotrophic factors (7, 9). The signaling receptors for MANF and CDNF remained undescribed. On the other hand, as shown in this study and earlier by Apostolou and co-workers (11), MANF (ARMET) possesses intracellular cytoprotective function. Endoplasmic reticulum stress, which is induced by tunicamycin or thapsigargin, triggers up-regulation of MANF to protect cells from this unfolded protein response-induced death (11). Our results support the intracellular mode of action of MANF. Indeed, we show that when expressed intracellularly, MANF can efficiently protect the apoptotic neurons. As the recombinant proteins directly injected to the cytoplasm also efficiently protected the neurons, we are confident that MANF indeed acts intracellularly in our experiments. Moreover, the plasmid-encoded C-MANF does not have the secretion signal and is therefore not secreted.
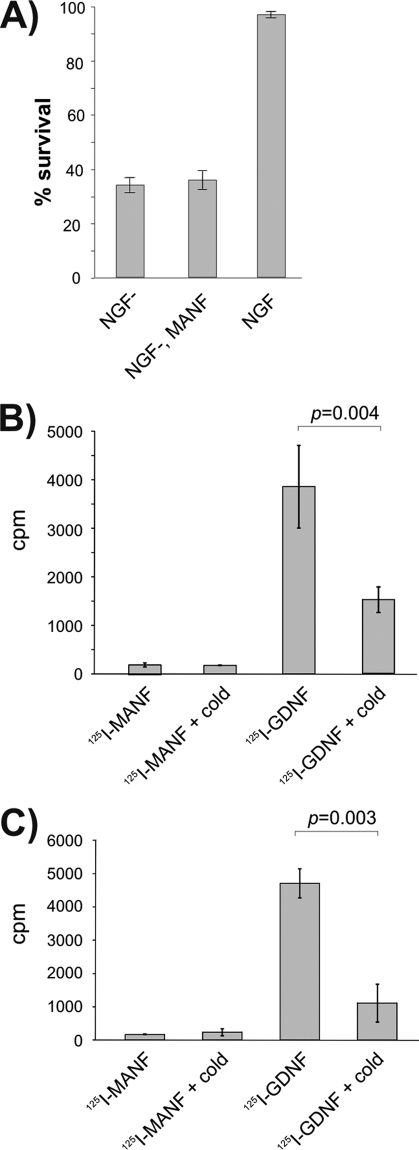
As our plasmid-expressed FL-MANF is partially retained in the cells and partially secreted (7), we cannot exclude that it could also protect the neurons in the autocrine/paracrine extracellular manner. This scenario seems, however, unlikely because extracellularly added MANF neither binds, enters, nor protects the cultured SCG neurons (Fig. 4), although the same MANF preparations, both 125I-labeled and unlabeled, were potently active in vivo (9). In addition, by our preliminary results, exogeneous MANF has also no activity on the cultured dopaminergic and cortical neurons, suggesting that differently from any other neurotrophic factor, it requires intact tissue environment.
FIGURE 4.
Exogeneous MANF does not affect SCG neurons in vitro. A, MANF protein in the culture medium did not protect SCG neurons. Newborn mouse SCG neurons were cultured with NGF for 5 days, and then NGF was removed (NGF−) and replaced or not replaced with recombinant MANF at 100 ng/ml. Living neurons were counted 3 days later and expressed as percent of initially counted neurons. The mean ± S.E. of four independent experiments is shown. B and C, 125I-labeled MANF is not bound or taken up by the cultured SCG neurons. 125I- MANF or 125I-GDNF was bound to the cultured SCG neurons in the presence or absence of 200-fold excess of respective unlabeled factors (cold). Bound (B) or internalized (C) radioactivity was measured. Shown are the cpm values from the representative experiments (means of three parallels ± S.E.). The means were compared by Student's t test, and the null hypothesis was rejected at p < 0.05. Qualitatively same results were obtained from three independent experiments for both assays.
In summary, MANF is an exceptional neurotrophic factor that can protect the cells both intracellularly (this study and Ref. 11) and in vivo extracellularly (7, 9, 10). The small C-terminal domain of MANF is responsible for the intracellular protection against the Bax-dependent apoptosis. Further studies are required to identify intracellular targets of MANF/C-MANF, which according to structural homology with the SAP domain family, may include e.g. Bax or transcription factors. Nevertheless, C-MANF plays an important role in protection of neurodegenerative, apoptotic, and stress conditions, which thereby makes it a highly potential therapeutic agent.
Supplementary Material
Acknowledgments
We thank Anne Hakonen and Elina Ahovuo for technical assistance.
This work was supported by the Academy of Finland Grants 122170 and 131144 (to P. P.) and Grants 11186236 and 1126735 (to M. S.), and a Sigrid Juselius Foundation grant (to M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Fig. S1, and additional references.
The atomic coordinates and structure factors (codes 2KVD and 2KVE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GDNF
- glial cell line-derived neurotrophic factor
- GFL
- GDNF family of ligands
- MANF
- mesencephalic astrocyte-derived neurotrophic factor
- C-MANF
- C-terminal domain of MANF
- NGF
- nerve growth factor
- CDNF
- cerebral dopamine neurotrophic factor
- SCG
- superior cervical ganglion
- FL-MANF
- full-length MANF
- PDB
- Protein Data Bank
- C-Ku70
- C-terminal domain of Ku70
- r.m.s.d.
- root mean square deviation
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1. Butte M. J. (2001) Cell. Mol. Life Sci. 58, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reichardt L. F. (2006) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 361, 1545–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parkash V., Leppänen V. M., Virtanen H., Jurvansuu J. M., Bespalov M. M., Sidorova Y. A., Runeberg-Roos P., Saarma M., Goldman A. (2008) J. Biol. Chem. 283, 35164–35172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang X., Lupardus P., Laporte S. L., Garcia K. C. (2009) Annu. Rev. Immunol. 27, 29–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dauer W., Przedborski S. (2003) Neuron 39, 889–909 [DOI] [PubMed] [Google Scholar]
- 6. Petrova P., Raibekas A., Pevsner J., Vigo N., Anafi M., Moore M. K., Peaire A. E., Shridhar V., Smith D. I., Kelly J., Durocher Y., Commissiong J. W. (2003) J. Mol. Neurosci. 20, 173–188 [DOI] [PubMed] [Google Scholar]
- 7. Lindholm P., Voutilainen M. H., Laurén J., Peränen J., Leppänen V. M., Andressoo J. O., Lindahl M., Janhunen S., Kalkkinen N., Timmusk T., Tuominen R. K., Saarma M. (2007) Nature 448, 73–77 [DOI] [PubMed] [Google Scholar]
- 8. Palgi M., Lindström R., Peränen J., Piepponen T. P., Saarma M., Heino T. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2429–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Voutilainen M. H., Bäck S., Pörsti E., Toppinen L., Lindgren L., Lindholm P., Peränen J., Saarma M., Tuominen R. K. (2009) J. Neurosci. 29, 9651–9659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Airavaara M., Shen H., Kuo C. C., Peränen J., Saarma M., Hoffer B., Wang Y. (2009) J. Comp. Neurol. 515, 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Apostolou A., Shen Y., Liang Y., Luo J., Fang S. (2008) Exp. Cell Res. 314, 2454–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parkash V., Lindholm P., Peränen J., Kalkkinen N., Oksanen E., Saarma M., Leppänen V. M., Goldman A. (2009) Protein Eng. Des. Sel. 22, 233–241 [DOI] [PubMed] [Google Scholar]
- 13. Sawada M., Sun W., Hayes P., Leskov K., Boothman D. A., Matsuyama S. (2003) Nat. Cell Biol. 5, 320–329 [DOI] [PubMed] [Google Scholar]
- 14. Hoseki J., Sasakawa H., Yamaguchi Y., Maeda M., Kubota H., Kato K., Nagata K. (2010) FEBS Lett. 584, 1536–1542 [DOI] [PubMed] [Google Scholar]
- 15. Hellman M., Peränen J., Saarma M., Permi P. (2010) Biomol. NMR Assign. 4, 215–217 [DOI] [PubMed] [Google Scholar]
- 16. Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 17. Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 18. Case D. A., Darden T. A., Cheatham T. E., III, Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Wang B., Pearlman D. A., Crowley M., Brozell S., Tsui V., Gohlke H., Mongan J., Hornak V., Cui G., Beroza P., Schafmeister C., Caldwell J. W., Ross W. S., Kollman P. A. (2004) AMBER 8, University of California, San Francisco [Google Scholar]
- 19. Yu L. Y., Jokitalo E., Sun Y. F., Mehlen P., Lindholm D., Saarma M., Arumae U. (2003) J. Cell Biol. 163, 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lindholm P., Peränen J., Andressoo J. O., Kalkkinen N., Kokaia Z., Lindvall O., Timmusk T., Saarma M. (2008) Mol. Cell. Neurosci. 39, 356–371 [DOI] [PubMed] [Google Scholar]
- 21. Mizobuchi N., Hoseki J., Kubota H., Toyokuni S., Nozaki J., Naitoh M., Koizumi A., Nagata K. (2007) Cell Struct. Funct. 32, 41–50 [DOI] [PubMed] [Google Scholar]
- 22. Bruhn H. A. (2005) Biochem. J. 389, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindholm P., Saarma M. (2010) Dev. Neurobiol. 70, 360–371 [DOI] [PubMed] [Google Scholar]
- 24. Holm L., Sander C. (1998) Nucleic Acids Res. 26, 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aravind L., Koonin E. V. (2000) Trends Biochem. Sci. 25, 112–114 [DOI] [PubMed] [Google Scholar]
- 26. Walker J. R., Corpina R. A., Goldberg J. (2001) Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 27. Tibbetts M. D., Shiozaki E. N., Gu L., McDonald E. R., 3rd, El-Deiry W. S., Shi Y. (2004) Structure 12, 2257–2263 [DOI] [PubMed] [Google Scholar]
- 28. Raaijmakers H., Törö I., Birkenbihl R., Kemper B., Suck D. (2001) J. Mol. Biol. 308, 311–323 [DOI] [PubMed] [Google Scholar]
- 29. Amsel A. D., Rathaus M., Kronman N., Cohen H. Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5117–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gama V., Gomez J. A., Mayo L. D., Jackson M. W., Danielpour D., Song K., Haas A. L., Laughlin M. J., Matsuyama S. (2009) Cell. Death Differ. 16, 758–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sawada M., Hayes P., Matsuyama S. (2003) Nat. Cell Biol. 5, 352–357 [DOI] [PubMed] [Google Scholar]
- 32. Deckwerth T. L., Elliott J. L., Knudson C. M., Johnson E. M., Jr., Snider W. D., Korsmeyer S. J. (1996) Neuron 17, 401–411 [DOI] [PubMed] [Google Scholar]
- 33. Vaughn A. E., Deshmukh M. (2007) Cell. Death Differ. 14, 973–981 [DOI] [PubMed] [Google Scholar]
- 34. Deleted in proof.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.