Abstract
The incidence and death rate of prostate cancer is increasing rapidly. In addition, the low sensitivity of prostate cancer to chemotherapy makes it difficult to treat this condition. The serine/threonine kinase Pim-1 plays an important role in cell cycle progression and apoptosis inhibition, resulting in prostate tumorigenesis. Therefore, Pim-1 inhibition has been expected to be an attractive target for developing new anti-cancer drugs. However, no small compounds targeting Pim-1 have progressed to clinical use because of their lack of specificity. Here, we have reported a new cell-permeable Pim-1 inhibitory p27Kip1 peptide that could interfere with the binding of Pim-1 to its substrates and act as an anti-cancer drug. The peptide could bind to Pim-1 and inhibit phosphorylation of endogenous p27Kip1 and Bad by Pim-1. Treatment of prostate cancer with the peptide induces G1 arrest and subsequently apoptosis in vitro. However, the peptide showed almost no growth inhibitory or apoptosis-inducing effects in normal cells. The peptide could inhibit tumor growth in in vivo prostate cancer xenograft models. Moreover, the peptide treatment could overcome resistance to taxol, one of the first line chemotherapeutic agents for prostate cancer, and a combination of the peptide with taxol synergistically inhibited prostate cancer growth in vivo. These results indicate that a Pim-1 inhibitory p27Kip1 peptide could be developed as an anti-cancer drug against prostate cancer.
Keywords: Anticancer Drug, Cancer Therapy, Crystal Structure, Peptide Interactions, Serine Threonine Protein Kinase, Pim-1, p27Kip1, Prostate Cancer
Introduction
Prostate cancer is the most common type of malignancy diagnosed in men in developed countries. Moreover, epidemiological studies indicate that the incidence and death rate of prostate cancer is increasing rapidly. In addition, the low sensitivity of prostate cancer to chemotherapy makes it more difficult to cure. Therefore, the development of new drugs against prostate cancer is an urgent requirement.
Recent evidences suggest that the proto-oncogene product Pim-1 kinase induces the initiation and progression of prostate cancer (1). The expression of Pim-1 has been reported to be up-regulated from the neoplastic state in the prostate and correlated with measures of clinical outcome in prostate cancer patients (2–4). Pim-1 could induce cell cycle progression by phosphorylating the cyclin-dependent kinase inhibitor p27Kip1 and inhibit apoptosis by phosphorylating the apoptosis-inducing factor Bad (5–7). Thus, Pim-1 seems to be an important drug target for cancer.
Several small compounds have been reported to inhibit Pim-1 (8–11). These compounds target the ATP-binding site of Pim-1, competing with the high intracellular concentration of ATP, though most of them cannot discriminate between the ATP-binding sites conserved in the protein kinase and other ATP-binding proteins. Their low specificity for Pim-1 may limit their clinical use, particularly if they have side effects. Thus, there is now an increasing interest in identifying a new type of Pim-1 kinase inhibitor that does not directly compete with ATP (12). Such inhibitors may potentially enable the selective inhibition of prostate cancer growth mediated by Pim-1 without showing severe physiological side effects.
Protein kinases have substrate preferences that are determined by the so-called recognition motif (13). This sequence represents a particular group of amino acids surrounding the phosphorylation site and is critical for substrate recognition by the protein kinase. Peptides that resemble this motif can act as substrate-competitive inhibitors. Some reports have shown that a synthetic peptide derived from a pseudo-substrate sequence had high selectivity with respect to inhibition (14–16). For example, peptides derived from the typical protein kinase A recognition motif or the GSK3 recognition motif were found to be potent selective inhibitors (14–16).
In our previous report (5), we identified the Pim-mediated p27Kip1 phosphorylation site, carboxyl-terminal Thr198 in human p27Kip1. To competitively inhibit the Pim binding to its substrates, we synthesized carboxyl terminus of human p27Kip1 containing a Pim recognition motif. We added a cell membrane-permeable polyarginine residue (Arg8) at the amino terminus of the peptide to transfer it into cells. This Arg8 peptide could inhibit Pim-1-mediated substrate phosphorylation and cell growth in vitro and in vivo. Moreover, the Arg8 peptide could overcome drug resistance in prostate cancer cells and sensitized them to taxol in vitro and in vivo. These results indicate that the peptide derived from carboxyl terminus of human p27Kip1 may be useful as a new Pim-1 kinase inhibitor, and the kinase-substrate binding sites may be promising targets for tumor treatment.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture Conditions
Human normal prostate epithelial RWPE-1 cells and human prostate carcinoma DU145 cells were purchased from ATCC. RWPE-1 cells were grown in keratinocyte serum-free medium supplemented with 0.05% bovine pituitary extract and 5 ng/ml of EGF (Invitrogen). DU145 and HEK293T cells were grown in RPMI 1640 medium (Nissui, Tokyo, Japan) supplemented with 10% FBS and 100 μg/ml of kanamycin in a humidified atmosphere of 5% CO2 and 95% air.
Transient Transfection, Immunoprecipitation, and Western Blot Analysis
Cells were transfected with the appropriate plasmids using Lipofectamine 2000 (Invitrogen). Cell lysates for immunoprecipitation and Western blot analysis and whole cell lysates were prepared as described previously (5). The nuclear and cytoplasmic fractions were separated using an NE-PER (Pierce). For immunoprecipitation, we used anti-GFP antibody (B-2; antibody sc-9996) (Santa Cruz Biotechnology) that had been conjugated with protein A-agarose. Then, the immunoprecipitated proteins or cell lysates were electrophoresed and blotted onto a nitrocellulose membrane. The membranes were incubated with antibodies to Pim-1 (12H8; antibody sc-13513), GFP (B-2; antibody sc-9996), and β-actin (C-2-HRP; antibody sc-8432 HRP) (Santa Cruz Biotechnology); p27Kip1 (610242), human Topoisomerase IIβ (611492), and Hsp90 (610418) (BD Biosciences); Bad (9292), phospho-Bad Ser112 (9290), caspase-3 (9662), and poly(ADP-ribose) polymerase (9542) (Cell Signaling); α-tubulin (YL1/2) (Serotec); phospho-Thr198 p27Kip1 (AF3994) (R&D Systems); FLAG tag M2 (A8592) (Sigma); and HA tag (3F10) (Roche Applied Science). After washing, the membranes were developed with an enhanced chemiluminescence system (GE Healthcare). Blots were scanned using Image Reader LAS-3000 mini (Fujifilm, Tokyo, Japan).
Immunostaining
DU145 cells were treated with the appropriate peptide (10 μm). After treatment for 24 h, cells were fixed with 4% formaldehyde in PBS. The endogenous p27Kip1 was detected by staining with anti-p27Kip1 antibody, following incubation with Alexa Fluor 568-conjugated anti-mouse antibody (Molecular Probes). Nuclei were also detected by staining with Hoechst 33342. Details of immunostaining have been described previously (5).
Flow Cytometric Analysis
DU145 cells were treated with synthetic fluorescein (FITC)-labeled R8 peptides (20 μm each). After treatment for 24 or 72 h, cells were stained with propidium iodide. Analyses were performed using a Cytomics 500 flow cytometer (Beckman Coulter). Details of cytometry have been described previously (5).
Estimation of Peptide Internalization
To examine internalization efficiencies of the synthetic peptides, 10 μm FITC-labeled peptides were added to the medium and incubation was continued for 4 h. The distribution of fluorescein-labeled peptides was visualized using a fluorescence microscope equipped with a CCD camera.
Estimation of Anti-tumor Activity
DU145 cells (3 × 106 cells/mouse) were implanted subcutaneously into the right flank of 5–6-week-old male BALB/c-nu/nu (nude) mice (Charles River). The mice were then euthanized, and tumors were resected and diced. A piece of tumor was sequentially injected subcutaneously into another 5–6-week-old male BALB/c nude mouse. When tumor volume was about 100 mm3, the mice were sorted into two groups. Then, the nude mice (six mice/group) were administered with the R8-T198wt peptide (50 μl of 10 mm) by intratumoral injection or the same volume of water as a control on days 0 and 3. In some experiments, taxol was also administered intravenously at doses of 60 mg/kg on day 0. Tumor mass was measured with a caliper, and tumor volumes were defined as (longest diameter) × (shortest diameter)2 divided by 2. Statistical evaluations were performed using Student's t test. All animal procedures were performed under specific pathogen-free conditions using protocols approved by the Japanese Foundation for Cancer Research Animal Care and Use Committee.
Small Interfering RNA Design and Transfection
Details of the two siRNA constructs for pim1 sequences can be found under the supplemental “Experimental Procedures.” Cells were transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen).
Statistical Analysis
All data were expressed by mean ± S.D. Student's t test or two-way analysis of variance was performed. p values <0.05 were considered statistically significant.
RESULTS
Cell-permeable T198WT Peptide Binds to Pim-1 and Suppresses Pim-1-mediated p27Kip1 Phosphorylation in Cells
Our previous study suggests that p27Kip1 is a novel substrate of Pim-1 and that Pim-1-mediated p27Kip1 phosphorylation is closely related to tumor malignancy (5). We also showed that Pim-1 strongly binds to p27Kip1 and phosphorylates the carboxyl-terminal Thr198 residue in human p27Kip1 (5). The one-dimensional sequence around p27Kip1 at the Thr198 residue (189KKPGLRRRQpT198; pT represents phosphorylated Thr) is quite different from the other identified Pim-1 phosphorylation sites (17, 18).
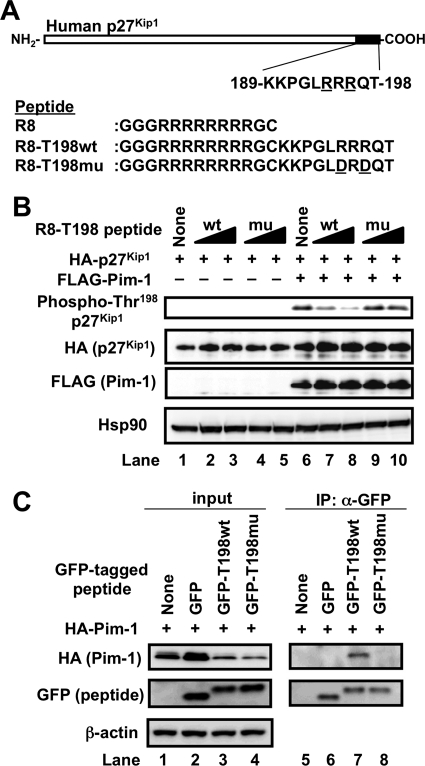
To examine the possibility that the peptide containing the Thr198 residue can act as a substrate-competitive Pim-1 inhibitor, we synthesized carboxyl-terminal p27Kip1 peptide (T198wt peptide; 189KKPGLRRRQT198). To transfer the T198WT peptide into cells, a cell membrane-permeable polyarginine residue (Arg8) was conjugated at the amino terminus of the T198wt peptide (R8-T198wt) (Fig. 1A). We also synthesized a R8-T198mu peptide in which basic amino acid (Arg) residues at Arg194 (−4) and Arg196 (−2) positions were substituted with acidic amino acid (Asp) residues because the Arg-X-Arg motif upstream of phosphorylation site was known to be important for Pim-1 recognition (Fig. 1A).
FIGURE 1.
Cell-permeable p27Kip1 peptide binds to and inhibits Pim-1. A, schematic representation of human p27Kip1 and its carboxyl-terminal T198wt peptide (amino acids 189–198). The amino acid sequences of cell-permeable Arg8, R8-T198wt, and R8-T198mu peptides used in the study are also shown. Amino acids are indicated by their single letter code. The mutated Asp194 and Asp196 residues in R8-T198mu are underlined. B, HEK293T cells were transfected with pHM6 encoding human p27Kip1 together with empty pc5FLAG (−, lanes 1–5) or pc5FLAG encoding Pim-1 (+, lanes 6–10). After transfection for 4 h, cells were incubated with 10 or 40 μm FITC-labeled R8-T198wt (wt), 10 or 40 μm FITC-labeled R8-T198mu (mu) peptide, or without any peptide (None) for 24 h. To make HA-p27Kip1 expression level equal, the amount of transfected pHM6-p27Kip1 has decreased gradually contrary to the amount of R8-T198wt peptide (lanes 2, 3, 7, and 8). Then, cell lysates were electrophoresed and immunoblotted with the indicated antibodies. C, HEK293T cells were not transfected (None) or were transfected with empty pEGFP-C2 vector (GFP), pEGFP-C2-T198wt (GFP-T198wt), or pEGFP-C2-T198mu (GFP-T198mu) together with the pHM6-Pim-1 plasmid (+). After transfection for 24 h, cells were lysed, and the GFP-tagged peptides were immunoprecipitated from the cell lysates with protein A-agarose-conjugated anti-GFP antibody. The immunoprecipitated proteins (IP) and cell lysates (input) were subjected to immunoblot analysis with the indicated antibodies.
We first examined the intracellular accumulation of FITC-labeled Arg8, R8-T198wt, and R8-T198mu in DU145 cells (supplemental Fig. S1A). Fluorescence microscopy revealed that Arg8 and Arg8-conjugated peptides had accumulated in cells. Moreover, flow cytometric analysis suggested that R8-T198wt and R8-T198mu peptides had accumulated almost equally in cells (supplemental Fig. S1B). Using these cell-permeable peptides, we investigated whether the peptides could inhibit Pim-1-mediated p27Kip1 phosphorylation. R8-T198wt treatment could enhance the HA-p27Kip1 expression level. To make HA-p27Kip1 expression level equal, the amount of transfected pHM6-p27Kip1 has decreased gradually contrary to the amount of R8-T198wt peptide (Fig. 1B, lanes 2, 3, 7, and 8). The carboxyl-terminal Thr198 residue in human p27Kip1 was phosphorylated by Pim-1 (Fig. 1B, lane 6). Phosphorylation was inhibited by the R8-T198wt peptide in a dose-dependent manner (Fig. 1B, lanes 7 and 8). In contrast, the R8-T198mu peptide could hardly suppress Pim-1-mediated p27Kip1 phosphorylation (Fig. 1B, lanes 9 and 10). Thr198 phosphorylation by Pim-1 could not be detected when the cells were cotransfected with Pim-1 and p27Kip1 T198A mutant (data not shown). These results indicate that the R8-T198wt peptide could inhibit p27Kip1 phosphorylation by Pim-1 in cells. We assumed that the inhibition was due to the blocking of the binding between Pim-1 and p27Kip1.
Next, we examined whether the T198wt peptide could form a complex with Pim-1 in cells. To study the binding, the T198wt peptide or T198mu peptide was fused with GFP (GFP-T198wt or GFP-T198mu, respectively). When these GFP-fused proteins were expressed with HA-Pim-1 in HEK293T cells, HA-Pim-1 was co-immunoprecipitated with GFP-T198wt but not with GFP-T198mu (Fig. 1C). This result suggests that in cells, Pim-1 forms a complex with T198wt but not with T198mu and that Arg194 and Arg196 residues seemed to be required for their binding. Moreover, phosphodefective T198A peptide, but not phosphomimic T198E and T198D peptides, could form a complex with Pim-1 (supplemental Fig. S2). Phosphorylation of T198wt peptide at the carboxyl-terminal Thr198 residue may down-regulate its ability to form a complex with Pim-1.
Co-crystallization of Pim-1 and T198wt Peptide
Although Pim-1 was reported to preferentially phosphorylate substrates containing basic residues, particularly arginine (Arg) at −5 and −3 positions, a basic amino acid is missing at the −5 position in human p27Kip1. Moreover, the Pim-1 phosphorylation site in human p27Kip1 loses an additional residue at the carboxyl terminus because of the phosphorylated Thr198 residue located at the end of p27Kip1 carboxyl terminus. Therefore, it remains unknown how Pim-1 binds to the p27Kip1 characteristic sequence around Thr198 and phosphorylates the Thr198 residue in human p27Kip1. To clarify the mechanism, we purified a recombinant human Pim-1 protein (amino acids 14–313) from Escherichia coli (supplemental Fig. S3A) and crystallized it with a T198wt peptide and a nonhydrolyzable ATP analog, AMP-PNP. As a result, we obtained needle-like crystals (supplemental Fig. S3B). The x-ray crystal structure of Pim-1 was determined as a complex with AMP-PNP at 1.6 Å resolution (supplemental Fig. S4A and supplemental Table S1; Protein Data Bank code 3A99). The electron density for the entire T198wt molecule was not obtained; however, clear and discontinuous electron density in the substrate-binding site indicated existence of some Pim-1-interacting residues of T198wt. When our Pim-1 structure in the presence of T198wt is superposed onto the previous one in complex with a consensus peptide substrate, pimtide (17), the electron density fits to the side chains of the two Arg residues at −5 and −3 positions of pimtide, which interact with acidic residues of Pim-1 (supplemental Fig. S4B) (17, 19). It is reasonable that, when we modeled T198wt using pimtide as a guide, the electron density is poor for the noninteracting main chain and side chain atoms, probably due to partial disorder. Thus, we predict that the T198wt binding of Pim-1 is principally mediated by the two Arg side chains, Arg194 and Arg196, which are located at −4 and −2 positions in the case of T198wt. This is consistent with our mutational analysis that GFP-T198mu could not form a complex with Pim-1 (Fig. 1C).
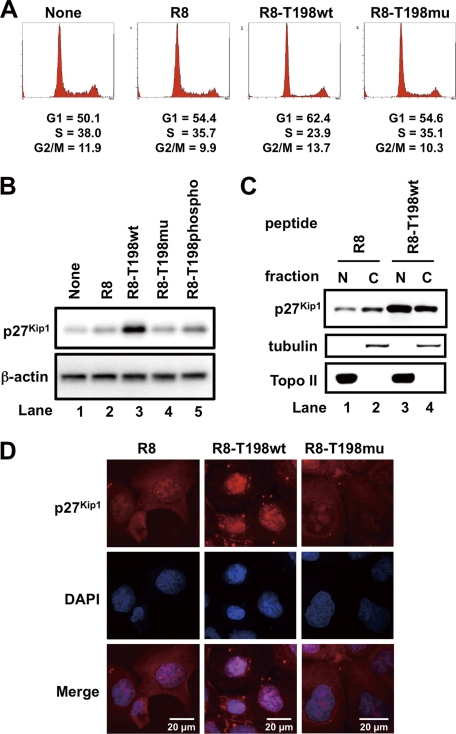
Short Term Treatment of Cells with R8-T198wt Peptide Induces G1 Cell Cycle Arrest
We previously reported that p27Kip1 phosphorylation at the Thr198 residue stimulates cytosolic localization and ubiquitin-proteasome-dependent degradation of p27Kip1 (5). Moreover, we also showed that nuclear accumulation of p27Kip1 induces cell cycle arrest at G1 phase (5). Thus, our observation that the R8-T198wt peptide attenuates Pim-1-mediated p27Kip1 phosphorylation (Fig. 1B) prompted us to investigate whether the T198wt peptide stimulates cell cycle arrest at G1 phase and nuclear accumulation of p27Kip1. Flow cytometric analysis revealed that increase in the number of cells in G1 phase was due to the treatment of DU145 cells with R8-T198wt, but not with Arg8 and R8-T198mu (Fig. 2A). Treatment of these cells with the R8-T198wt peptide but not R8-T198mu enhances the expression of p27Kip1 (Fig. 2B) and the nuclear accumulation of p27Kip1 (Fig. 2, C and D). Because phosphorylated peptide R8-T198phospho slightly enhanced the expression of p27Kip1 (Fig. 2B), we tried to prove direct interactions between p27Kip1 peptides and Pim-1 kinase by SPR analysis. As shown in supplemental Fig. S5, R8-T198wt peptide (KD = 323 nm) but not R8-T198mu peptide (KD = 4.25 μm) showed significant affinity for Pim-1. R8-T198phospho showed weak but significant binding ability to Pim-1 (KD = 968 nm), indicating that R8-T198wt peptide may partially bind to Pim-1 even after Thr198 phosphorylation. These results suggest that the peptide inhibit Pim-1 kinase activity through substrate competition. In these experiments, we treated DU145 cells with 20 μm peptides for 24 h so that the cyclin-dependent kinase inhibitory effect of p27Kip1 could be easily detected. These results suggest that short term treatment of cells with the R8-T198wt peptide up-regulates p27Kip1 protein and induces G1 arrest.
FIGURE 2.
Short term treatment of DU145 cells with cell-permeable p27Kip1 peptide up-regulates p27Kip1 protein and induces G1 arrest. A, DU145 cells were treated with 20 μm FITC-labeled cell-permeable Arg8, R8-T198wt, or R8-T198mu peptide, or not treated with any peptide (None). After incubation for 24 h, cells were stained with propidium iodide. Analyses were performed using a flow cytometer. B, expression levels of endogenous p27Kip1 and β-actin proteins in cells treated with 20 μm FITC-labeled cell-permeable Arg8, R8-T198wt, R8-T198mu, or R8-T198-phosphopeptide, or not treated with any peptide (None). Expression of p27Kip1 and β-actin were confirmed by immunoblotting. C, DU145 cells were treated with 20 μm FITC-labeled Arg8 or R8-T198wt peptide. After incubation for 24 h, cells were harvested and lysed. The cytoplasmic (C) and nuclear (N) fractions were separated, electrophoresed, and immunoblotted with the indicated antibodies. Topo II, Topoisomerase II. D, localization of endogenous p27Kip1 protein in cells treated as in C was confirmed by immunostaining with anti-p27Kip1 antibody. Nuclei were detected by staining with Hoechst 33342.
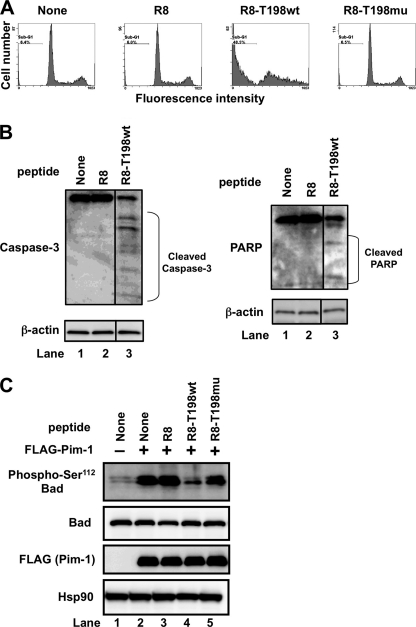
Long Term Treatment of Cells with R8-T198wt Peptide Induces Apoptotic Cell Death
We also checked the morphological changes in DU145 cells after treatment with 20 μm peptides for 72 h. Under the microscope, we observed the apoptotic cell death of DU145 cells treated with R8-T198wt. Flow cytometric analysis revealed an increase in the sub-G1 fraction of cells treated with the R8-T198wt peptide but not with R8-T198mu (Fig. 3A). Treatment of these cells with R8-T198wt induces caspase-3 activation and increases the amount of cleaved poly(ADP-ribose) polymerase fragments (Fig. 3B). These results suggest that the R8-T198wt peptide could induce apoptosis in DU145 cells.
FIGURE 3.
Long term treatment of DU145 cells with cell-permeable p27Kip1 peptide induces apoptosis. A, DU145 cells were treated with 20 μm FITC-labeled cell-permeable Arg8, R8-T198wt, or R8-T198mu peptide or not treated with any peptide (None). After incubation for 72 h, cells were stained with propidium iodide. Analyses were performed using a flow cytometer. B, expression levels of endogenous caspase-3, poly(ADP-ribose) polymerase, and β-actin proteins in cells treated as in A were confirmed by immunoblotting. C, HEK293T cells were cultured in serum-free medium for 24 h. Cells were then transfected with empty pc5FLAG alone (−) or pc5FLAG encoding Pim-1 (+). After transfection for 4 h, medium was replaced with serum-free medium containing none (None) or 40 μm FITC-labeled Arg8, R8-T198wt, or R8-T198mu peptide and incubated for an additional 24 h. Then, cell lysates were electrophoresed and immunoblotted with the indicated antibodies.
The apoptosis-inducing factor Bad was reported to be an important substrate of Pim-1 (6, 7). Pim-1 phosphorylates Bad at the Ser112 residue and inhibits its apoptosis-inducing activity (Fig. 3C, lane 2). Phosphorylation was inhibited by addition of R8-T198wt peptide (Fig. 3C, lane 4) but not by Arg8 or R8-T198mu peptide (Fig. 3C, lanes 3 and 5, respectively). These results suggest that the R8-T198wt peptide triggers apoptotic cell death after G1 arrest by suppressing Bad phosphorylation.
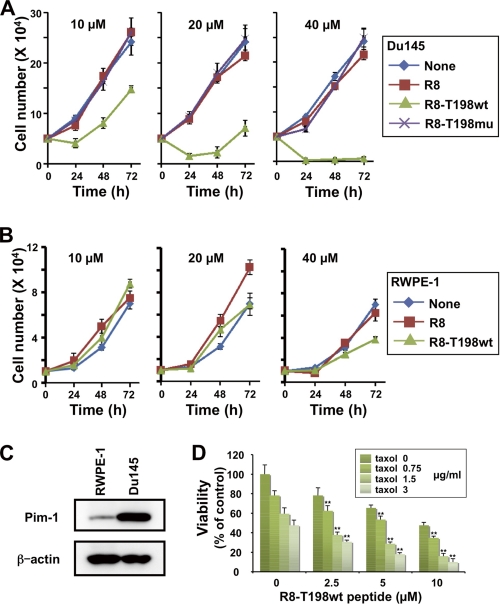
R8-T198wt Peptide Inhibits Growth of Prostate Cancer Cells
Because R8-T198wt peptide induces G1 arrest and apoptosis, we investigated the effect of this peptide on the growth of prostate cancer cells. In this experiment, we used prostate cancer DU145 cells because their growth strongly depended on Pim-1 activity (20). Consistent with the results shown in Figs. 2A and 3A, treatment with the R8-T198wt peptide inhibited the growth of DU145 cells in a dose-dependent manner (Fig. 4A), whereas Arg8 and R8-T198mu peptides had no effect on the growth of DU145 cells (Fig. 4A). We also tested the growth inhibitory effects of the R8-T198wt peptide in normal prostate epithelial RWPE-1 cells. As shown in Fig. 4B, 10 and 20 μm R8-T198wt peptide did not affect the growth of RWPE-1 cells, whereas 40 μm R8-T198wt peptide affected the growth of these cells but to a lesser extent. Thus, the R8-T198wt peptide inhibited the growth of prostate cancer cells more than that of normal prostate cells.
FIGURE 4.
Growth inhibitory effect of cell-permeable p27Kip1 peptide. A, DU145 cells were treated with the indicated concentrations of FITC-labeled cell-permeable Arg8, R8-T198wt, or R8-T198mu peptide. Viable cells were counted after incubation for 24, 48, and 72 h. Each vertical bar represents the mean ± S.D. of three independent experiments. B, RWPE-1 cells were treated as in A. Viable cells were counted after incubation for 24, 48, and 72 h. Each bar represents the mean ± S.D. of three independent experiments. C, total cell lysates of RWPE-1 and DU145 cells were electrophoresed and immunoblotted with the indicated antibodies. D, DU145 cells were incubated in a medium containing the indicated concentrations of the FITC-labeled R8-T198wt peptide (0–10 μm) together with the indicated concentrations of taxol (0–3 μg/ml). Viable DU145 cells were counted by the trypan blue dye exclusion method. The number of viable untreated control cells was normalized to 100%. Each vertical bar represents the mean ± S.D. of three independent experiments. The interaction of various concentrations of the R8-T198wt peptide and taxol was subjected to two-way analysis of variance. *, p < 0.05; **, p < 0.01.
To confirm that cell growth inhibition by the R8-T198wt peptide depended on Pim-1, we first compared Pim-1 expression levels between DU145 and RWPE-1 cells by Western blot analysis. As shown in Fig. 4C, the expression of Pim-1 protein in DU145 cells was significantly higher than that in RWPE-1 cells. Moreover, Pim-1 knockdown attenuates R8-T198wt peptide-mediated growth inhibition of DU145 cells (supplemental Fig. S6). These results indicate that the R8-T198wt peptide suppresses cell growth and induces cell cycle arrest and apoptosis by inhibiting Pim-1 in DU145 cells.
Taxol is an important chemotherapeutic drug used in the treatment of patients with hormone-resistant prostate cancer. Pim-1 expression was reported to be up-regulated by docetaxel, and it served as an inhibitor of drug-induced apoptosis (21). Therefore, we investigated whether the R8-T198wt peptide could restore the sensitivity of prostate cancer DU145 cells to taxol. Two-way analysis of variance is frequently used to statistically determine whether these two agents act synergistically when eliciting a biological response (20). In this experiment, we used the same strategy to examine the effect of a combination of R8-T198wt peptide and taxol on growth inhibition of DU145 cells. As shown in Fig. 4D, the interaction among various combinations of R8-T198wt and taxol was examined and analyzed statistically by two-way analysis of variance based on the results of DU145 cell viability. A statistically significant synergistic interaction (**, p < 0.01) was seen with the combination of R8-T198wt and taxol. Our results indicate that the R8-T198wt peptide can overcome taxol resistance in DU145 cells by inhibiting Pim-1, which leads to suppression of growth of prostate cancer cells.
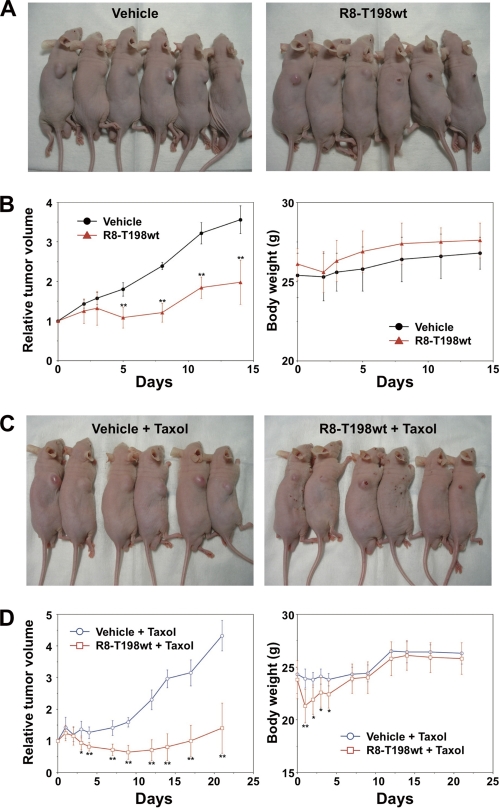
In Vivo Anti-tumor Activity of R8-T198wt Peptide in Combination with a Chemotherapeutic Agent in Human Prostate Cancer DU145 Xenografts
To test whether the R8-T198wt peptide could inhibit tumor growth and increase the sensitivity of such tumor cells to the chemotherapeutic agent taxol in vivo, we developed DU145 xenografts in nude mice. Mice were treated with vehicle or R8-T198wt peptide (Fig. 5, A and B) and taxol alone or taxol and R8-T198wt peptide (Fig. 5, C and D). Tumor size was significantly smaller in the group treated with the R8-T198wt peptide compared with the vehicle-treated group, with a reduction of 40 and 43% tumor volume on days 5 and 14, respectively (n = 6, p < 0.05) (Fig. 5, A and B, left). No toxic death or significant body weight change was observed throughout these experiments (Fig. 5B, right). Co-treatment with taxol and R8-T198wt peptide showed a marked inhibitory effect on the development of DU145 xenografts compared with taxol alone, with a reduction of 52 and 61% tumor volume on days 9 and 28, respectively (n = 6, p < 0.05) (Fig. 5, C and D, left). Although toxic death was not observed in this experiment, a slight decrease in body weight was observed in the R8-T198wt peptide and taxol group (Fig. 5D, right). In this experiment, both taxol and the vehicle when given alone did not show an inhibitory effect (data not shown). These results suggest that the R8-T198wt peptide has in vivo anti-tumor activity and can increase sensitivity to taxol with minimal toxicity in mice.
FIGURE 5.
In vivo anti-tumor activity of p27Kip1 peptide. A and B, DU145 cells were subcutaneously implanted into the right flank of BALB/c nude mice. Therapeutic experiments (six mice/group) were started (day 0) when the tumor reached a volume of ∼100 mm3. The R8-T198wt peptide was administered by intratumoral injection in 50 μl of 10 mm solution on days 0 and 3. The control group received the same volume of vehicle on days 0 and 3. A, representative surface tumor morphology of the mice on day 8 after initiation of treatment (left, vehicle; right, R8-T198wt peptide). B, relative tumor volume and body weight. C and D, R8-T198wt peptide (50 μl of 10 mm) or the same volume of vehicle was administered intratumorally on days 0 and 5. All DU145-bearing mice were intravenously administered 60 mg/kg taxol on day 0. C, representative surface tumor morphology on day 14 after initiation of treatment (left, vehicle and taxol; right, R8-T198wt peptide and taxol). D, relative tumor volume and body weight. *, p < 0.05; **, p < 0.01.
DISCUSSION
We report here the identification and characterization of a peptide derived from carboxyl-terminal human p27Kip1 that contained the Pim-1 phosphorylation site at the Thr198 residue (T198wt peptide). The peptide induces cell cycle arrest at G1 phase and subsequently apoptosis by inhibiting Pim-1 activity. We previously identified p27Kip1 as a novel substrate of Pim-1 (5). Pim-1 formed a complex with p27Kip1 and phosphorylated the carboxyl-terminal Thr198 residue more preferentially than the other p27Kip1 residues in cells (5). We also reported that the primary sequence around the Thr198 residue was different from the previously reported Pim-1 consensus sequence, in which Arg residues were located at −5 and −3 positions around the phosphorylation site (17, 19). In addition, the phosphorylated Thr198 residue in human p27Kip1 lacked the additional residue at the carboxyl terminus found in the Pim-1 consensus sequence (17, 19). Moreover, the one-dimensional sequence around the Thr198 residue in the human p27Kip1 (KKPGLRRRQpT198; pT represents phosphorylated Thr) was different from that of other low affinity substrates, such as p21Cip1 (RKRRQTpSMTD; pS represents phosphorylated Ser) and PAP1 (KKRKHKApSKSS), which were phosphorylated by Pim-1 (17). From these results, it could be assumed that Pim-1 preferentially binds to the sequence around the carboxyl terminus of human p27Kip1. However, it is not known how Pim-1 binds to and phosphorylates the Thr198 residue in human p27Kip1. We then investigated the crystal structure of Pim-1 in the presence of 10 carboxyl-terminal amino acids of the human p27Kip1 peptide containing the Thr198 residue (T198wt). Although the entire electron density of T198wt could not be observed because of partial disorder in the Pim-1 structure, a possible recognition of the T198wt Arg194 and Arg196 side chains (−4 and −2 positions) by Pim-1 was suggested. This result was different from the previous reports in that Arg residues at −5 and −3 positions around the phosphorylation site proved to be important for Pim-1-mediated phosphorylation (17, 19). We speculate that substrate binding of Pim-1 largely depends on two Arg residues but allows some structural flexibility around the phosphorylation site.
On the basis of the fact that arginine-rich peptides have efficient translocation activity through cell membranes (22), we synthesized the T198wt peptide and conjugated it with eight arginine residues (Arg8) at the amino terminus to form R8-T198wt. The R8-T198wt peptide interacted with Pim-1 (Fig. 1B) and inhibited phosphorylation of p27Kip1 and Bad by Pim-1 (Figs. 1B and 3C, respectively) in cells. Moreover, the R8-T198wt peptide induced cell cycle arrest at G1 phase (Fig. 2A) and apoptosis (Fig. 3A). The G1 arrest may depend on the nuclear accumulation of p27Kip1 (Fig. 2, C and D), and the apoptosis may depend on the loss of Bad phosphorylation, leading to the activation of proapoptotic Bad (Fig. 3C). Although the expression of cell cycle inhibitor p27Kip1 increased, the apoptotic signal via inhibiting Bad was not observed after 24 h of treatment. Thus, we mainly detected G1 arrest phenotype (Fig. 2A). Indeed, we could not detect the caspase-3 activation at 24 h (data not shown). After treatment for 48 h, cells exhibited apoptotic phenotype (Fig. 3A). It might due to the caspase-3 activation (Fig. 3B). Late caspase-3 activation might generate such different cellular responses to R8-T198wt peptide.
Because pimtide itself could not be transferred into cells, it could not induce G1 arrest or caspase activation. Adding membrane-permeable polyarginine residue to pimtide may improve its transfer into cells and exhibit the similar effects on the cell cycle arrest and apoptosis like the R8-T198wt peptide.
The R8-T198wt peptide strongly inhibited the in vitro and in vivo proliferation of prostate cancer DU145 cells, whose growth was dependent on Pim-1 activity (20, 23) (Fig. 4). Because the pim1 knockdown abrogated the inhibitory effect of the R8-T198wt peptide in DU145 cells (supplemental Fig. S6), the R8-T198 peptide suppresses the growth of DU145 cells by inhibiting Pim-1 activity. This notion was supported by the fact that the R8-T198wt peptide showed little inhibitory effect on the growth of normal RWPE-1 cells in which the Pim-1 expression level was low (Fig. 4C). The high selectivity might depend on the survival addiction to the proto-oncogene Pim-1 in DU145 cells but not in RWPE-1 cells. In addition, pim1 transfection reduced the growth inhibitory effect of the R8-T198wt peptide (data not shown). These facts indicate the specificity of the R8-T198wt peptide to Pim-1. These results suggest that the treatment with the R8-T198wt peptide could have a broad safety margin between normal and tumor cells. However, we could not exclude the possibility that this peptide inhibited other kinases such as Akt and RSK that could phosphorylate human p27Kip1 at the Thr198 residue (24, 25). Some modifications of the R8-T198wt peptide may improve and increase the specificity for Pim-1, as in the case of GSK3β (16).
The R8-T198wt peptide synergistically increases the cytotoxicity of the chemotherapeutic agent taxol in prostate cancer DU145 cells (Figs. 4D and 5). The emergence of hormone-resistant prostate cancer is a serious problem because of the lack of an effective therapeutic agent or method. Drug resistance is a major obstacle in the management of prostate cancer. Pim-1 up-regulation was frequently observed after cytotoxic drug treatment, and this might be one of the mechanisms of drug resistance (21). Therefore, Pim-1 inhibition by the R8-T198wt peptide could overcome this drug resistance.
We have shown the potency of the R8-T198wt peptide as a selective inhibitor of Pim-1 kinase. However, it is possible that this peptide can also inhibit other kinases of the Pim family, such as Pim-2 and Pim-3. We have previously shown that not only Pim-1 but also Pim-2 and Pim-3 can directly phosphorylate p27Kip1 at the Thr198 residue (5). Moreover, Bullock et al. (17, 19) reported that pimtide could bind to Pim-1, Pim-2, and Pim-3 with high affinity. Therefore, we surmised that the peptide might regress leukemia, lymphoma, and pancreatic cancer, the development of which depends on Pim-2 and Pim-3.
Supplementary Material
Acknowledgments
We thank Drs. Masaki Yamamoto (BL26B2, SPring-8), Go Ueno (BL26B2, SPring-8), and Shunji Goto (SPring-8/Japan Synchrotron Radiation Research Institute) for providing the opportunity to collect diffraction data using the beamline optics of the newly developed dynamic sagittal monochromator. We also thank GE Healthcare for SPR data analysis.
Footnotes
This work was supported in part by a grant-in-aid for Challenging Exploratory Research from the Japan Society for the Promotion of Science, by Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology, Japan and by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, Japan (to N. F.).

The on-line version of this article (available at http://www.jbc.org) contains “Experimental Procedures,” Tables S1 and S2, Figs. S1–S6, and additional references.
The atomic coordinates and structure factors (code 3A99) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
REFERENCES
- 1. Dhanasekaran S. M., Barrette T. R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2001) Nature 412, 822–826 [DOI] [PubMed] [Google Scholar]
- 2. Xu Y., Zhang T., Tang H., Zhang S., Liu M., Ren D., Niu Y. (2005) J. Surg. Oncol. 92, 326–330 [DOI] [PubMed] [Google Scholar]
- 3. Cibull T. L., Jones T. D., Li L., Eble J. N., Ann Baldridge L., Malott S. R., Luo Y., Cheng L. (2006) J. Clin. Pathol. 59, 285–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Valdman A., Fang X., Pang S. T., Ekman P., Egevad L. (2004) Prostate 60, 367–371 [DOI] [PubMed] [Google Scholar]
- 5. Morishita D., Katayama R., Sekimizu K., Tsuruo T., Fujita N. (2008) Cancer Res. 68, 5076–5085 [DOI] [PubMed] [Google Scholar]
- 6. Aho T. L., Sandholm J., Peltola K. J., Mankonen H. P., Lilly M., Koskinen P. J. (2004) FEBS Lett. 571, 43–49 [DOI] [PubMed] [Google Scholar]
- 7. Macdonald A., Campbell D. G., Toth R., McLauchlan H., Hastie C. J., Arthur J. S. (2006) BMC Cell. Biol. 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bullock A. N., Debreczeni J. E., Fedorov O. Y., Nelson A., Marsden B. D., Knapp S. (2005) J. Med. Chem. 48, 7604–7614 [DOI] [PubMed] [Google Scholar]
- 9. Cheney I. W., Yan S., Appleby T., Walker H., Vo T., Yao N., Hamatake R., Hong Z., Wu J. Z. (2007) Bioorg. Med. Chem Lett. 17, 1679–1683 [DOI] [PubMed] [Google Scholar]
- 10. Holder S., Lilly M., Brown M. L. (2007) Bioorg. Med. Chem. 15, 6463–6473 [DOI] [PubMed] [Google Scholar]
- 11. Holder S., Zemskova M., Zhang C., Tabrizizad M., Bremer R., Neidigh J. W., Lilly M. B. (2007) Mol. Cancer Ther. 6, 163–172 [DOI] [PubMed] [Google Scholar]
- 12. Bogoyevitch M. A., Fairlie D. P. (2007) Drug Discovery Today 12, 622–633 [DOI] [PubMed] [Google Scholar]
- 13. Kaidanovich-Beilin O., Eldar-Finkelman H. (2006) Physiology 21, 411–418 [DOI] [PubMed] [Google Scholar]
- 14. Glass D. B., Cheng H. C., Kemp B. E., Walsh D. A. (1986) J. Biol. Chem. 261, 12166–12171 [PubMed] [Google Scholar]
- 15. Cheng H. C., Kemp B. E., Pearson R. B., Smith A. J., Misconi L., Van Patten S. M., Walsh D. A. (1986) J. Biol. Chem. 261, 989–992 [PubMed] [Google Scholar]
- 16. Plotkin B., Kaidanovich O., Talior I., Eldar-Finkelman H. (2003) J. Pharmacol. Exp. Ther. 305, 974–980 [DOI] [PubMed] [Google Scholar]
- 17. Bullock A. N., Debreczeni J., Amos A. L., Knapp S., Turk B. E. (2005) J. Biol. Chem. 280, 41675–41682 [DOI] [PubMed] [Google Scholar]
- 18. Pogacic V., Bullock A. N., Fedorov O., Filippakopoulos P., Gasser C., Biondi A., Meyer-Monard S., Knapp S., Schwaller J. (2007) Cancer Res. 67, 6916–6924 [DOI] [PubMed] [Google Scholar]
- 19. Peng C., Knebel A., Morrice N. A., Li X., Barringer K., Li J., Jakes S., Werneburg B., Wang L. (2007) J. Biochem. 141, 353–362 [DOI] [PubMed] [Google Scholar]
- 20. Hu X. F., Li J., Vandervalk S., Wang Z., Magnuson N. S., Xing P. X. (2009) J. Clin. Invest. 119, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zemskova M., Sahakian E., Bashkirova S., Lilly M. (2008) J. Biol. Chem. 283, 20635–20644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang L., Mashima T., Sato S., Mochizuki M., Sakamoto H., Yamori T., Oh-Hara T., Tsuruo T. (2003) Cancer Res. 63, 831–837 [PubMed] [Google Scholar]
- 23. Chen W. W., Chan D. C., Donald C., Lilly M. B., Kraft A. S. (2005) Mol. Cancer Res. 3, 443–451 [DOI] [PubMed] [Google Scholar]
- 24. Fujita N., Sato S., Katayama K., Tsuruo T. (2002) J. Biol. Chem. 277, 28706–28713 [DOI] [PubMed] [Google Scholar]
- 25. Fujita N., Sato S., Tsuruo T. (2003) J. Biol. Chem. 278, 49254–49260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.