Abstract
Heat shock factor 1 (HSF1) mediates the cellular response to stress to increase the production of heat shock protein (HSP) chaperones for proper protein folding, trafficking, and degradation; failure of this homeostatic mechanism likely contributes to neurodegeneration. We show that the neuroprotective drug riluzole increased the amount of HSF1 in NG108-15 neuroprogenitor cells by slowing the specific turnover of HSF1 and supporting a more robust and sustained activation of HSF1. Using Hsp70-luciferase as a functional readout of the activity of HSF1, we show that riluzole amplified the heat shock induction of the reporter gene with an optimal increase at 1 μm. Immunocytochemical staining and Western blot quantitation of HSP70 in NG108-15 neuroprogenitor cells and embryonic spinal cord neurons provided corroborative evidence that riluzole amplified the HSF1-dependent regulation of HSP70 expression. Parallel studies on the GLT1 glutamate transporter showed that riluzole increased GLT1-reporter and GLT1 protein expression and that the increase was enhanced by heat shock and coincident with the increased expression of HSP70 and HSP90. This result is consistent with the anti-glutamatergic profile of riluzole and the presence of multiple heat shock elements on the GLT1 gene promoter, suggesting that riluzole may modulate GLT1 expression through HSF1. The increased HSP chaperones and GLT1 transporter blunted glutamate-induced and N-methyl d-aspartate receptor-mediated excitotoxic death. In summary, we show that riluzole increased the amount and activity of HSF1 to boost the expression of HSPs and GLT1 for neuroprotection under stress.
Keywords: Amyotropic Lateral Sclerosis (Lou Gehrig Disease), Heat Shock Protein, Neurodegeneration, Neuron, Neurotransmitter Transport, Protein Turnover
Introduction
Induction of the heat shock response (HSR2; also known as stress response) is a universally important quality control mechanism in protein homeostasis; it is a primary and evolutionarily conserved response to diverse stressors, mediated by activation of the HSF1 transcription factor, culminating in the induction of a family of heat shock proteins (HSPs) that function as chaperones to assist in the proper folding of non-native proteins and proteases to help in the degradation of damaged proteins for the protection and recovery from cellular damages (1–3). The biological importance of HSR is underscored by genetic evidence linking stress and protein homeostasis with the health and life span of the organism (4–6) and by the increasing appreciation that problems in protein folding and aggregation likely contribute to the pathogenesis of a number of age-dependent neurodegenerative diseases that include amyotrophic lateral sclerosis (ALS), Huntington, Parkinson, Alzheimer, and prion diseases (6–10).
We are interested in harnessing the cytoprotective activity of HSR/HSF1 to promote cell survival under stress. In particular, we are interested in identifying agents that are not overtly proteotoxic and would not by themselves induce a robust HSR but would amplify the HSR and enhance the expression of HSPs. Riluzole is the only Food and Drug Administration-approved drug for ALS. The mode of action appears to be antiglutamatergic, but the detailed mechanism is not clear as some other anti-glutamate drugs that have not been shown to be effective in the treatment of ALS (11). In a previous study on HeLa cells, we showed that riluzole increases the amount of HSF1 for cytoprotection (12). Here, we present results of the effect of riluzole on HSF1 in neuronal cell systems and show that in addition to boosting expression of HSPs, HSF1 may mediate the induction of GLT1 glutamate transporter. The increased HSPs and GLT1 support neuronal cell survival under stress.
EXPERIMENTAL PROCEDURES
Materials
Riluzole was obtained from Sigma and AB Chem Technologies. Both sources of riluzole gave similar results, although the Sigma compound appeared to have a greater cytotoxic effect at concentrations >10 μm. The Hsp70-firefly luciferase reporter gene construct was as described previously (13). The human GLT1-firefly luciferase was a generous gift from Dr. Jeffrey Rothstein, The Johns Hopkins Medical School, and it was constructed by inserting a 2472-bp human GLT1 promoter into the KpnI/HindIII site of the pGL4 luciferase reporter vector from Promega (14). Lipofectamine 2000 reagent used for DNA transfection was from Invitrogen. Humanized Renilla DNA (phRLSV40), Dual-Glo® luciferase assay reagent (E2920), CellTiter-Glo® (G7571), and Caspase-Glo®3/7 (G8091) assay reagents were from Promega. Rabbit polyclonal antibody against HSF1, either the RTG88 antibody (1:10,000 dilution) that we generated against a recombinant histidine-tagged human HSF1 protein (12, 15) or obtained from Cell Signaling Technology (1:1000 dilution; catalog no. 4356), was used in immuno-Western blot detection and quantitation of HSF1. Other antibodies used include a rabbit polyclonal antibody against the heat-inducible HSP72 protein (1:10,000 for Western blot and 1:200 for immunocytochemistry; catalog no. SPA812, StressGen), a rabbit monoclonal antibody against HSP90 (Cell Signaling 4877), a rabbit polyclonal antibody against GLT1 (also known as EAAT2; catalog no. sc-15317, Santa Cruz Biotechnology), and a mouse monoclonal antibody against EAAT2 (catalog no. ab61859, Abcam). The composition of radioimmunoprecipitation assay (RIPA) buffer for cell extract preparation was as follows: 150 mm NaCl, 10 mm Tris, pH 7.2, 0.1% SDS, 1.0% Triton X-100, 1% deoxycholate, 5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin, 100 μm sodium orthovanadate. [35S]Methionine/cysteine was from PerkinElmer Life Sciences (NEG7720). All other biochemical and chemicals were of molecular biology or reagent grade.
Cell Culture and Conditions of Riluzole Treatment and Heat Shock
The NG108-15 and N2a cell lines were used as neuroprogenitor cell models as these cells can differentiate into neuron-like cells and acquire neuron-specific phenotypes, including voltage-gated sodium channels and the ability to synapse with co-cultured muscle cells (15–17). Cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Mediatech Inc.) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Inc.). Cells were subcultured at or near confluency by minimal trypsinization (0.25% trypsin; Mediatech Inc.) and dispersion into single cell suspension in new growth medium and plating onto new growing surfaces. To induce neuronal differentiation, cells were plated into DMEM containing 2% FBS supplemented with 1 mm dibutyryl cAMP (15–17). Unless indicated otherwise, cells in early stationary phase of growth were used as follows: confluent undifferentiated cells or differentiated cells 2–3 days after dibutyryl cAMP treatment.
Primary embryonic spinal cord neuron cultures were prepared as described (18). Briefly, spinal cords were dissected from embryonic day 16 (E16) rat embryos. Meninges were removed, and tissues were dissociated with gentle trituration. Cells were plated at a density of 60,000–100,000 neurons/cm2 in Neurobasal medium (Invitrogen) supplemented with B-27, penicillin, and streptomycin. Experiments were done using neurons at 7–10 days in vitro.
For riluzole treatment, a 100 mm stock in dimethyl sulfoxide (DMSO) was serially diluted with either Dulbecco's phosphate-buffered saline or DMEM to a 10× working stock and added to cell culture medium and incubated at 37 °C for the time indicated. An equivalent volume of the vehicle was added to the control. Unless indicated otherwise, heat shock was at 42 °C for a specified time period beginning at 12–18 h after the addition of riluzole. Cells were either harvested immediately after heat shock for Western blot detection of HSF1 or allowed to recover at 37 °C for time periods indicated for analysis of the induction of Hsp70-firefly luciferase reporter gene and HSP70/HSP90/GLT1 protein expression.
Western Blot and Immunocytochemistry
Cells were pelleted by centrifugation, and extracts were prepared using either RIPA buffer or lysed and solubilized directly and completely in a 2× SDS-PAGE sample buffer. For Western blot, aliquots of the cell extracts containing the same amount protein (∼10 μg/lane) were size-fractionated using an 8 or a 6–15% gradient SDS-polyacrylamide gel, and proteins on the gel were transferred to a PVDF membrane. Western blot detection of HSF1 and HSP70 was done as described previously (12, 15, 16). Detection of the HSP90 protein was performed using a rabbit monoclonal antibody (1:1000 dilution; catalog no. 4877, Cell Signaling). GLT1 was probed with either sc-15317 (1:200 dilution; SCBT) or ab61859 (1:500 dilution; Abcam); both gave similar results, although the Abcam antibody gave a lower background. To affirm even loading of protein, membrane was co-probed for actin (42 kDa) or Sirt1 (110 kDa), and select PVDF membrane was stained with Coomassie Blue after Western blot detection.
For immunocytochemical staining of the 72-kDa heat-inducible HSP70, we used SPA-812 rabbit polyclonal antibody from StressGen at 1:200 dilution and incubation at 4 °C for 1 h, followed by FITC-conjugated goat anti-rabbit IgG secondary antibody for 60 min at 4 °C. Cells were viewed using a Nikon Diaphot 300 microscope, and images were captured with a SPOT camera system (Diagnostic Instruments, Inc., Sterling Heights, MI).
Determination of the Turnover of HSF1 and General Protein by [35S]Methionine Labeling and Chase
NG108-15 cells were labeled with 300 μCi/ml of [35S]methionine and -cysteine (Amersham Biosciences Pro-Mix, a 70:30% mixture of [35S]methionine and [35S]cysteine) for 2 h at 37 °C. At the end of this labeling period, cells were rinsed extensively and refurbished with DMEM containing 2 mm each of cysteine and methionine to initiate chase. To test for the effects of riluzole on the turnover of 35S-labeled HSF1, it was added to designated plates to a final concentration of 2 μm at the beginning of the chase. Cells were harvested at 0, 4, 8, 12, and 24 h after initiation of the chase in the absence versus the presence of 2 μm riluzole. The cell pellets were resuspended in 200 μl of RIPA buffer, and an aliquot (∼50 μl) of the RIPA extract was used for the immunoprecipitation of HSF1. For this, anti-HSF1 antibody was added to the cell lysate and incubated at 4 °C overnight. The antigen-antibody complex was precipitated by the addition of insoluble protein A and incubation at 25 °C for 2 h. The immunoprecipitate was collected by centrifugation and washed three times each with 200 μl of RIPA buffer. The amount of radioactivity present in the immunoprecipitate was determined by liquid scintillation counting. Statistical analysis was by one-way ANOVA using the GraphPad InStat program, with p > 0.05 as not significant, between 0.01 and 0.05 as significant, and <0.01 being highly significant.
To assess turnover of total cellular protein, cells were labeled with 10 μCi/ml [35S]methionine/cysteine at 37 °C for 48 h. Cells were then rinsed extensively and refurbished with DMEM containing 2 mm each of cysteine and methionine to initiate the chase. To test for the effects of riluzole on the turnover/decay of the 35S-labeled cellular proteins, it was added to designated plates to a final concentration of 2 μm at the beginning of the chase. Aliquots (in quadruplicate) of the cell culture medium were removed at 2, 4, 6, and 24 h after initiation of the chase, and the amount of trichloroacetic acid soluble radioactivity was determined according to published methods (19).
Reporter Gene Assay
The two reporter genes used in this study are Hsp70-firefly luciferase and GLT1-firefly luciferase. Reporter gene activity was assayed using standard transient transfection protocol; the reporter DNA was transfected along with the internal control of phRLSV40 (synthetic humanized Renilla luciferase DNA). Unless indicated otherwise, freshly plated 80–90% confluent cells were used, and the amount of each DNA was 0.5 μg/35-mm plate or 1.5 μg/60-mm plate, and the amount of Lipofectamine 2000 (in μl) was three times that of the total amount of DNA (in micrograms). 6 h after DNA transfection, cells were trypsinized and aliquoted into individual wells of a 96 StripwellTM plate (Corning/Costar 9102); these identically transfected cells allowed for testing of the effects of riluzole and heat shock on reporter gene expression. To evaluate the effects of heat shock on reporter gene activity, strips of eight wells or designated wells of cells were placed in a 42 °C incubator for 2 h followed by recovery at 37 °C for 4 h (or as indicated otherwise) prior to harvesting. Riluzole, when used, was added 16 h prior to heat shock unless indicated otherwise. Result is the average ± S.D. of four separate determinations from the same pool of transfected cells. Within each experiment, the reporter gene signal was tight with an ∼10% sample-to-sample variation. Between experiments, there is a range in the magnitude of heat shock induction of reporter gene activity as we have previously reported for the hsp70-reporter (heat shock/control from 20- to >100-fold) (12, 15, 16). Variation in the basal HSF1 activity (20) and differences in the cell status (confluency, days in culture, etc.) likely contributed to this variation.
The Dual-Glo® luciferase assay reagent system from Promega (E2920) was used to assay for first the firefly and then the Renilla luciferase activity according to manufacturer's instructions. Luciferase activity was measured using the PerkinElmer Life Sciences Victor 2 multiplate reader equipped with dual injectors. Where indicated, results of the Hsp70-firefly luciferase activity (in relative luminescence units) was normalized against that of the Renilla luciferase (relative luminescence units), and to facilitate comparison across experiments for statistical analysis across experiments, this ratio was set at 1 for the control (without heat shock or riluzole treatment). By normalizing the Hsp70-firefly luciferase activity against that of the Renilla luciferase internal control, we effectively negated experimental variables such as differences in cell number, as well as nonselective effects of the treatment conditions/reagents on gene expression.
Glutamatergic Stress and Assay for Cell Viability
Cells in 96-well plates were used. Riluzole was added to individual wells to final concentrations as indicated and incubated at 37 °C for 16 h. For conditioning heat shock, designated strips/wells of cells were heat-shocked at 42 °C for 2 h followed by recovery at 37 °C for 8 h. To test for cell survival under conditions of glutamatergic challenge, specified concentrations of glutamate along with 50 μm glycine was added to designated wells and incubated at 37 °C for 12–24 h prior to microscopic assessment of cell morphology and assay for cell viability. The N-methyl d-aspartate receptor antagonist MK801 (21), when used, was added to a final concentration of 1 μm at 60 min prior to the addition of glutamate and glycine to the cells. Viability of the cells was determined using the CellTiter-Glo® (G7571, Promega) (13). The result on cell viability signal of three independent measurements was averaged and normalized against that of the untreated control of 100%.
Data and Statistical Analysis
Quantitation of HSP70 staining intensity was done by ImageJ. The mean staining intensity of 30–40 representative cells was scored and averaged, and the result of this ± S.D. is shown. Student's t test was for unpaired data assuming equal variance and two-tailed distribution.
For quantitation of protein band intensity on Western blot, the integrated density of specified protein bands determined by the ImageJ program was used. Statistical analysis of the significance of a specific treatment, e.g. effect of riluzole or heat shock on HSF1 or GLT1, was done by paired Student's t test with two-tailed distribution. Repeated measures of ANOVA was done using the GraphPad Instat statistical analysis program. We have also used the ratio t test method to assess the significance of difference between paired samples, e.g. HSF1 intensity without versus with riluzole treatment. For this, the paired t test of the logarithms of integrated densities of specified protein bands was performed using the GraphPad InStat statistical analysis program. An example of this is included in supplemental Fig. 1.
Statistical analysis of reporter gene and cell viability was done using Student's t test (paired and unpaired as indicated) assuming equal variance and two-tailed distribution. The calculated probability of difference p > 0.05 is defined as not significant, between 0.01 and 0.05 as significant (*), <0.01 as very significant (**), and <0.001 as extremely significant (***).
RESULTS
Effects of Riluzole on the Regulation and Function of HSF1
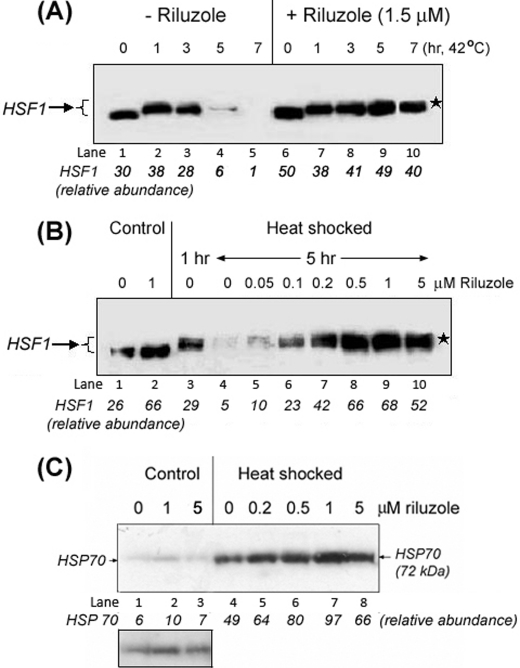
Induction of the HSR is mediated by activation of the HSF1 transcription factor; stress acutely converts the constitutively expressed dormant, monomeric HSF1 in the cytosol to a nuclearly localized, post-translationally modified HSF1 trimer that binds and transactivates the heat shock genes (22, 23). We used immuno-Western blot techniques to evaluate the effects of riluzole on the expression and duration of activation of HSF1 in NG108-15 cells. As seen in Fig. 1A, heat shock at 42 °C promoted the activation and post-translational modification of HSF1, as indicated by the upward mobility shift of HSF1 in SDS-gel electrophoresis (“supershift”); this activation was transient such that after 5 or 7 h of heat shock little HSF1 signal could be detected (Fig. 1A, lanes 4 and 5). Whether this was due to degradation, modification, and/or sequestration of HSF1 into entities not readily extractable or detectable by the methods used was not entirely clear. Previous studies showed that heat shock promoted the sequestration of HSF1 into subnuclear structures known as stress granules, and this HSF1 was extractable with Triton X-100 (24–26). Treatment of cells with 1.5 μm riluzole for 16 h at 37 °C increased the amount of latent HSF1 (compare lanes 1 versus 6 of Fig. 1A, lanes 1 versus lane 2 of B) and supported a sustained activation of HSF1, which remained supershifted after 7 h of heat shock at 42 °C, the longest time point examined (Fig. 1A, lanes 7–10). This effect of riluzole was dose-dependent as preincubation of cells with increasing concentrations of riluzole supported a sustained (>5 h) activation of HSF1 with an optimal effect observed at ∼1 μm (Fig. 1B). The effect of riluzole of increasing the HSF1 reserve was reproducible over the course of a 2-year study. Statistical analysis of the significance of difference in the abundance of HSF1 of control versus riluzole-treated neuroprogenitor cells of 10 independent measurements is shown in supplemental Fig. 1. The calculated p value associated with a Student's paired t test with a two-tailed distribution was <0.001 (extremely significant difference) with an average of 1.86 ± 0.399-fold increase in HSF1 in the riluzole-treated samples over that of the untreated controls. A similar p value of <0.001 was obtained using the ratio t test method to gauge the significance of difference between control and riluzole-treated cells (supplemental Fig. 1).
FIGURE 1.
Effects of riluzole on the amount and duration of activation of HSF1 and induction of HSP70 in NG108-15 cells. A, riluzole increased the amount of HSF1 and supported a more sustained activation of HSF1. NG108-15 cells, without and with riluzole pretreatment (1.5 μm, 16 h), were heat-shocked at 42 °C for times of 1, 3, 5, and 7 h. Cells incubated at 37 °C were used as control (i.e. 0 h at 42 °C). The position of the activated, and post-translationally modified HSF1 is indicated by a ★. B, dose-dependent effect of riluzole for a sustained activation of HSF1. The time of heat shock of the NG108-15 cells at 42 °C was as indicated. The activated and post-translationally modified form of HSF1 is indicated by an ★. C, dose-dependent effect of riluzole on expression of the HSP70 protein under control (37 °C) and heat-shocked (42 °C) conditions. Cells were incubated with specified concentrations of riluzole for 16 h at 37 °C. The condition of heat shock was 42 °C for 2 h followed by recovery at 37 °C for 6 h. Abundance of the heat-inducible 72-kDa HSP70 is as indicated at the bottom of the figure. A darker exposure of lanes 1–3 from control (37 °C) samples was included for better visualization of the effect of riluzole on the basal expression of HSP70.
The combined effect of an increase in the amount of latent HSF1 (HSF1 reserve) and a more sustained activation of HSF1 allowed for more robust induction of HSPs. In Fig. 1C, we show that riluzole gave a dose-dependent enhancement in induction of the 72-kDa HSP70 protein by heat shock. At the optimal concentration of 1 μm, riluzole boosted the heat shock induction of HSP70 by ∼2 times. Although the level of HSP70 in the control 37 °C samples was low when compared with that of the heat-shocked samples, we show in Fig. 1C, with a darker film exposure, an effect of riluzole in increasing the amount of HSP70 in cells maintained at 37 °C.
Riluzole Slows the Turnover of HSF1
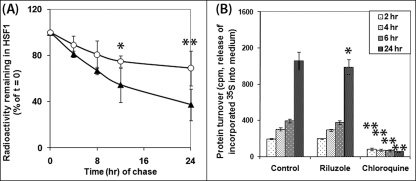
The effect of riluzole on HSF1 could be due to either an increase in the expression or a decrease in the turnover of HSF1. A previous study from our laboratory suggests that riluzole has little effect on the transcription and abundance of the mRNAhsf1 (12). We evaluated the effect of riluzole treatment on the turnover/degradation of HSF1 and total/long lived cellular proteins using a “pulse-chase” protocol where proteins were first labeled with [35S]methionine followed by removal of the radioactivity and “chase” of the 35S-labeled protein in the absence and presence of riluzole. Samples were harvested, and HSF1 was immunoprecipitated at various times after the beginning of chase. In Fig. 2A, we showed that riluzole had a statistically significant effect in slowing the rate of decay of 35S-labeled HSF1 at the 12-h (p between 0.01 and 0.05) and 24-h (p < 0.01) time points of chase. The fractional rate of decay of HSF1 (Kd = 2.303(a/b)) was calculated to be 0.041 and 0.014/h for the control and riluzole-treated NG108-15 cells at the early stationary phase of growth. This result suggests that riluzole slowed the turnover of HSF1 to increase the steady state level of the protein. Importantly, riluzole and chloroquine had different effects on the turnover of long lived cellular proteins (Fig. 2B). Chloroquine had a statistically significant effect in blunting (p < 0.1) the time-dependent protein turnover as assessed by the release/conversion of trichloroacetic acid-insoluble 35S-labeled proteins in cells into trichloroacetic acid-soluble radioactivity in the cell culture medium, whereas riluzole had no significant effect at 2, 4, and 6 h of chase and had a small but statistically significant effect at 24 h when compared with that of the untreated control, suggesting that the mode of action of riluzole is different from bulk inhibition of lysosome function by chloroquine.
FIGURE 2.
Effects of riluzole on the turnover of HSF1 and long lived cellular proteins. A, pulse-chase analysis of HSF1 turnover in control versus riluzole-treated NG108-15 cells. NG108-15 cells were labeled with [35S]methionine/cysteine and then chased in the absence (control, solid symbol) or presence of 2 μm riluzole at 37 °C (open symbol) for 0, 4, 8, 12, and 24 h. HSF1 was immunoprecipitated using protein A for pulldown by centrifugation. Result on the amount of radioactivity remaining with the HSF1 immunoprecipitate, relative to that of the t = 0 control, is plotted against the time of chase. Result is the average of four separate determinations ± S.D. B, effects of riluzole and chloroquine on the turnover of long lived cellular proteins. Cells were labeled for 48 h. Riluzole (2 μm) and chloroquine (0.2 mm) were added at the beginning of the chase. Aliquots (in quadruplicate) of the cell culture medium were removed at 2, 4, 6, and 24 h after initiation of the chase to determine the amount of radioactivity released into the medium. Result is the average ± S.D. of four separate determinations.
Riluzole Enhances Basal and Heat-induced hsp70-luciferase Reporter Gene in NG108-15 Cells
To assess the functionality of the increased HSF1, we determined the effects of riluzole on Hsp70 promoter-driven luciferase reporter gene expression in undifferentiated and differentiated NG108-15 cells incubated under basal (37 °C) and heat shock (42 °C) conditions.
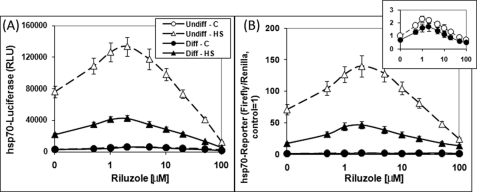
Preincubation of the cells with riluzole for 16 h at 37 °C followed by heat shock gave a riluzole dose-dependent enhancement of the heat shock induction of Hsp70-firefly luciferase reporter gene expression (Fig. 3, A and B). At the optimal riluzole concentration of 1–2 μm, the heat shock-induced Hsp70-reporter gene activity was ∼2 times higher than that of heat shock alone (without riluzole). Concentrations of riluzole >10 μm caused a significant drop in the Hsp70-reporter gene activity, suggesting cytotoxic effects, and this was later validated in cell viability assay. For subsequent experiments, we set 10 μm as the highest concentration of riluzole used. Analysis of the effects of riluzole on the basal level of expression of Hsp70-luciferase (i.e. 37 °C; inset of Fig. 3B, open and closed circles for undifferentiated and differentiated NG108-15 cells, respectively) revealed a qualitatively similar effect both in terms of the optimal concentration of riluzole (∼1–2 μm) and fold of enhancement. Repeated measures ANOVA of the four groups of data gave p < 0.0001 indicating extremely significant differences. Results presented in our previous study show that both the basal (37 °C) and the induced (42 °C) reporter gene expression requires a functional HSF1 as genetic ablation of the hsf1 gene voided both (12). The effect of riluzole in boosting Hsp70-reporter gene expression required preincubation because the addition of riluzole immediately prior to heat shock had little or no effect (see supplemental Fig. 2). We note that a minimal preincubation period of 4 h with riluzole was necessary for a reproducible and statistically significant increase in the Hsp70-reporter gene activity.
FIGURE 3.
Dose-response effects of riluzole on the basal and heat shock-induced Hsp70-luciferase reporter gene expression in the undifferentiated and differentiated NG108-15 cells. Undifferentiated (Undiff) and differentiated (Diff) NG108-15 cells were transfected with the Hsp70-firefly luciferase reporter DNA and the internal control Renilla luciferase DNA. A, raw Hsp70-luciferase reporter gene activity. The conditions of riluzole pretreatment and heat shock (HS) were as described. C, control. Result on Hsp70-firefly luciferase reporter activity (in relative luminescence units, RLU) is the average ± S.D. of four determinations, and this is plotted against riluzole concentration in log scale. Repeated measures ANOVA of the four groups of data gave p < 0.0001 indicating extremely significant difference. B, normalized Hsp70-reporter gene activity. The Hsp70-firefly luciferase activity shown in A was normalized against that of the internal control, Renilla luciferase, and this ratio was set at 1 for the 37 °C undifferentiated control (no riluzole, no heat shock). Repeated measures ANOVA of the four groups of data gave p < 0.0001 indicating extremely significant difference.
Comparison of Hsp70-reporter gene expression in undifferentiated and differentiated NG108-15 cells shows a significant reduction in basal and heat shock-induced reporter gene activity in differentiated cells (Fig. 3, A and B), a result consistent with our previous observations (15, 16). Nonetheless, riluzole had a qualitatively similar effect, in dose and magnitude, in differentiated and undifferentiated NG108-15 cells. The basal Hsp70-reporter gene activity without and with 2 μm riluzole was 1.0 and 2.2 for the undifferentiated cells and 0.8 and 2.1 for the differentiated cells. Heat-induced reporter gene activity ± 2 μm riluzole was 71 and 139 for the undifferentiated cells, and 28 and 63 for the differentiated cells (Fig. 3B; all values normalized against the undifferentiated control of 1).
Immunocytochemical Detection and Quantitation of HSP70
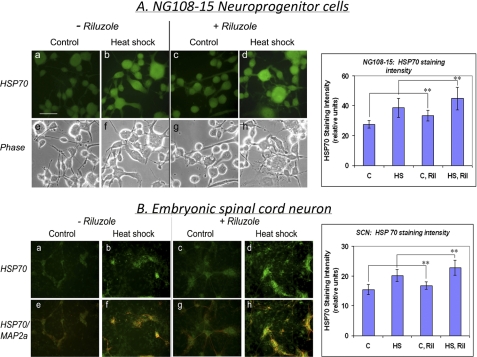
The effects of riluzole and of heat shock on the abundance of HSP70 protein were determined by immunocytochemical staining (Fig. 4). Visual inspection as well as counter-staining of the dendritic processes of SCN with anti-MAP2 antibody (in red) show that HSP70 was primarily localized to the cell body (Fig. 4B, panels e–h). HSP70 staining intensity was increased by heat shock (42 °C for 2 h followed by recovery for 6 h), and pretreatment of cells with riluzole (1 μm, 16 h) enhanced the HSP70 staining intensity under both control and heat shock conditions. Student's t test of the probability of difference in staining intensity confirmed that riluzole gave statistically significant increases in the basal and the heat-induced HSP70 staining in both the NG108-15 neuroprogenitors and SCN.
FIGURE 4.
Effects of heat shock and riluzole treatment on the immunocytochemical staining pattern of HSP70 in NG108-15 cells and rat embryonic spinal cord neurons. A, differentiated NG108-15 cells. Fluorescence (panels a–d) and phase (panels e–h) photomicrographs were captured with a SPOT camera system (Diagnostic Instruments, Inc., Sterling Heights, MI). Scale bar, 50 μm. For quantitation of staining intensity, ImageJ program was used to score the “brightness” of 20 cells/fields (mean density) to obtain average ± S.D., and statistical analysis was done using the GraphPad InStat program. The asterisks indicate statistically significant difference (p < 0.01) in staining intensity of untreated versus riluzole-treated (Ril) cells under both control (C) and heat shock (HS) conditions. ANOVA of the HSP70 staining intensity of the four groups of samples gave p < 0.0001, indicating significant difference. B, E16 rat spinal cord neurons at 10 days in vitro were used. Spinal cord neurons were double-stained for HSP70 (green; panels a–d) and MAP2 (red; panels e–h: merged images). Student's t test of the staining intensity of control versus riluzole-treated cells gave p < 0.001 under both control and heat shock conditions. ANOVA gave p < 0.0001, indicating significant difference.
Effects of Riluzole and Heat Shock on the GLT1 Glutamate Transporter
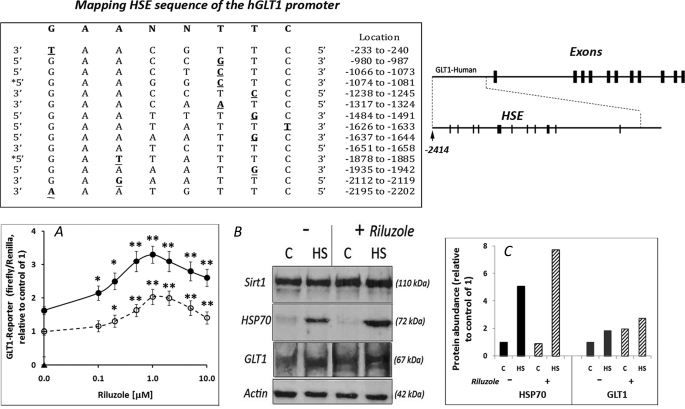
Riluzole has an anti-glutamatergic profile, and its ability to increase glutamate uptake provides a mechanism to mitigate the neurotoxic effects of excess glutamate (27, 28). Of the two major glutamate transporters, GLT1 (excitatory amino transporter 2, EAAT2) accounts for ∼95% of the glutamate transport activity, and loss of GLT1 is sufficient to induce a phenotype of motor neuron degeneration (29). We asked whether the effect of riluzole on glutamate uptake may be mediated, at least in part, through HSF1 as in silico analysis of the GLT1 promoter revealed a number of heat shock elements (HSE) for the binding of HSF1 (top of Fig. 5). We assessed the effects of riluzole on GLT1-firefly luciferase reporter gene expression and abundance of the GLT1 protein. We show in Fig. 5A that riluzole gave dose-dependent increase in GLT1-reporter gene activity under both control (37 °C) and heat shock (42 °C) conditions. The effect of heat shock on GLT1-reporter was modest (60–80% increase) when compare with the increase in hsp70-reporter shown in Fig. 3. Paired t test with two-tailed distribution gave a p value <0.001 for the difference in GLT1-reporter gene activity between the control versus heat shock cells. Western blot analysis of GLT1 in Fig. 5B showed that both riluzole and heat shock increased the expression of GLT1, although the effect of heat shock on GLT1 expression was modest when compared with that of HSP70.
FIGURE 5.
Effects of riluzole and heat shock on GLT1 glutamate transporter expression. Top, schematic representation of the human GLT1 gene and the in silico mapping of HSE in the promoter. The location and sequence of putative HSE are as indicated with nucleotide deviation from the consensus of GAANNTTC highlighted in bold. A, dose-dependent effect of riluzole on GLT1-firefly luciferase reporter gene activity under control (37 °C, open symbol) and heat shock (HS) (42 °C, filled symbol) conditions. Cells were transfected with the GLT1-firefly luciferase and Renilla luciferase DNA constructs (12, 15, 16). Result on the GLT1-firefly luciferase was normalized against Renilla luciferase, and this ratio was set at 1 for the control (C) (no heat shock and no riluzole). * and ** indicate significance of difference by two-tailed t test assuming equal variance of riluzole-treated versus control samples. Statistical analysis of the difference in GLT1-reporter activity between control and heat shock samples by paired t test with a two-tailed distribution gave p < 0.001, indicating extremely significant difference. B, cells without and with 1 μm riluzole pretreatment were incubated under control and heat shock conditions (2 h 42 °C and recovery at 37 °C for 6 h). Sirt1 was used as a loading control for the GLT1 membrane and actin was for HSP70. C, quantitation of result on HSP70 and GLT1. Intensity of the HSP70 and GLT1 bands in B was quantitated by ImageJ and the result, relative to control of 1, is shown.
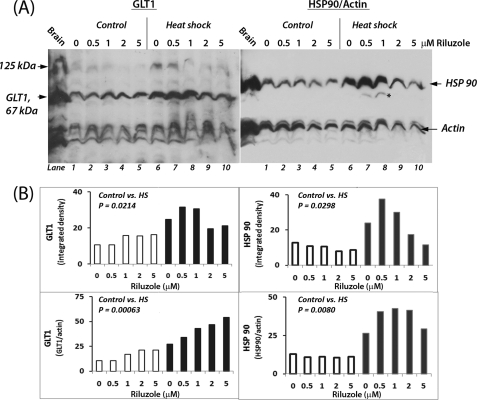
We further determined the dose-dependent effect of riluzole, without and with heat shock, on GLT1 expression (Fig. 6). For comparison and validation, the same membrane was reprobed for HSP90 and actin (HSP90 rather than HSP70 was used to assess the regulation of HSPs as HSP70 was not well resolved from GLT1). Quantitation of GLT1 band signal and normalization of this against that of the loading control actin showed that incubation with 0, 0.5, 1, 2, and 5 μm riluzole increased the abundance of GLT1, and the increase was augmented by heat shock. In the GLT1 immunoblot, we also detected a high molecular weight (∼125 kDa) as well as several low molecular weight (≤40 kDa) bands in addition to the 67-kDa GLT1 protein. Whether the 125-kDa entity represents a dimer of the 67-kDa GLT1 protein as suggested previously is not known (30). Reprobing of the membrane for HSP90 and actin showed that riluzole and heat shock gave synergistic induction of HSP90; an optimal effect was observed at 1 μm, and further increasing the concentration of riluzole to 5 μm actually caused a drop in the induction of HSP90. Pairwise statistical analysis of the intensity of GLT1 and HSP90 showed that heat shock had statistically significant effects in increasing the abundance of both. Our results are consistent with previous observations that HSF1 may target many cellular genes other than HSPs (31–36) and suggest that riluzole may harness various cytoprotective mechanisms targeted by HSF1, induction of HSPs and GLT1 included.
FIGURE 6.
Dose-dependent effects of riluzole on GLT1 and HSP90 expression under control and heat shock conditions. A, cells, pretreated with the indicated concentrations of riluzole, were incubated under control (37 °C, lanes 1–5) and heat shock conditions (42 °C, lanes 6–10). Immuno-Western blot detection of the 67-kDa GLT1 was done using antibody ab61859 from Abcam at 1:500 dilution. An aliquot of E18 rat brain extract served as control. The membrane was reprobed with antibodies against HSP90 (1:1000 dilution; catalog no. 4877, Cell Signaling), and actin (1:10,000 dilution; Sigma). The two images were aligned, and the carried over GLT1 signal in the HSP90/actin immunoblot is indicated by an asterisk (see lanes 7 and 8). B, quantitation of the relative abundance of GLT1 and HSP90. ImageJ was used to determine the intensities of individual bands of the immunoblots shown in A, with open bars representing control samples (i.e. 37 °C) and filled bars representing heat shock (HS) samples (42 °C). The signal intensity of GLT1 and HSP90 bands is presented “as is” at the top, and signal normalized against that of actin as a loading control is shown at the bottom. Results on Student's t test (paired, two tailed) of control (open bars) versus heat shock (filled bars) samples are as shown in each of the quadrants.
Riluzole and Conditioning Heat Shock Promote Neuronal Cell Survival under Glutamatergic Stress
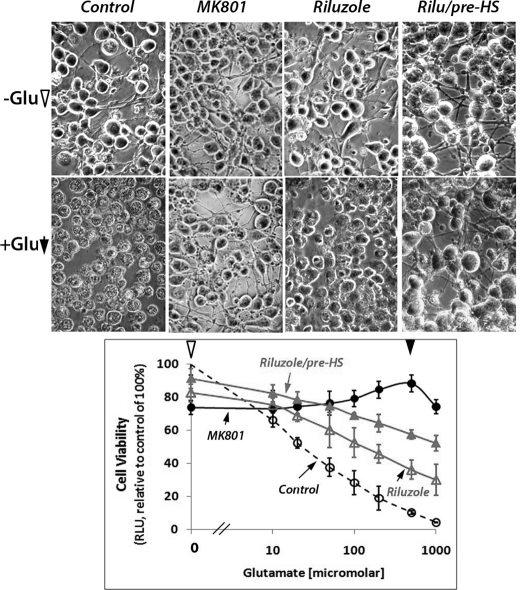
Increased expression of HSP and GLT1 can provide general and neuron-specific protective mechanisms for survival under stress. In Fig. 7, we examined the effects of riluzole without and with conditioning heat shock on the ability of differentiated NG108-15 cells to survive excitotoxic glutamate/glycine challenge. Phase contrast photomicrographs of cells without and with 1 mm glutamate, 50 μm glycine challenge were as illustrated at the top of Fig. 7, and the result on cell viability was plotted as a function of the dose of glutamate used. We showed that glutamate had a dose-dependent cell kill effect, and this cytotoxicity was negated by the prior addition of MK801, a noncompetitive N-methyl d-aspartate receptor antagonist (21). Indeed, photomicrographs showed that cells appeared dead in the glutamate-challenged NG108-15 cells, and this was mitigated by pretreatment with MK801. Pretreatment of cells with 1 μm riluzole partially protected cells from this glutamate-induced excitotoxicity, and this protective effect was boosted by conditioning heat shock prior to the glutamate challenge. Statistical analysis by paired t tests with two-tailed distribution yielded results of extremely significant differences in cell viability of control versus riluzole-treated, control versus riluzole/pre-HS, and riluzole versus riluzole/pre-HS, respectively. Repeated measures ANOVA gave p value of <0.001, indicating an extremely significant difference between the groups.
FIGURE 7.
Riluzole promoted neuronal cell survival under glutamate excitotoxic stress. Cells were pretreated with 1 μm riluzole (Rilu) followed by conditioning heat shock (HS) as described in the text. Cells were challenged with the indicated concentrations of glutamate (0, 10, 20, 50, 100, 200, and 500 μm and 1 mm) with 50 μm glycine for 12 h at 37 °C. The N-methyl d-aspartate receptor antagonist, MK801, was added 1 h prior to the addition of glutamate. Open circle, control, cells challenged with glutamate and glycine without pretreatment; filled circle, MK801-treated; open triangle, riluzole-treated; filled triangle, riluzole-treated and pre-heat-shocked. Phase contrast photomicrographs of cells without and with 1 mm glutamate, 50 μm glycine were captured with a SPOT camera system (Diagnostic Instruments, Inc., Sterling Heights, MI). Repeated measures ANOVA of the four groups of cell viability data gave p < 0.0001, indicating extremely significant differences between the groups.
DISCUSSION
Riluzole (2-amino-6(trifluoromethoxy)benzothiazole; brand name Rilutek®, Sanofi-Aventis Inc.) is the first and only drug approved by the Food and Drug Administration for the treatment of ALS (Lou Gehrig disease). Although generally considered an antiglutamatergic drug, the mode of action of riluzole is not entirely clear. We suggest, on the basis of the following considerations, that the ability of riluzole to up-regulate the amount of HSF1 to boost the expression of HSP chaperones and the GLT1 glutamate transporter contribute to the clinical efficacy of riluzole as a neuroprotective drug as follows. 1) Riluzole gave similar dose-dependent effects in increasing the amount and duration of activation of HSF1, in boosting HSP70 and GLT1 expression (both reporter gene and protein abundance), and in conferring protection for survival of neuronal cells under glutamate excitotoxic challenge. Such correlation is consistent with a cause-effect relationship of the HSF1-initiated signaling events and the improved survival under stress. The cytoprotective activity of riluzole requires a functional HSF1 as we showed that genetic deletion of hsf1 (37, 38) negated this effect (12). 2) The effective concentration of 1–2 μm riluzole as an HSR amplifier in our in vitro assays is coincident with the 1.62 μm plasma concentration of riluzole detected in healthy human volunteers 1 h after administration of the prescribed 100 mg daily dose of riluzole (39, 40). 3) The effect of riluzole on HSF1 appeared not to be cell type-specific as the effect is observed in different cell types, including HeLa cells (12), NG108-15, and N2a neuroprogenitor cells and embryonic neurons of spinal cord, hippocampus, and cortex. This is consistent with observations that the palliative effect of riluzole is not limited to diseased motor neurons in ALS; riluzole confers neuroprotection in spinal cord injury and cortical ischemia (41–44), retards huntingtin aggregate formation in a cell-free system and hippocampi organ culture (45), slows the progression of multiple sclerosis in human subjects (46), and retards neuromuscular dysfunction in wobbler mouse motor neuron disease (47). The apparently ubiquitous effect of riluzole on HSF1/HSR likely contributes to its efficacy in the different diseases/pathologies.
An important finding of this study is that riluzole increases GLT1 promoter activity and GLT1 protein expression, and the increase is enhanced by heat shock. These results together with in silico identification of HSE sequence in both the human and mouse GLT1 promoters suggest that GLT1 may be regulated by HSF1 and is consistent with experimental observations that the activities of HSF1 extend far beyond the induction of HSPs. In yeast, HSF1 regulates ∼3% of the genome, including genes in energy production, signal transduction, and vesicle transport (48). In higher eukaryotes, microarray and chromatin immunoprecipitation analysis reveal a complex transcriptional program regulated directly and indirectly by HSF1 (31–36). Like GLT1, heat shock has a modest effect on these non-HSP genes. We suggest that riluzole, by increasing HSF1, may mobilize a number of mechanisms, induction of HSPs and GLT1 included to promote neuron survival under stress.
In the context of HSF1/HSP/GLT1 and neuroprotection, there are two noteworthy considerations of the effects of riluzole. First, given that riluzole increases the amount of latent HSF1, it is likely to have a biased beneficial effect in diseased neurons burdened with conformationally challenged proteins. This is because the activation of HSF1 is inextricably tied to the dynamics of substrate/chaperone in cells (6). In normal cells, an increase in HSF1 reserve is expected to be phenotypically silent until cells are stressed and HSF1 becomes activated. In diseased neurons burdened with misfolded proteins, the increased HSF1 reserve would be activated in proportion to “need” to re-balance the substrate/chaperone dynamics. Second, the neuroprotective effects of riluzole may be directed as much toward glia as neurons as GLT1 glutamate transporter has a predominantly glial distribution (29) and riluzole has been shown to enhance the activity of glial GLAST and GLT1 to protect against glutamate neurotoxicity (28, 49).
Both heat shock and riluzole treatment appear to have a greater effect in boosting HSP expression when determined by either Western blot (Fig. 1) and reporter gene assay (Fig. 3) than by immunocytochemistry (Fig. 4). This may be due to a high background staining in our immunocytochemistry protocol where the antibody was used at a 50× higher concentration compared with Western blot. In a previous study on spinal cord motor neurons, little HSP signal was detected after heat shock, and expression of a wild-type HSF1 failed to resurrect the mortified HSR (50). One factor may be the difference in “age” of neurons used; we used SCN at 7–10 days in vitro when the neurons are less mature/differentiated than those neurons used in the other study after a 4–8-week in vitro culture period (50). We know, from our previous studies, that there is a definite negative correlation of neuronal differentiation and the ability to mount the HSR and induce HSP chaperones under stress (15).
The mechanism by which riluzole effects an increase in HSF1 has yet to be elucidated. It is not likely to involve transcriptional activation of the hsf1 gene as RT-PCR analysis failed to reveal a significant difference in the amount of hsf1 transcripts in the control versus riluzole-treated cells (12). Our result demonstrates that riluzole slowed the turnover of HSF1. Consistent with a role of protein turnover in this regulation and changes in protein turnover as a function of the growth conditions of cells (51), the effect of riluzole on HSF1 is influenced by serum/nutrient status and confluency of the cells, and unless indicated otherwise, experiments shown here were done using cells at the early stationary phase of growth. We further note that long incubation of cells with high concentrations of riluzole (>5–10 μm) had detrimental effects on cell viability and hsp70-reporter gene expression. Whether this may be related to changes in protein degradation mechanism is not entirely clear.
Riluzole may be the prototype of a family of drugs that amplify the HSR by increasing HSF1 reserve. Several hydroxylamine compounds, arimoclomo, iroxanadine, and bimoclomol, function as HSR co-inducers and prolong the activation of HSF1 to enhance the production of HSPs (52). Treatment with arimoclomol improves behavioral phenotypes of ALS mice, prevents neuronal loss, and extends survival rate by 22%. The possibility that these compounds may increase the amount of HSF1 reserve remains to be investigated. In a recent study, celastrol, a natural product derived from the Celastraceae family of plants that acts as “proteostasis regulator,” was shown to increase HSF1 reserve in a cellular model of Gaucher disease to restore function to mutant lysosomal enzymes and ameliorate cellular pathobiology caused by the misfolding of mutant proteins (53). We suggest that drugs that up-regulate the expression of HSF1, riluzole included, and small molecule inducer(s) of the HSR may have synergistic beneficial effects in the prevention and treatment of protein misfolding diseases.
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey Rothstein of The Johns Hopkins Medical School for the hGLT1-luciferase reporter DNA. We thank Diana J. Liu for discussion and a careful reading of the manuscript.
This work was supported in part by grants from the Busch Biomedical Research Grant Program, National Science Foundation Grant MCB0240009 (to A. Y. C. L.), and New Jersey Commission on Spinal Cord Research Grants 05-3037-SCR-E-0 (to A. Y. C. L.) and 07A-019-SCR1 (to B. L. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- HSR
- heat shock response
- HSP
- heat shock protein
- ALS
- amyotrophic lateral sclerosis
- ANOVA
- analysis of variance
- RIPA
- radioimmunoprecipitation assay
- HSE
- heat shock element
- SCN
- spinal cord neuron.
REFERENCES
- 1. Morimoto R. I. (1993) Science 259, 1409–1410 [DOI] [PubMed] [Google Scholar]
- 2. Westerheide S. D., Morimoto R. I. (2005) J. Biol. Chem. 280, 33097–33100 [DOI] [PubMed] [Google Scholar]
- 3. Morimoto R. I. (2006) N. Engl. J. Med. 355, 2254–2255 [DOI] [PubMed] [Google Scholar]
- 4. Morley J. F., Morimoto R. I. (2004) Mol. Biol. Cell 15, 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rea S. L., Wu D., Cypser J. R., Vaupel J. W., Johnson T. E. (2005) Nat. Genet. 37, 894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morimoto R. I. (2008) Genes Dev. 22, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soto C. (2003) Nat. Rev. Neurosci. 4, 49–60 [DOI] [PubMed] [Google Scholar]
- 8. Forman M. S., Trojanowski J. Q., Lee V. M. (2004) Nat. Med. 10, 1055–1063 [DOI] [PubMed] [Google Scholar]
- 9. Lansbury P. T., Lashuel H. A. (2006) Nature 443, 774–779 [DOI] [PubMed] [Google Scholar]
- 10. Muchowski P. J., Wacker J. L. (2005) Nat. Rev. Neurosci. 6, 11–22 [DOI] [PubMed] [Google Scholar]
- 11. Nirmalananthan N., Greensmith L. (2005) Curr. Opin. Neurol. 18, 712–719 [DOI] [PubMed] [Google Scholar]
- 12. Yang J., Bridges K., Chen K. Y., Liu A. Y. (2008) PLoS ONE 3, e2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khalil S., Luciano J., Chen W., Liu A. Y. (2006) J. Cell. Physiol. 207, 562–569 [DOI] [PubMed] [Google Scholar]
- 14. Rothstein J. D., Patel S., Regan M. R., Haenggeli C., Huang Y. H., Bergles D. E., Jin L., Dykes Hoberg M., Vidensky S., Chung D. S., Toan S. V., Bruijn L. I., Su Z. Z., Gupta P., Fisher P. B. (2005) Nature 433, 73–77 [DOI] [PubMed] [Google Scholar]
- 15. Yang J., Oza J., Bridges K., Chen K. Y., Liu A. Y. (2008) Brain Res. 1203, 39–50 [DOI] [PubMed] [Google Scholar]
- 16. Oza J., Yang J., Chen K. Y., Liu A. Y. (2008) Cell Stress Chaperones 13, 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nirenberg M., Wilson S., Higashida H., Rotter A., Krueger K., Busis N., Ray R., Kenimer J. G., Adler M. (1983) Science 222, 794–799 [DOI] [PubMed] [Google Scholar]
- 18. Du Y., Chen C. P., Tseng C. Y., Eisenberg Y., Firestein B. L. (2007) Glia 55, 463–472 [DOI] [PubMed] [Google Scholar]
- 19. Auteri J. S., Okada A., Bochaki V., Dice J. F. (1983) J. Cell. Physiol. 115, 167–174 [DOI] [PubMed] [Google Scholar]
- 20. Rabindran S. K., Haroun R. I., Clos J., Wisniewski J., Wu C. (1993) Science 259, 230–234 [DOI] [PubMed] [Google Scholar]
- 21. Wong E. H., Kemp J. A., Priestley T., Knight A. R., Woodruff G. N., Iversen L. L. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 7104–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morimoto R. I., Sarge K. D., Abravaya K. (1992) J. Biol. Chem. 267, 21987–21990 [PubMed] [Google Scholar]
- 23. Voellmy R. (1994) Crit. Rev. Eukaryot. Gene Expr. 4, 357–401 [PubMed] [Google Scholar]
- 24. Cotto J., Fox S., Morimoto R. (1997) J. Cell Sci. 110, 2925–2934 [DOI] [PubMed] [Google Scholar]
- 25. Jolly C., Morimoto R., Robert-Nicoud M., Vourc'h C. (1997) J. Cell Sci. 110, 2935–2941 [DOI] [PubMed] [Google Scholar]
- 26. Jolly C., Usson Y., Morimoto R. I. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6769–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frizzo M. E., Dall'Onder L. P., Dalcin K. B., Souza D. O. (2004) Cell. Mol. Neurobiol. 24, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fumagalli E., Funicello M., Rauen T., Gobbi M., Mennini T. (2008) Eur. J. Pharmacol. 578, 171–176 [DOI] [PubMed] [Google Scholar]
- 29. Maragakis N. J., Rothstein J. D. (2001) Arch. Neurol. 58, 365–370 [DOI] [PubMed] [Google Scholar]
- 30. Chen W., Aoki C., Mahadomrongkul V., Gruber C. E., Wang G. J., Blitzblau R., Irwin N., Rosenberg P. A. (2002) J. Neurosci. 22, 2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eastmond D. L., Nelson H. C. (2006) J. Biol. Chem. 281, 32909–32921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jensen L. T., Nielsen M. M., Loeschcke V. (2008) Cell Stress Chaperones 13, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Page T. J., Sikder D., Yang L., Pluta L., Wolfinger R. D., Kodadek T., Thomas R. S. (2006) Mol. Biosyst. 2, 627–639 [DOI] [PubMed] [Google Scholar]
- 34. Pirkkala L., Nykänen P., Sistonen L. (2001) FASEB J. 15, 1118–1131 [DOI] [PubMed] [Google Scholar]
- 35. Sakurai H., Takemori Y. (2007) J. Biol. Chem. 282, 13334–13341 [DOI] [PubMed] [Google Scholar]
- 36. Westwood J. T., Clos J., Wu C. (1991) Nature 353, 822–827 [DOI] [PubMed] [Google Scholar]
- 37. McMillan D. R., Xiao X., Shao L., Graves K., Benjamin I. J. (1998) J. Biol. Chem. 273, 7523–7528 [DOI] [PubMed] [Google Scholar]
- 38. Xiao X., Zuo X., Davis A. A., McMillan D. R., Curry B. B., Richardson J. A., Benjamin I. J. (1999) EMBO J. 18, 5943–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bryson H. M., Fulton B., Benfield P. (1996) Drugs 52, 549–563 [DOI] [PubMed] [Google Scholar]
- 40. Song J. H., Huang C. S., Nagata K., Yeh J. Z., Narahashi T. (1997) J. Pharmacol. Exp. Ther. 282, 707–714 [PubMed] [Google Scholar]
- 41. Heurteaux C., Laigle C., Blondeau N., Jarretou G., Lazdunski M. (2006) Neuroscience 137, 241–251 [DOI] [PubMed] [Google Scholar]
- 42. Lips J., de Haan P., Bodewits P., Vanicky I., Dzoljic M., Jacobs M. J., Kalkman C. J. (2000) Anesthesiology 93, 1303–1311 [DOI] [PubMed] [Google Scholar]
- 43. Schwartz G., Fehlings M. G. (2001) J. Neurosurg. 94, Suppl. 2, 245–256 [DOI] [PubMed] [Google Scholar]
- 44. Stover J. F., Beyer T. F., Unterberg A. W. (2000) J. Neurotrauma. 17, 1171–1178 [DOI] [PubMed] [Google Scholar]
- 45. Heiser V., Engemann S., Bröcker W., Dunkel I., Boeddrich A., Waelter S., Nordhoff E., Lurz R., Schugardt N., Rautenberg S., Herhaus C., Barnickel G., Böttcher H., Lehrach H., Wanker E. E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, Suppl., 4, 16400–16406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Killestein J., Kalkers N. F., Polman C. H. (2005) J. Neurol. Sci. 233, 113–115 [DOI] [PubMed] [Google Scholar]
- 47. Ishiyama T., Okada R., Nishibe H., Mitsumoto H., Nakayama C. (2004) Brain Res. 1019, 226–236 [DOI] [PubMed] [Google Scholar]
- 48. Hahn J. S., Hu Z., Thiele D. J., Iyer V. R. (2004) Mol. Cell. Biol. 24, 5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Estevez A. G., Stutzmann J. M., Barbeito L. (1995) Eur. J. Pharmacol. 280, 47–53 [DOI] [PubMed] [Google Scholar]
- 50. Batulan Z., Shinder G. A., Minotti S., He B. P., Doroudchi M. M., Nalbantoglu J., Strong M. J., Durham H. D. (2003) J. Neurosci. 23, 5789–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fuertes G., Martín, De Llano J. J., Villarroya A., Rivett A. J., Knecht E. (2003) Biochem. J. 375, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kieran D., Kalmar B., Dick J. R., Riddoch-Contreras J., Burnstock G., Greensmith L. (2004) Nat. Med. 10, 402–405 [DOI] [PubMed] [Google Scholar]
- 53. Mu T. W., Ong D. S., Wang Y. J., Balch W. E., Yates J. R., 3rd., Segatori L., Kelly J. W. (2008) Cell 134, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.