Abstract
Precise control of the timing of translational activation of dormant mRNAs stored in oocytes is required for normal progression of oocyte maturation. We previously showed that Pumilio1 (Pum1) is specifically involved in the translational control of cyclin B1 mRNA during Xenopus oocyte maturation, in cooperation with cytoplasmic polyadenylation element-binding protein (CPEB). It was reported that another Pumilio, Pumilio2 (Pum2), exists in Xenopus oocytes and that this protein regulates the translation of RINGO mRNA, together with Deleted in Azoospermia-like protein (DAZL). In this study, we characterized Pum1 and Pum2 biochemically by using newly produced antibodies that discriminate between them. Pum1 and Pum2 are bound to several key proteins involved in translational control of dormant mRNAs, including CPEB and DAZL, in immature oocytes. However, Pum1 and Pum2 themselves have no physical interaction. Injection of anti-Pum1 or anti-Pum2 antibody accelerated CPEB phosphorylation, cyclin B1 translation, and oocyte maturation. Pum1 phosphorylation coincides with the dissociation of CPEB from Pum1 and the translational activation of cyclin B1 mRNA, a target of Pum1, whereas Pum2 phosphorylation occurred at timing earlier than that for Pum1. Some, but not all, of cyclin B1 mRNAs release the deadenylase PARN during oocyte maturation, whereas Pum1 remains associated with the mRNA. On the basis of these findings, we discuss the functions of Pum1 and Pum2 in translational control of mRNAs during oocyte maturation.
Keywords: Cyclins, mRNA, Oocyte, Protein-Protein Interactions, RNA-binding Protein, Translation Control, Xenopus
Introduction
In many species, oocytes stop their meiotic cell cycle at the first prophase, during which time they grow by accumulation of proteins and mRNAs necessary for subsequent developmental processes. In response to hormonal stimulation, e.g. progesterone in frogs, the immature oocytes resume meiosis to mature, a process called oocyte maturation (1). The final inducer of oocyte maturation is the maturation-promoting factor (MPF),2 which consists of cyclin B-bound cdc2. In Xenopus, MPF is stockpiled in immature oocytes as an inactive form called pre-MPF, and its activation induces oocyte maturation. In addition to the activation of pre-MPF, the normal progression of oocyte maturation, including the transition from meiosis I to meiosis II without intervening DNA synthesis, is ensured by the continued synthesis of cyclin B1 protein, which depends on the translational activation of cyclin B1 mRNA stored in the oocytes (2). One of the mechanisms that regulate the translational activation of mRNA is cytoplasmic polyadenylation, in which cytoplasmic polyadenylation element (CPE) in the 3′-untranslated region (UTR) of mRNA plays a critical role (3, 4). In fact, the translational activation of cyclin B1 mRNA during oocyte maturation is under the CPE-mediated control, which involves dynamic changes in interactions among several key proteins, including CPE-binding protein (CPEB), Symplekin, Maskin, cleavage, and polyadenylation specificity factor (CPSF), PARN, and GLD2 (4–6).
Besides cyclin B1 mRNA, mos and wee1 mRNAs also carry the CPE sequence in the 3′-UTR, and their translation is regulated by cytoplasmic polyadenylation. However, the timings of translational activation are different in the mRNAs during oocyte maturation (7–10). Accordingly, oocytes must have additional mechanisms that define the precise timing of translational activation of each CPE-containing mRNA during oocyte maturation. Indeed, it has been shown that the Musashi-binding element (11), the Pumilio-binding element (10, 12), and the translational control sequence (13) in the 3′-UTR play important roles as cis-elements in the temporal control of translation of mos, cyclin B1, and wee1 mRNA, respectively, during Xenopus oocyte maturation (14). These cis-elements are recognized by trans-acting proteins specific to each element as follows: Musashi for Musashi-binding element, Pumilio for Pumilio-binding element, and an unidentified protein for translational control sequence. However, these proteins have not yet been fully characterized biochemically.
Pumilio homologs have been identified in various organisms, from yeast to humans, and constitute a large and evolutionarily conserved protein family, the Puf family (15). In vertebrates, the Puf family consists of two members, Pumilio1 (Pum1) and Pumilio2 (Pum2) (16–19). Because a Pumilio homolog that we previously identified in Xenopus (12) resembles human and mouse Pum1 rather than Pum2, it should be Xenopus Pum1. Xenopus Pum1 is bound to cyclin B1 mRNA in immature oocytes (12). Injection of anti-Pum1 antibody or overexpression of Pum1 protein has a significant impact on the timing of oocyte maturation and cyclin B1 synthesis specifically, suggesting the involvement of Pum1 in translational control of cyclin B1 mRNA (10). In addition to Pum1, Xenopus oocytes contain another member of the Puf family, Pum2, which was reported to regulate the translation of mRNA encoding rapid inducer of G2-M in oocytes (RINGO), a cyclin B1-like molecule responsible for inducing oocyte maturation, in cooperation with Deleted in Azoospermia-like protein (DAZL) (20). It is therefore likely that Pum1 and Pum2 contribute to oocyte maturation by controlling the translation of different target mRNAs. However, little is known about their biochemical characteristics, including expression patterns, their biochemical modifications, and their interacting proteins during oocyte maturation. In this study, we biochemically characterized Pum1 and Pum2 in Xenopus oocytes by using newly produced antibodies that discriminate between Pum1 and Pum2.
EXPERIMENTAL PROCEDURES
Animals and Oocytes
All animal experiments in this study were approved by the Committee on Animal Experimentation, Hokkaido University. Xenopus laevis frogs were purchased from a dealer and maintained in laboratory aquaria at 20 °C. Oocytes were isolated from ovaries and induced to mature, as described previously (10). Oocyte maturation was assessed by the appearance of a white spot at the animal pole, which indicates the occurrence of germinal vesicle breakdown (GVBD).
Cloning of cDNA Encoding X. laevis Pum2
Because the entire amino acid sequence of X. laevis Pum2 was unknown, we first intended to isolate a full-length cDNA. Following isolation of total RNA from an ovary with Isogen (Nippon Gene, Tokyo, Japan), cDNAs were produced with a first strand cDNA kit (Invitrogen). A cDNA fragment encoding an open reading frame (ORF) of X. laevis Pum2 was isolated by RT-PCR using a primer set, Pum2-ORF-F and Pum2-ORF-R (supplemental Table S1), which was designed according to a sequence found by a BLAST search in the Xenopus tropicalis genome data base, using mouse Pum2 as a query.
RT-PCR yielded a cDNA fragment of 3510 bp (DDBJ/EMBL/GenBankTM accession number AB565475), which encodes 1170 amino acids including the Puf domain, the hallmark sequence of the Puf family (supplemental Fig. S1) (10). The deduced amino acid sequence showed 68.1% identity to X. laevis Pum1 (AB091091), 67.1% to human Pum1 (AF315592), 81.3% to human Pum2 (AF315591), 67.4% to mouse Pum1 (AF321909), and 79.8% to mouse Pum2 (AF315590). We have therefore identified the cDNA as X. laevis Pum2 cDNA.
Production of Full-length Pum1 and Pum2 Proteins
ORFs of X. laevis Pum1 and Pum2 were amplified by PCR using primer sets, Pum1N-F/Pum1F-R and Pum2N-F/Pum2F-R, respectively (supplemental Table S1), and inserted into pENTR/D-TOPO Gateway vector (Invitrogen). The resulting plasmids were recombined with the destination vectors pDEST15 and pET161-DEST using the Gateway cloning system (Invitrogen) to produce proteins with a glutathione S-transferase (GST) tag at the N terminus and a polyhistidine (His) tag at the C terminus, respectively. Pum1-His and Pum2-His were expressed in Escherichia coli and purified by SDS-PAGE followed by electroelution in Tris-glycine buffer without SDS, according to the method described previously (21). The purified proteins were concentrated by Centricon (YM-100) (Millipore, Tokyo, Japan), and their concentration was determined with a protein assay kit (Bio-Rad) using bovine serum albumin as a standard. Purified proteins with known concentrations were used to confirm the specificity of antibodies and to measure the protein contents of endogenous Pum1 and Pum2 in oocytes.
Production of Antibodies
We produced new antibodies that differentially recognize Pum1 and Pum2. Anti-Pum1 antibody was produced as follows. An N-terminal fragment of Pum1 (Pum1N) (supplemental Fig. S1) was amplified by PCR using a Pum1N-F/Pum1N-R primer set (supplemental Table S1) and inserted into pDEST15. E. coli-expressed GST-Pum1N was gel-purified, dialyzed against 1 mm HEPES (pH 7.5), and lyophilized for use in affinity purification. A guinea pig antibody produced against an N-terminal fragment of Pum1 (PumNN) (10) was affinity-purified with GST-Pum1N electroblotted onto an Immobilon membrane (Millipore, Tokyo, Japan) and then absorbed with GST-Pum2N, which was produced as described below.
A cDNA fragment encoding Pum2N (supplemental Fig. S1) was amplified with Pum2N-F/Pum2N-R (supplemental Table S1) and inserted into pDEST15 and pET161-DEST. Pum2N-His proteins were gel-purified and injected into mice to produce antibodies as described previously (22). The antisera were affinity-purified with GST-Pum2N and absorbed with GST-Pum1N. The specificity of anti-Pum1N and anti-Pum2N antibodies was confirmed using Pum1-His and Pum2-His proteins by immunoblotting (Fig. 1A).
FIGURE 1.
Characterization of antibodies and detection of Pum1 and Pum2 in oocytes. A, E. coli-produced Pum1-His (10 ng) (lanes 1, 3, and 5) and Pum2-His (10 ng) (lanes 2, 4, and 6) were probed with anti-His antibody (lanes 1 and 2), anti-Pum1N antibody (lanes 3 and 4), and anti-Pum2N antibody (lanes 5 and 6). B, anti-Pum1N and anti-Pum2N immunoblots of crude extracts from immature (Im) and mature (M) oocytes. C, phosphorylation of Pum1 and Pum2 accompanied by changes in electrophoretic mobility. Extracts from [γ-32P]ATP-injected immature (Im) and mature (M) oocytes were immunoprecipitated with anti-Pum1 or anti-Pum2 antibody, treated with (+) or without (−) λ-phosphatase (λ-PPase), immunoblotted (IB) with the same antibodies as those used for IP, and subjected to autoradiography (1-month exposure for Pum1 and 1-week exposure for Pum2) to detect the label of 32P (32P). Extracts before IP were also immunoblotted (Initial). D and E, quantification of the protein contents of endogenous Pum1 (D) and Pum2 (E) in one oocyte. One microliter of immature oocyte extracts (Im), which is equivalent to one oocyte, was immunoblotted with anti-Pum1N and anti-Pum2N antibodies with quantified recombinant Pum1-His and Pum2-His. The signal intensity of individual bands of Pum1 and Pum2 on the immunoblots is expressed as arbitrary units. WB, Western blot.
To produce antibodies against X. laevis Symplekin, CPSF100 (100-kDa subunit of CPSF), Maskin, and GLD2, an N-terminal fragment of Symplekin, and ORFs of CPSF100, Maskin, and GLD2 were amplified by RT-PCR using primer sets designed according to the published sequences, BC047265, AF139986, AF200212, and AY655140, respectively (supplemental Table S1). The PCR products were ligated into pET21 (Novagen, Madison, WI). Recombinant His-tagged proteins expressed in E. coli were gel-purified, and the resulting Symplekin, CPSF100, and Maskin antigens were injected into mice, and GLD2 antigen was injected into guinea pigs to produce antibodies. Similarly, mouse and guinea pig antibodies against GST were also produced.
Oocyte Extraction
Oocytes were washed three times with ice-cold extraction buffer (EB: 100 mm β-glycerophosphate, 1% Triton X-100, 20 mm HEPES, 15 mm MgCl2, 5 mm EGTA, 1 mm dithiothreitol, 100 μm (p-amidinophenyl)methanesulfonyl fluoride, 3 μg/ml leupeptin (pH 7.5)). For RNA analysis, RNasin Plus RNase inhibitor (100 units/ml at the final concentration, Promega) was added to EB. After the final wash, EB was removed with filter paper, and 1 μl/oocyte of new EB was added. The oocytes were homogenized and centrifuged at 8,000 × g for 5 min at 4 °C. The supernatant was collected and stored at −80 °C until use.
Immunoprecipitation (IP) and Immunoblotting
To immunoprecipitate Pum1 and Pum2, oocyte extracts (10 μl) were incubated with either anti-Pum1N antibody or anti-Pum2N antibody in the presence of protein A- or protein G-Sepharose beads (GE Healthcare), respectively. To examine the dependence of protein interaction on RNA, 1 μl of 10 mg/ml RNase A was added to the oocyte extracts prior to IP. After washing five times in EB, the immunoprecipitates were separated by SDS-PAGE, blotted onto an Immobilon membrane, and probed with anti-Pum1N, anti-Pum2N, anti-CPEB (12), anti-CPSF100, and anti-DAZL (23) antibodies. Similarly, anti-Symplekin, anti-Maskin, anti-CPEB, anti-DAZL, and anti-PARN (24) immunoprecipitates were blotted and probed with appropriate antibodies.
Anti-His G-18 antibody (Santa Cruz Biotechnology) was used to detect Pum1-His and Pum2-His. Anti-MAPK1/2 antibody (Upstate) and anti-phospho-p44/42 MAPK (Thr-202/Tyr-204) antibody (Cell Signaling Technology) were used to detect MAPK and phospho-MAPK, respectively. Cyclin B1-cdc2 complex was detected by immunoblotting Suc1 precipitates from crude oocyte extracts (10 μl) with anti-cdc2 (MC2–21) (25) and anti-cyclin B1 (Bufo 2F5) (26) monoclonal antibodies, as described previously (27). Monoclonal anti-T7 antibody (Novagen) was used to immunoprecipitate T7-GLD2 and to detect T7-GST.
In Vitro Synthesis of mRNAs
mRNAs encoding GST-tagged full-length Pum1 and Pum2 were synthesized from the corresponding cDNAs in pDEST15 with an mMESSAGE mMACHINE T7 ultra kit (Ambion). T7-tagged GST mRNAs were synthesized from the cDNA in pET21 (10). To produce mRNA encoding T7-tagged GLD2, an ORF was amplified by PCR using a primer set, GLD2-F and GLD2-R (supplemental Table S1), and the PCR products were ligated into pET21. Using the resulting plasmid as a template, T7-GLD2 mRNA was synthesized with an mMESSAGE mMACHINE T7 ultra kit.
Oocyte Microinjection
Oocytes were manually isolated from the ovary with forceps and injected with 50 nl of 20 mm sodium phosphate buffer (pH 7.0) containing either 2 ng of anti-Pum1N guinea pig antibody or 0.75 ng of anti-Pum2N mouse antibody. The same volume of 2 ng of anti-GST guinea pig antibody or 0.75 ng of anti-GST mouse antibody was injected as a control. The same volume of sodium phosphate buffer without antibody was also injected as a vehicle control. The injected oocytes were incubated overnight at 18 °C and treated with progesterone. The rate of GVBD was monitored, and oocytes were extracted at appropriate intervals. Oocytes injected with in vitro synthesized mRNAs encoding GST, GST-Pum1, GST-Pum2, and T7-GLD2 were subjected to GST pulldown assay with glutathione (GSH)-Sepharose 4B (GE Healthcare) or to co-IP assay with anti-PARN, anti-GLD2, and anti-T7 antibodies.
Oocytes were injected with 100 nl of [γ-32P]ATP (6000 Ci/mmol, PerkinElmer Life Sciences) to detect phosphorylation. The injected oocytes were incubated for 2 h and induced to mature with progesterone. Immature and mature oocyte extracts were immunoprecipitated with anti-Pum1N or anti-Pum2N antibody, and the resulting precipitates were treated with λ-phosphatase (200 units, New England Biolabs) for 1 h at 30 °C. The samples were immunoblotted with the same antibodies as those used for IP and subjected to autoradiography to detect the incorporation of 32P into Pumilio proteins.
RT-PCR Analysis following IP (IP/RT-PCR)
Immature and mature oocyte extracts (50 μl) were precleared with protein A- or protein G-Sepharose beads for 1 h at 4 °C. The extracts were then incubated with anti-CPEB or anti-Pum1N antibodies for 3 h at 4 °C and washed five times. Following extraction of mRNAs from the beads, RT-PCR was performed using primer sets specific to cyclin B1, RINGO, mos, wee1, D7, G10, cyclin A1, and β-actin mRNA (supplemental Table S1). Similarly, mRNAs in GST pulldown samples from oocytes overexpressing GST-Pum1 were analyzed.
RESULTS
Characterization of Antibodies and Detection of Pum1 and Pum2 in Oocytes
We previously raised a monoclonal antibody (XPum 2A5) against the Puf domain of X. laevis Pum1 (12), but this domain is highly homologous to that of Pum2 (supplemental Fig. S1). Indeed, immunoblotting analysis revealed that the antibody reacts to E. coli-produced full-length Pum2, as well as Pum1 (data not shown). In this study, we therefore produced new antibodies to detect X. laevis Pum1 and Pum2 differentially. The newly produced anti-Pum1N antibody recognized E. coli-produced full-length Pum1 but not Pum2, whereas the anti-Pum2N antibody recognized Pum2 but not Pum1 (Fig. 1A), confirming that these antibodies discriminate between Pum1 and Pum2.
Anti-Pum1N and anti-Pum2N antibodies recognized a 137- and a 129-kDa band, respectively, in immature oocyte extracts (Fig. 1B). Because apparent molecular masses of these bands correspond to the values estimated from the cDNA sequences, we conclude that these bands are X. laevis Pum1 and Pum2. Pum1 and Pum2 in mature oocytes showed a slower electrophoretic mobility than those in immature oocytes (Fig. 1B). Anti-Pumilio IP from oocytes injected with [γ-32P]ATP showed that the slower mobility forms of Pum1 and Pum2 were labeled with 32P and that treatment with λ-phosphatase restored the mobility and removed the label (Fig. 1C). When Pumilio proteins fused with GST were expressed in oocytes, they also underwent the phosphorylation-induced mobility shift during maturation (supplemental Fig. S2). These results indicate that Pum1 and Pum2 are phosphorylated, in accordance with the change in electrophoretic mobility, during oocyte maturation.
The protein contents of endogenous Pum1 and Pum2 in Xenopus oocytes were quantified by using purified Pum1-His and Pum2-His as a standard, respectively. A full-grown immature oocyte (its diameter being 1.2 mm) was estimated to contain 300 pg (2.6 nm) of Pum1 and 2500 pg (21.7 nm) of Pum2 (Fig. 1, D and E).
Behaviors of Pum1 and Pum2 during Oocyte Maturation
Pum1 and Pum2 were phosphorylated in mature oocytes (Fig. 1C). We then examined the time course of Pumilio phosphorylation during oocyte maturation, in comparison with CPEB phosphorylation, MAPK activation, and translational activation of cyclin B1 mRNA (Fig. 2A and supplemental Fig. S3A). Although the time course varied in batches of oocytes, in most cases, Pum1 phosphorylation was initiated 8 h after progesterone treatment (soon after the onset of GVBD), at which time the translation of cyclin B1 mRNA, a target of Pum1, was also initiated as indicated by the appearance of cyclin B1 proteins. On the other hand, Pum2 phosphorylation was initiated at timing earlier than the translational activation of cyclin B1 mRNA (6 h after progesterone treatment) but later than the activation of MAPK (as indicated by the appearance of bands recognized by anti-phospho-MAPK antibody; Fig. 2A, P-MAPK), which occurred 2–4 h after progesterone treatment, before the onset of GVBD. A phosphorylated form of CPEB (a band with slow electrophoretic mobility) began to appear at various time points according to oocyte batches, and the protein contents of both phosphorylated and unphosphorylated forms of CPEB were then gradually decreased (Fig. 2A and supplemental Fig. S3A).
FIGURE 2.
Pum1, Pum2, CPEB, cyclin B1, and MAPK during oocyte maturation. A, immature oocytes were treated with progesterone, collected at intervals of 2 h, extracted, and immunoblotted with antibodies against Pum1, Pum2, CPEB, cyclin B1, cdc2, phospho-MAPK (P-MAPK) and MAPK. Asterisks indicate the time points at which phosphorylation of Pum1 and Pum2 occurs, and an arrowhead indicates the time point at which synthesis of cyclin B1 protein occurs. The progression of oocyte maturation is indicated by the percentage of GVBD. B, oocyte extracts used in A were immunoprecipitated by anti-Pum1N (α-Pum1N-IP) or anti-Pum2N antibody (α-Pum2N-IP) and immunoblotted with antibodies against CPEB, Pum1, and Pum2. Asterisks indicate phosphorylated Pum1 and Pum2. Immunoglobulins (Ig) of the antibody used for IP are also indicated. Three independent experiments were performed using different batches of oocytes, and representative results are shown (for results of the remaining two experiments, see supplemental Fig. S3). C, quantification of the contents of CPEB in crude oocyte extracts (Total CPEB), anti-Pum1N immunoprecipitates (Pum1-associated CPEB), and anti-Pum2N immunoprecipitates (Pum2-associated CPEB). The signal intensity of individual bands of CPEB, Pum1, and Pum2 on immunoblots was measured by using ImageJ software. Because the efficiency of IP was not constant, the value of Pum1- or Pum2-associated CPEB was divided by that of Pum1 or Pum2 in each immunoprecipitate to standardize the values. The content of CPEB at each time point is expressed as a value relative to that at time 0. Data are shown as means ± S.E. for three independent experiments using different batches of oocytes (A and B and supplemental Fig. S3). The timings of Pum1 phosphorylation, Pum2 phosphorylation, and cyclin B1 protein synthesis are also indicated in the figure.
We also examined changes in the interaction between Pumilio and CPEB during oocyte maturation by IP with anti-Pumilio antibodies followed by immunoblotting with anti-CPEB antibody (Fig. 2B and supplemental Fig. S3B). Consistent with the results of our previous study (10), Pum1 formed a complex with CPEB in immature oocytes. However, the content of CPEB associated with Pum1 obviously decreased 8 h after progesterone treatment, the timing of which was consistent with that of Pum1 phosphorylation (Fig. 2C). The onset of cyclin B1 protein synthesis (translational activation of cyclin B1 mRNA) also occurred at timing similar to that of Pum1 phosphorylation (Fig. 2A). Similar to Pum1-associated CPEB, the content of Pum2-associated CPEB decreased 8 h after progesterone treatment, but no significant correlation was found between the dissociation of CPEB from Pum2, and the timing of Pum2 phosphorylation (Fig. 2C). Despite the fact that a phosphorylated form of CPEB was found in crude oocyte extracts (Fig. 2A and supplemental Fig. S3A), it was not detected in anti-Pumilio immunoprecipitates (Fig. 2B and supplemental Fig. S3B), suggesting that the phosphorylated CPEB is immediately released from Pumilio.
Effects of Anti-Pum1 and Anti-Pum2 Antibodies on Oocyte Maturation
We previously reported that injection of an antibody against the Puf domain of Pum1 (XPum 2A5 monoclonal antibody) into oocytes accelerates progesterone-induced oocyte maturation by the enhancement of cyclin B1 mRNA translation (10). Padmanabhan and Richter (20) reported that injection of an antibody against human Pum2 accelerates oocyte maturation through the promotion of translational activation of RINGO mRNA, which, in turn, induces CPEB destruction and cyclin B1 mRNA translation. We examined whether our newly produced antibodies, anti-Pum1N and anti-Pum2N antibodies, have similar effects on oocyte maturation. Oocytes injected with anti-Pum1N antibody (Fig. 3, A and B) or anti-Pum2N antibody (Fig. 3, D and E) underwent GVBD more rapidly than did control oocytes injected with anti-GST antibody or buffer, although neither anti-Pum1N antibody nor anti-Pum2N antibody induced oocyte maturation without progesterone. The injection of anti-Pum1N antibody (Fig. 3C) or anti-Pum2N antibody (Fig. 3F) accelerated the phosphorylation and degradation of CPEB and the translational activation of cyclin B1 mRNA during oocyte maturation. These results are consistent with those of previous studies (10, 20), at least superficially (see “Discussion”).
FIGURE 3.
Acceleration of oocyte maturation by anti-Pumilio antibodies. Oocytes were injected with buffer, anti-GST, anti-Pum1N, or anti-Pum2N antibody. The injected oocytes were incubated overnight and induced to mature with progesterone. A and D, time course of GVBD in oocytes injected with buffer, anti-GST, anti-Pum1N (A), or anti-Pum2N antibody (D). Representative results are shown. B and E, percentage of GVBD in the control buffer- or GST-injected oocytes and in the anti-Pum1N (B) or anti-Pum2N (E) antibody-injected oocytes. Values are means ± S.E. of three independent experiments. To clearly show the difference in the kinetics of GVBD, the experiments were stopped when the control oocytes (GST-injected oocytes) had undergone ∼25% GVBD. Both Pum1N antibody and Pum2N antibody accelerated progesterone-induced GVBD significantly (p < 0.05, Student's t test). C and F, immunoblots of extracts from oocytes injected with buffer, anti-GST, anti-Pum1N (C), or anti-Pum2N (F) antibody, indicating expression patterns of cyclin B1, cdc2, and CPEB during oocyte maturation. Note that the synthesis of cyclin B1 and the phosphorylation and destruction of CPEB are initiated at earlier timing in oocytes injected with anti-Pum1 or anti-Pum2 antibody than in oocytes injected with buffer or anti-GST antibody.
Target mRNAs of Pum1
We demonstrated that Pum1 is involved in the translational control of cyclin B1 mRNA but not mos mRNA (12). However, it remains uncertain whether Pum1 regulates mRNAs other than cyclin B1. To identify target mRNAs of Pum1, immature oocytes that overexpress GST-Pum1 were subjected to a GST pulldown assay, and mRNAs associated with the GST-tagged proteins were analyzed by RT-PCR. We selected seven mRNAs that are known to undergo cytoplasmic polyadenylation during Xenopus oocyte maturation, cyclin B1, RINGO, mos, wee1, D7, G10, and cyclin A1 (20, 28), together with β-actin mRNA that exhibits no polyadenylation as a control (Fig. 4A). Consistent with our previous report (12), GST-Pum1 precipitates contained cyclin B1 mRNA but not mos mRNA. We also found that RINGO and cyclin A1 mRNAs were present in the GST-Pum1 precipitates (Fig. 4A), suggesting that they are targets of Pum1.
FIGURE 4.
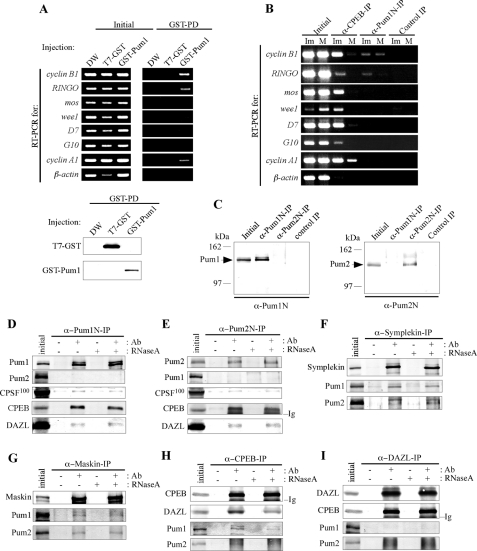
Interaction of Pumilio with mRNAs and other proteins. A, oocytes overexpressing GST-Pum1 were subjected to GST pulldown (GST-PD) followed by RT-PCR to detect mRNAs indicated in the figure. As controls, oocytes injected with distilled water (DW) or GST mRNA were also examined. RT-PCR was also carried out using total RNAs before GST pulldown (Initial, RNAs corresponding to 5% of those subjected to GST pulldown were used) to confirm the presence of indicated mRNAs in the initial samples. Lower figure shows results of an experiment to verify the specific precipitation of GST and GST-Pum1 by GST pulldown. B, immature (Im) and mature (M) oocyte extracts were immunoprecipitated with anti-CPEB (α-CPEB-IP) or anti-Pum1N (α-Pum1N-IP) antibody or without an antibody (Control IP), and the resulting precipitates were subjected to RT-PCR to detect mRNAs indicated in the figure. RT-PCR was also carried out using samples before IP (Initial) to confirm the presence of indicated mRNAs in the initial samples. C, immature oocyte extracts incubated with anti-Pum1N (α-Pum1N-IP) or anti-Pum2N antibody (α-Pum2N-IP) or without an antibody (Control IP) were precipitated with protein A- or protein G-Sepharose beads and immunoblotted with anti-Pum1N antibody (left) and anti-Pum2N antibody (right). Extracts before IP were also immunoblotted to confirm the presence of Pum1 and Pum2 (Initial). D, immature oocytes extracts were treated with (+) or without (−) RNase A, incubated in the presence (+) or absence (−) of anti-Pum1N antibody (Ab), and subjected to IP followed by immunoblotting with antibodies against Pum1, Pum2, CPSF100, CPEB, and DAZL. E, similar experiments to those in D using anti-Pum2 antibody for IP. Immunoglobulins (Ig) of the antibody used for IP are indicated. F, similar experiments to those in D using anti-Symplekin antibody for IP and Symplekin and Pum1 and Pum2 antibodies for immunoblotting. G, similar experiments to those in F using anti-Maskin antibody for IP. H, similar experiments to those in D using anti-CPEB antibody for IP and CPEB, DAZL, Pum1, and Pum2 antibodies for immunoblotting. I, similar experiments to those in H using anti-DAZL antibody for IP. Indistinct bands found in anti-Pum2 immunoblots of anti-DAZL immunoprecipitates are derived from immunoglobulins of the antibody used for IP but not from Pum2. The absence of Pum1 and Pum2 in the anti-DAZL immunoprecipitates is probably due to the failure of antibody in accessing the epitope that was buried in the Pumilio-DAZL complex.
Because it was uncertain whether overexpressed GST-Pum1 precisely mimics endogenous Pum1, we searched for target mRNAs of Pum1 by a different method, IP/RT-PCR using anti-Pum1N antibody, which enables the interaction of endogenous Pum1 with endogenous mRNAs. In agreement with the results from GST pulldown experiments (Fig. 4A), the IP/RT-PCR assay revealed that the anti-Pum1 immunoprecipitates from immature oocytes contained cyclin B1 and RINGO mRNAs (Fig. 4B). However, cyclin A1 mRNA was not present in the anti-Pum1 immunoprecipitates. A possible explanation for the discrepancy between results of GST pulldown and IP experiments is that the Pum1-cyclin A1 mRNA complex was disrupted during IP because the epitope of anti-Pum1 antibody is involved in the interaction of Pum1 with cyclin A1 mRNA. The immunoprecipitates from mature oocytes contained cyclin B1 mRNA as those from immature oocytes, but they did not contain RINGO mRNA (Fig. 4B). This indicates that the interaction of RINGO mRNA with Pum1 disappeared during maturation, in striking contrast to the interaction between cyclin B1 mRNA and Pum1, which was maintained even in mature oocytes.
We also performed IP/RT-PCR using anti-CPEB antibody to examine the interaction between CPEB and mRNAs. Except for β-actin mRNA, all of the mRNAs examined were detected in anti-CPEB immunoprecipitates from immature oocytes (Fig. 4B). However, their contents were significantly decreased in the precipitates from mature oocytes, probably due to the destruction of CPEB during oocyte maturation. These results support the notion that CPEB and its destruction are involved in the translational control of these mRNA. The existence of RINGO mRNA in the anti-CPEB immunoprecipitates indicates interaction of RINGO mRNA with CPEB. Although RINGO mRNA has a CPE in its 3′-UTR, this finding is inconsistent with the results of a previous study that RINGO mRNA is not bound to CPEB (20). Because the content of RINGO mRNA in anti-CPEB immunoprecipitates from immature oocyte extracts was very low as compared with those of cyclin B1, mos, D7 and cyclin A1 mRNAs (Fig. 4B), the previous study might fail to detect the interaction between CPEB and RINGO mRNA.
Interaction of Pumilio with Other Proteins Responsible for Translational Control
Although it has been reported that Pum1 controls cyclin B1 mRNA translation in association with CPEB (10, 12) and that Pum2 controls RINGO mRNA translation in association with DAZL (20), it is not known whether Pum1 and Pum2 interact with each other or whether they interact with other proteins involved in the translational control, including Symplekin, CPSF, DAZL, and Maskin. We thus examined the binding between Pum1 and Pum2 as well as their binding to other proteins by co-IP analysis. Anti-Pum1N immunoprecipitates from immature oocyte extracts did not contain Pum2 and, consistent with this, anti-Pum2N immunoprecipitates did not contain Pum1 (Fig. 4C). These results clearly indicate that there are no physical interactions between Pum1 and Pum2.
CPEB, CPSF100, and DAZL were detected in anti-Pum1 and anti-Pum2 immunoprecipitates from immature oocyte extracts (Fig. 4, D and E). Although Symplekin and Maskin was not found in the anti-Pum1 and Pum2 immunoprecipitates (probably due to the failure of antibodies in accessing the epitopes that were buried in the Pum1-Symplekin, Pum1-Maskin, Pum2-Symplekin, or Pum2-Maskin complex) (data not shown), anti-Symplekin and anti-Maskin immunoprecipitates included Pum1 and Pum2 (Fig. 4, F and G). These findings indicate that Pum1 and Pum2 have almost the same binding partners in immature oocytes. We also noticed interaction between CPEB and DAZL in immature oocytes by an IP assay using anti-CPEB and anti-DAZL antibodies (Fig. 4, H and I). Treatment of the samples with RNase prior to IP provided essentially the same results (Fig. 4, D–I), indicating that the interactions are due to direct binding of proteins but not to mRNA-mediated binding.
In addition to Symplekin, CPSF, CPEB, DAZL, and Maskin, the deadenylase PARN, and the poly(A) polymerase GLD2 are known to play crucial roles in CPE-mediated translational control as key components of the translation control machinery (29). In immature oocytes, both PARN and GLD2 are active, but the poly(A) tail of CPE-containing mRNA remains short because PARN is more active. In mature oocytes, however, PARN is dissociated from the control machinery, which allows GLD2 to elongate the poly(A) tail of CPE-containing mRNA.
We thus attempted to examine the interaction of Pumilio with PARN and GLD2 by a co-IP assay with anti-Pum1 and anti-Pum2 antibodies, which can assess the interaction of endogenous proteins. Unfortunately, however, the co-IP assay did not work, probably due to the inability of antibodies to access their target epitopes when Pumilio is bound to PARN and GLD2. We therefore analyzed the interaction with the aid of GST-tagged recombinant proteins that were overexpressed in oocytes by mRNA injection. i.e. we examined the presence of GST-Pum1 or GST-Pum2 in anti-PARN immunoprecipitates and of PARN in GST pulldown samples from oocytes overexpressing GST-Pum1 or GST-Pum2. GST-Pum1 (Fig. 5A) and GST-Pum2 (Fig. 5B) were found in anti-PARN immunoprecipitates from immature and mature oocytes. The possibility that GST-Pum1 and GST-Pum2 were precipitated by nonspecific binding of anti-PARN antibody to the GST tag was excluded by a control experiment that demonstrated the absence of GST in anti-PARN immunoprecipitates (supplemental Fig. S4). The interaction of PARN with Pum1 or Pum2 was not influenced by RNase treatment (Fig. 5, C and D). Consistent with the results from the co-IP assay, the GST pulldown assay showed that PARN was present in the precipitates, although its protein content was very low (Fig. 5, A and B); the inefficiency in PARN precipitation by GST pulldown might be due to the difficulty of GSH-Sepharose in binding to the complex of GST-Pum1 and PARN or GST-Pum2 and PARN. These results showed that PARN interacts with Pum1 and Pum2 via direct binding of proteins in immature and mature oocytes.
FIGURE 5.
Interaction of Pumilio with PARN and GLD2. A, oocytes were injected with mRNAs encoding GST-Pum1 or T7-tagged GST and incubated with or without progesterone to induce maturation. Extracts from immature (Im) and mature (M) oocytes were incubated with GSH-Sepharose for GST pulldown assay (GST-PD) or incubated in the presence (+) or absence (−) of anti-PARN antibody for IP (α-PARN-IP). The resulting precipitates were immunoblotted with antibodies (Ab) against Pum1, PARN, and T7 tag (for T7-GST in GST-PD). Extracts before IP were also immunoblotted (Initial). B, similar experiments to those in A using oocytes overexpressing GST-Pum2. C, oocyte extracts used in A were treated with (+) or without (−) RNase A and incubated in the presence (+) or absence (−) of anti-PARN antibody for IP. The resulting precipitates were immunoblotted with antibodies against Pum1 and PARN. Extracts before IP were also immunoblotted (Initial). D, similar experiments to those in C using oocyte extracts used in B. E, oocytes were injected with mRNAs encoding T7-GLD2 and GST-Pum1 and incubated with or without progesterone to induce maturation. Oocytes injected with GST mRNA were used as a control. Extracts from immature (Im) and mature (M) oocytes were subjected to GST pulldown assay (GST-PD) or incubated in the presence (+) or absence (−) of anti-T7 antibody for IP (α-T7-IP). The resulting precipitates were immunoblotted with antibodies against Pum1, T7 tag (for T7-GLD2 and T7-GST in GST-PD), and GLD2 (for T7-GLD2 in α-T7-IP to avoid visualization of the antibody used for IP). F, similar experiments to those in E using oocytes overexpressing GST-Pum2. G, oocyte extracts used in E were treated with (+) or without (−) RNase A and incubated in the presence (+) or absence (−) of anti-T7 antibody for IP. The resulting precipitates were immunoblotted with antibodies against Pum1 and T7 tag (for T7-GLD2). H, similar experiments to those in G using oocyte extracts used in F. I, immature (Im) and mature (M) oocyte extracts were incubated with anti-CPEB (α-CPEB-IP), anti-Pum1N (α-Pum1N-IP), or anti-PARN antibody (α-PARN-IP) or incubated without an antibody (Control IP). Following IP, the samples were subjected to RT-PCR to detect cyclin B1 and β-actin mRNAs. Samples before IP (Initial, RNAs corresponding to 5% of those subjected to IP were used) were also analyzed to confirm the presence of cyclin B1 and β-actin mRNAs. J, oocytes overexpressing T7-GLD2 were treated with or without progesterone to induce maturation. Immature (Im) and mature (M) oocyte extracts were incubated with anti-T7 antibody (α-T7(GLD2)-IP) to precipitate T7-GLD2 or incubated without an antibody as a control (Control IP). Samples were subjected to RT-PCR to detect cyclin B1 and β-actin mRNAs.
The interaction between Pumilio and GLD2 was examined by a co-precipitation assay of oocytes overexpressing recombinant Pumilio and GLD2 proteins, because the anti-GLD2 antibody used in this study was not available for IP. GST pulldown and anti-T7 IP from immature and mature oocytes overexpressing T7-GLD2 and GST-Pum1 or GST-Pum2 showed that the pulldown samples contained T7-GLD2 and that the anti-T7 immunoprecipitates contained GST-Pum1 and GST-Pum2 (Fig. 5, E and F). These results revealed that Pum1 and Pum2 interacted with GLD2 in immature and mature oocytes. The interaction of T7-GLD2 with GST-Pum1 or GST-Pum2 was not affected by RNase (Fig. 5, G and H), indicating direct binding of proteins. The interaction between Pumilio and GLD2 was not detected in a previous report (6), in which GST or HA was used to tag GLD2, instead of T7 used in this study.
Pum1, CPEB, PARN, and GLD2 Residing on Cyclin B1 mRNA
We found that Pum1 and Pum2 retain interaction with PARN and GLD2 even in mature oocytes (Fig. 5, A–H). However, this situation seems to conflict with a model showing that PARN is dissociated from the translation control machinery during oocyte maturation (29). Because GST pulldown and co-IP assays are unable to distinguish the complex residing on a specific mRNA from those on other mRNAs, we specifically examined the complex of Pum1, CPEB, PARN, and GLD2 on cyclin B1 mRNA by IP/RT-PCR, in which anti-Pum1N, anti-CPEB, or anti-PARN immunoprecipitates from immature and mature oocytes (Fig. 5I) and anti-T7 immunoprecipitates from oocytes overexpressing T7-GLD2 (Fig. 5J) were subjected to RT-RCR for cyclin B1 mRNA. Except for the anti-CPEB immunoprecipitate from mature oocytes, cyclin B1 mRNA was detected in all of the immunoprecipitates, indicating that the complex of Pum1, CPEB, PARN, and GLD2 exists on cyclin B1 mRNA in immature oocytes but that CPEB is removed from the mRNA (probably by its destruction) during oocyte maturation.
It should be noted that the content of cyclin B1 mRNA in the anti-PARN immunoprecipitate from mature oocytes was clearly lower than that in the immunoprecipitate from immature oocytes despite the fact that the contents of cyclin B1 mRNAs in the anti-Pum1N and anti-T7 (GLD2) immunoprecipitates from immature and mature oocytes were almost the same (Fig. 5, I and J). The most likely explanation for this finding is that some, but not all, of cyclin B1 mRNAs release PARN during oocyte maturation.
DISCUSSION
Using newly produced antibodies that can discriminate between Pum1 and Pum2, we biochemically characterized these proteins in Xenopus oocytes. Co-IP experiments revealed that both Pum1 and Pum2 interact with other key proteins responsible for cytoplasmic polyadenylation and translational control, including CPEB, Maskin, Symplekin, CPSF, PARN, GLD2, and DAZL, in immature oocytes in an RNA-independent manner (Figs. 4, D–I, and 5, A–H) but that Pum1 and Pum2 do not form a complex (Fig. 4, C–E). These findings indicate that oocytes have at least two types of protein complex that act as a machinery for controlling translation of mRNAs in oocytes; one type contains Pum1 and the other contains Pum2, both of which include CPEB, Maskin, Symplekin, CPSF, PARN, GLD2, and DAZL as common components. Pum1 and Pum2 may confer the ability to select mRNAs on each machinery, enabling the Pum1-containing machinery to control cyclin B1 mRNA and the Pum2-containing machinery to control RINGO mRNA. In this context, it should be noted that Pum2 was reported to regulate RINGO mRNA in collaboration with DAZL but not with CPEB (20). In disagreement with this, we found in this study that Pum2 interacts with CPEB and DAZL (Figs. 2B and 4E). Moreover, we found that Pum1 binds to RINGO mRNA in immature oocytes by experiments using GST pulldown and IP followed by RT-PCR (Fig. 4, A and B), whereas we were unable to detect the interaction of Pum2 with RINGO mRNA by similar experiments with oocytes overexpressing GST-Pum2 for GST pulldown and with anti-Pum2 antibody for IP.3 Thus, our finding that Pum1, but not Pum2, interacts with RINGO mRNA apparently conflicts with the notion that Pum2 regulates the translation of RINGO mRNA (20). The discrepancy might be due to the difference in antibodies and tags used in the two studies, anti-human Pum2 antibody and Myc-tagged Pum2 in the previous study versus anti-Xenopus Pum2 antibody and GST-tagged Pum2 in our study.
Consistent with previous reports (10, 20), anti-Pum1N and anti-Pum2N antibodies accelerated progesterone-induced GVBD when injected into oocytes (Fig. 3). A previous explanation was that anti-Pum1 antibody and anti-Pum2 antibody affected the translation of cyclin B1 mRNA and RINGO mRNA, respectively. However, we found that Pum2 is not associated with RINGO mRNA in immature oocytes as opposed to a previous finding that Pum2 is a partner of RINGO mRNA (20). Our finding raises the possibility that anti-Pum2 antibody accelerates oocyte maturation by enhancing the translational activation of an unidentified mRNA(s) other than RINGO mRNA.
Pum1 is phosphorylated during oocyte maturation (Fig. 1C). Although its biological meaning is uncertain, the coincidence of Pum1 phosphorylation, CPEB dissociation from Pum1, and cyclin B1 synthesis (Fig. 2C) suggests that Pum1 phosphorylation induces a conformational change in the complex consisting of Pum1 and CPEB that targets CPEB for dissociation and degradation, the process required for translational activation of cyclin B1 mRNA (30).
Besides cyclin B1 mRNA, RINGO mRNA is assumed to be a target of Pum1 (Fig. 4, A and B). However, the timing of translational activation of RINGO is apparently earlier than cyclin B1 during oocyte maturation (20). Then, how is Pum1 involved in the translational activation that occurs at different timings? A hint to resolve this issue might be the finding that the interaction between Pum1 and RINGO mRNA existing in immature oocytes was aborted in mature oocytes, a situation apparently different from the case of Pum1 and cyclin B1 mRNA, in which the interaction is sustained during oocyte maturation (Fig. 4B). It is conceivable that Pum1 contributes to the selection of mRNAs for the translation control machinery but that the mechanisms controlling target mRNAs vary from mRNA to mRNA; the translation of cyclin B1 mRNA is controlled with continuous association of Pum1 with the mRNA during oocyte maturation, whereas the translation of RINGO mRNA is controlled via the dissociation of Pum1 from the mRNA.
Pum2 is phosphorylated and dissociates from CPEB during oocyte maturation (Figs. 1C and 2, A and B), but the relationship between the two events is unclear. Thus, the biological significance of Pum2 phosphorylation is completely unknown at present. Nevertheless, the fact that Pum2 phosphorylation occurs at earlier timing than Pum1 phosphorylation suggests that mRNAs regulated by Pum2-containing translation control machinery are translated earlier than cyclin B1 mRNA regulated by Pum1-containing machinery. Some mRNAs, including mos, D7, and G10, have a CPE in their 3′-UTR and undergo polyadenylation and translational activation earlier than cyclin B1 mRNA during oocyte maturation (9). Therefore, we investigated the interaction of these mRNAs with Pum2 by GST pulldown assay and IP/RT-PCR assay, but we were unable to detect an interaction. We also attempted to identify Pum2-associated mRNAs by sequencing cDNA clones in a library constructed from anti-Pum2 immunoprecipitates, and we discovered cyclin A2 and cdc20/fizzy, both of which harbor a CPE and a Pumilio-binding element in their 3′-UTRs, in the library; however, their association with Pum2 in oocytes was not confirmed by the IP/RT-PCR or UV cross-linking assay.3 In mammals, Pum2 has been reported to regulate several mRNAs, including those encoding Erk2 and p38α (members of the MAPK family), in human embryonic stem cells (31) and those encoding the initiation factor eIF4E and the voltage-gated sodium channel Scn1a in mouse neurons (32). Nonetheless, a gene homologous to Scn1a is not found in the X. laevis genome. Erk2 and eIF4E are continuously expressed during oocyte maturation (33, 34), and p38α does not play a role in Xenopus oocyte maturation (35). Accordingly, it is unlikely that the translation of mRNAs encoding Erk2, p38α, eIF4E, and Scn1a are regulated by Pum2 in Xenopus oocytes, in contrast to mammalian cells. Although RINGO mRNA has been reported to be a target of Pum2 in Xenopus oocytes (20), our findings seem to contradict this idea as already noted. Therefore, it remains a mystery what mRNAs are regulated by the Pum2-containing machinery in Xenopus. In humans, 61 mRNAs have been reported to be potential targets of Pum2 and DAZL (36). Some of them might be regulated by Pum2 also in frogs. Identification of target mRNAs of Pum2 and evaluation of the biological significance of Pumilio phosphorylation in translational activation of their target mRNAs should provide further insights into the mechanisms underlying the temporal control of translational activation of mRNAs during Xenopus oocyte maturation.
In this study, we obtained results strongly suggesting the release of PARN from some, but not all, of cyclin B1 mRNAs during oocyte maturation (Fig. 5I). This finding implies that cyclin B1 mRNAs stockpiled in oocytes are not uniform and that their translation is controlled differently during the processes of oocyte maturation, fertilization, and embryogenesis. Indeed, cyclin B1 mRNA has been reported to be enriched in the mitotic spindle pole (37), with which Maskin and CPEB are also associated (38). The spindle-associated cyclin B1 mRNAs may be controlled differently from those dispersed in the cytoplasm or those localized to other cytoskeletal components. For example, oocyte maturation includes several morphological changes accompanying the progression of meiosis from prophase I to metaphase II, such as GVBD, chromosome condensation, and spindle formation (39). It is plausible that these events are induced by spatially controlled translation of cyclin B1 mRNAs localized to the nuclear envelope, chromosomes, and spindle microtubules, respectively. Further studies are required to disclose the molecular mechanisms that enable the translational activation of localized mRNAs at specific timings (40).
Supplementary Material
Acknowledgments
We are deeply grateful to Dr. Michael Wormington (University of Virginia) for providing anti-PARN antiserum. We also thank Dr. Koichi Mita (Tokushima Bunri University) and Dr. Toshiharu Iwai (Ehime University) for technical advice.
This work was supported in part by research fellowships from the Japan Society for the Promotion of Science for Young Scientists (to R. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
The nucleotide sequence reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) AB565475.
R. Ota, T. Kotani, and M. Yamashita, unpublished data.
- MPF
- maturation-promoting factor
- CPE
- cytoplasmic polyadenylation element
- CPEB
- CPE-binding protein
- CPSF
- cleavage and polyadenylation specificity factor
- DAZL
- Deleted in Azoospermia-like protein
- EB
- extraction buffer
- GVBD
- germinal vesicle breakdown
- IP
- immunoprecipitation
- RINGO
- rapid inducer of G2-M in oocytes.
REFERENCES
- 1. Nagahama Y., Yamashita M. (2008) Dev. Growth Differ. 50, S195–S219 [DOI] [PubMed] [Google Scholar]
- 2. Belloc E., Méndez R. (2008) Nature 452, 1017–1021 [DOI] [PubMed] [Google Scholar]
- 3. Mendez R., Richter J. D. (2001) Nat. Rev. Mol. Cell Biol. 2, 521–529 [DOI] [PubMed] [Google Scholar]
- 4. Richter J. D. (2007) Trends Biochem. Sci. 32, 279–285 [DOI] [PubMed] [Google Scholar]
- 5. Radford H. E., Meijer H. A., de Moor C. H. (2008) Biochim. Biophys. Acta 1779, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rouhana L., Wang L., Buter N., Kwak J. E., Schiltz C. A., Gonzalez T., Kelley A. E., Landry C. F., Wickens M. (2005) RNA 11, 1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Moor C. H., Richter J. D. (1997) Mol. Cell. Biol. 17, 6419–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Charlesworth A., Welk J., MacNicol A. M. (2000) Dev. Biol. 227, 706–719 [DOI] [PubMed] [Google Scholar]
- 9. Charlesworth A., Cox L. L., MacNicol A. M. (2004) J. Biol. Chem. 279, 17650–17659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakahata S., Kotani T., Mita K., Kawasaki T., Katsu Y., Nagahama Y., Yamashita M. (2003) Mech. Dev. 120, 865–880 [DOI] [PubMed] [Google Scholar]
- 11. Arumugam K., Wang Y., Hardy L. L., MacNicol M. C., MacNicol A. M. (2010) EMBO J. 29, 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakahata S., Katsu Y., Mita K., Inoue K., Nagahama Y., Yamashita M. (2001) J. Biol. Chem. 276, 20945–20953 [DOI] [PubMed] [Google Scholar]
- 13. Wang Y. Y., Charlesworth A., Byrd S. M., Gregerson R., MacNicol M. C., MacNicol A. M. (2008) Dev. Biol. 317, 454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. MacNicol M. C., MacNicol A. M. (2010) Mol. Reprod. Dev. 77, 662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wickens M., Bernstein D. S., Kimble J., Parker R. (2002) Trends Genet. 18, 150–157 [DOI] [PubMed] [Google Scholar]
- 16. Spassov D. S., Jurecic R. (2002) Gene 299, 195–204 [DOI] [PubMed] [Google Scholar]
- 17. Spassov D. S., Jurecic R. (2003) Blood Cells Mol. Dis. 30, 55–69 [DOI] [PubMed] [Google Scholar]
- 18. Kurisaki I., Iwai T., Yamashita M., Kobayashi M., Ito E., Matsuoka I. (2007) Cell Tissue Res. 327, 33–42 [DOI] [PubMed] [Google Scholar]
- 19. Lee J. Y., Lim J. M., Kim D. K., Zheng Y. H., Moon S., Han B. K., Song K. D., Kim H., Han J. Y. (2008) Mol. Reprod. Dev. 75, 184–190 [DOI] [PubMed] [Google Scholar]
- 20. Padmanabhan K., Richter J. D. (2006) Genes Dev. 20, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirai T., Yamashita M., Yoshikuni M., Lou Y. H., Nagahama Y. (1992) Mol. Reprod. Dev. 33, 131–140 [DOI] [PubMed] [Google Scholar]
- 22. Ota R., Suwa K., Kotani T., Mita K., Yamashita M. (2008) Zool. Sci. 25, 773–781 [DOI] [PubMed] [Google Scholar]
- 23. Mita K., Yamashita M. (2000) Mech. Dev. 94, 251–255 [DOI] [PubMed] [Google Scholar]
- 24. Körner C. G., Wormington M., Muckenthaler M., Schneider S., Dehlin E., Wahle E. (1998) EMBO J. 17, 5427–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tanaka T., Yamashita M. (1995) Dev. Growth Differ. 37, 387–393 [DOI] [PubMed] [Google Scholar]
- 26. Sakamoto I., Takahara K., Yamashita M., Iwao Y. (1998) Dev. Biol. 195, 60–69 [DOI] [PubMed] [Google Scholar]
- 27. Yamashita M., Yoshikuni M., Hirai T., Fukada S., Nagahama Y. (1991) Dev. Growth Differ. 33, 617–624 [DOI] [PubMed] [Google Scholar]
- 28. Ballantyne S., Daniel D. L., Jr., Wickens M. (1997) Mol. Biol. Cell 8, 1633–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim J. H., Richter J. D. (2006) Mol. Cell 24, 173–183 [DOI] [PubMed] [Google Scholar]
- 30. Mendez R., Barnard D., Richter J. D. (2002) EMBO J. 21, 1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee M. H., Hook B., Pan G., Kershner A. M., Merritt C., Seydoux G., Thomson J. A., Wickens M., Kimble J. (2007) PLoS Genet. 3, e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vessey J. P., Schoderboeck L., Gingl E., Luzi E., Riefler J., Di Leva F., Karra D., Thomas S., Kiebler M. A., Macchi P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Git A., Allison R., Perdiguero E., Nebreda A. R., Houliston E., Standart N. (2009) RNA 15, 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cao Q., Richter J. D. (2002) EMBO J. 21, 3852–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perdiguero E., Pillaire M. J., Bodart J. F., Hennersdorf F., Frödin M., Duesbery N. S., Alonso G., Nebreda A. R. (2003) EMBO J. 22, 5746–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fox M., Urano J., Reijo Pera R. A. (2005) Genomics 85, 92–105 [DOI] [PubMed] [Google Scholar]
- 37. Blower M. D., Feric E., Weis K., Heald R. (2007) J. Cell Biol. 179, 1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barnard D. C., Cao Q., Richter J. D. (2005) Mol. Cell Biol. 25, 7605–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kotani T., Yamashita M. (2002) Dev. Biol. 252, 271–286 [DOI] [PubMed] [Google Scholar]
- 40. Yasuda K., Kotani T., Ota R., Yamashita M. (2010) Dev. Biol. 348, 76–86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.