Abstract
Endocrine therapy with tamoxifen (TAM) significantly improves outcomes for patients with estrogen receptor-positive breast cancer. However, intrinsic (de novo) or acquired resistance to TAM occurs in a significant proportion of treated patients. To identify genes involved in resistance to TAM, we introduced full-length cDNA expression library into estrogen receptor-positive MCF7 cells and exposed them to a cytotoxic dose of 4-hydroxytamoxifen (4OHTAM). Four different library inserts were isolated from surviving clones. Re-introduction of the genes individually into naive MCF7 cells made them resistant to 4OHTAM. Cells overexpressing these genes had an increase in acidic autophagic vacuoles induced by 4OHTAM, suggesting their role in autophagy. One of them, prolylcarboxypeptidase (PRCP), was investigated further. Overexpression of PRCP increased cell proliferation, boosted several established markers of autophagy, including expression of LC3-2, sequestration of monodansylcadaverine, and proteolysis of BSA in an ER-α dependent manner, and increased resistance to 4OHTAM. Conversely, knockdown of endogenous PRCP in MCF7 cells increased cell sensitivity to 4OHTAM and at the same time decreased cell proliferation and expression of LC3-2, sequestration of monodansylcadaverine, and proteolysis of BSA. Inhibition of enzymatic activity of PRCP enhanced 4OHTAM-induced cytotoxicity in MCF7 cells. Cells with acquired resistance to 4OHTAM exhibited increased PRCP activity, although inhibition of PRCP prevented development of 4OHTAM resistance in parental MCF7 cells and restored response to 4OHTAM in MCF7 cells with acquired resistance to 4OHTAM. Thus, we have for the first time identified PRCP as a resistance factor for 4OHTAM resistance in estrogen receptor-positive breast cancer cells.
Keywords: Anticancer Drug, Autophagy, Breast Cancer, Cancer Therapy, Cell Death, Estrogen, 4-Hydroxytamoxifen, ER-positive Breast Cancer, Prolylcarboxypeptidase, Tamoxifen Resistance
Introduction
Efforts to develop new chemotherapeutic drugs can be thwarted by the emergence of drug resistance. Insights into the mechanisms of resistance can greatly facilitate development of better, more potent drugs. Knowledge of resistance pathways paves the way to their targeted manipulation to minimize the effects of resistance or to reverse its course. Regulatory pathways involved in drug resistance might also reveal predictive biological markers for resistance, thus allowing rational adjustments of the therapeutic regimen before growth of resistant tumors becomes clinically obvious.
Tamoxifen (TAM),2 a selective estrogen receptor modulator, is probably one of the most efficient therapeutic agents for breast cancer, and many patients have benefited from TAM therapy over the years (1). Initially thought to act as an estrogen receptor (ER) antagonist and inhibit growth of ER-positive tumors, TAM proved to be active against some ER-negative tumors as well, while failing to achieve even temporary remission in other tumors with expressed ER (2). Dual activity of TAM as an ER inhibitor and a cytotoxic agent explained its effects against ER-negative cells (3), but the mechanism(s) of intrinsic as well as acquired resistance to TAM turned out to be complex and is not yet fully understood (4). Recently, aromatase inhibitors are used as the second-line therapy to block the synthesis of estrogen from precursors in ER-positive postmenopausal women with breast cancer (5). These drugs can block ER better than selective estrogen receptor modulators, but emerging resistance remains a challenge to breast cancer treatment (6).
One of the biological responses of ER-positive MCF7 breast cancer cells to 4OHTAM is autophagy (7–9), a multistep process that begins with formation of autophagosomes followed by their fusion with lysosomes to produce autolysosomes (10). Initially, autophagy induced by a cytotoxic dose of 4OHTAM in MCF7 cells was thought to lead to cell death (7). Recent studies have identified autophagy as a mechanism for cell survival in response to 4OHTAM treatment. Disruption of the autophagic process by inhibition of autophagosome formation or autolysosomal function led to increased 4OHTAM-induced cell death in TAM-sensitive and TAM-resistant MCF7 cells (8, 9). These findings have redefined the role of autophagy as a mechanism of cell survival in response to 4OHTAM, which is similar to its established role in cell survival after genomic injury, endoplasmic reticulum stress, oxidative stress, nutrient insufficiency, and infection (11–14).
In this study, we applied functional selection techniques to identify genes that protect MCF7 cells against the cytotoxic action of 4OHTAM. A full-length cDNA expression library was delivered into MCF7 cells, which were then selected for survival in the presence of the drug. Selection conditions were chosen to approximate clinical settings, with a certain level of estrogen present during TAM therapy. Serum proteins in the medium, including albumin, could sequester TAM (15), thereby reducing its effective concentration. To compensate, concentrations higher than (although comparable with) clinically achievable in tumors were used during functional selection to ensure elimination of unprotected cells (16). Four genes were selected, and one of them, prolylcarboxypeptidase (PRCP), was characterized in detail. PRCP appears to regulate proliferation and autophagy in an ER-α-dependent manner while protecting MCF7 cells against 4OHTAM-induced cytotoxicity.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
Goat anti-PRCP antibody has been described (17). Mouse polyclonal anti-PRCP antibody, rabbit anti-LC3 antibody, and rabbit anti-Beclin 1 antibody was from Abcam, Inc. (Cambridge, MA). Mouse anti-β-actin antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Mouse anti-ER-α antibody was from Vector Laboratories (Burlingame, CA). 4OHTAM, MTT, and monodansylcadaverine (MDC) were from Sigma. Z-Pro-Prolinal (ZPP) was from Biomol (Plymouth Meeting, PA). High molecular weight kininogen and prekallikrein (PK) were from Enzyme Research Laboratory (South Bend, IN). HD-Pro-Phe-Arg-paranitroaniline (S2302) was from DiaPharma (Franklin, OH). Ala-Pro-paranitroaniline was from Bachem (Torrance, CA). DQ-Red-BSA, MitoTracker Red CMXRos, Mitofluor 589, LysoTracker Blue DND-22, and PicoGreen were from Invitrogen.
Cell
MCF7 (ATCC, HTB-22) cells were grown in Dulbecco's modified Eagle's medium, 2 mm glutamine, 0.1 mm nonessential amino acids, 10 units/ml penicillin, 10 μg/ml streptomycin (all from Invitrogen), supplemented with 10% fetal bovine serum (Sigma), 6 μg/ml insulin (Sigma), 30 μg/ml fungin, and 10 μg/ml plasmocin (both from InvivoGen, San Diego). 4OHTAM (Sigma) was used as 10 mm stock solution in ethanol and stored at −20 °C. MCF7.beclin cells expressing Beclin 1 from a tetracycline-repressible promoter (pTRE/FLAG-Beclin 1) (18) were provided by Dr. Beth Levine (University of Texas Southwestern Medical Center, Dallas).
cDNA Expression Library
ViraPort® fetal human brain full-length cDNA expression library in pFB vector (Stratagene, CA) was amplified once on solid support. To monitor the efficiency of retroviral infection, a pFB vector with enhanced green fluorescent protein (EGFP, Invitrogen) was used.
Retroviral and Lentiviral Infection
VSVg-pseudotyped retroviral supernatant for the cDNA library expression was prepared after transient transfection of 293FT cells by Dr. A. Miyanohara (Program in Human Gene Therapy, University of California, San Diego, La Jolla, CA) using a 10:1 mixture of cDNA library- and EGFP-containing constructs. For library transduction, MCF7 cells were plated at 106 per 100-mm plate 24 h prior to infection. Polybrene (1 μg/ml final concentration) was added to the viral supernatant, which was filtered through a 0.45-μm filter and added to MCF7 for 24 h. Cells were allowed to recover for 24 h, collected, and frozen in aliquots of 106 cells. An aliquot was used to determine the fraction of cells that expressed GFP, and an estimate of library coverage was made. Re-infection experiments with individual clones were performed similarly. The lentiviral pLKO-PRCP shRNA and ER-α shRNA sets were purchased from OpenBiosystems (clone ID TRCN0000050808 for PRCP shRNA1, TRC0000050809 for PRCP shRNA2, and clone ID TRCN0000003300 for ER-α shRNA). The lentiviral packaging and envelop vectors psPAX2 and pMD2.G (Addgene plasmid 12260 and 12259 deposited by Dr. Didier Trono) and the pLKO-control shRNA (Addgene plasmid 1864 deposited by Dr. David M. Sabatini) (19) were obtained from Addgene plasmid repository. Lentiviral supernatants for the expression of shRNAs were generated from 293FT cells by co-transfection of shRNA constructs with psPAX2 and pMD2.G packaging and envelope vectors according to the OpenBiosystems protocol. MCF7 cells were infected and selected with puromycin for 2 weeks to establish polyclonal lines. The retroviral EGFP-LC3 construct was generated by two-step PCR amplification. LC3 was first amplified using the pEGFP-LC3 plasmid DNA (Addgene plasmid 24920 deposited by Dr. T. Finkel) (20) as template and subcloned into the pEGFP-C1 vector using engineered BglII and EcoRI sites. The EGFP-LC3 was subsequently amplified and subcloned into pMSCV-pac retroviral vector with the engineered XhoI and EcoRI restriction sites. Retroviral supernatant was prepared to infect MCF7.beclin cells using the method described previously (21).
Selection of Resistant Clones from MCF7 Cells Transduced with cDNA Library with 4OHTAM
cells were plated in Peel-Off tissue culture flasks (Sigma) at 106 cells per 150-cm2 flask 24 h prior to selection with 4OHTAM (20 μΜ final concentration); selection continued for 14 days with media replacement every 2 days. Surviving cells were expanded in drug-free media. The screen was performed twice with independent infections.
DNA and RNA Isolation
Genomic DNA was prepared using DNeasy tissue kit (Qiagen, CA); total RNA was prepared using RNAqueous-4PCR kit (Ambion, TX); the first DNA strand was synthesized using RETROscript kit (Ambion, TX). Manufacturers' protocols were followed in each case.
PCR, Cloning, and Sequencing
Advantage-2 polymerase (Clontech) was used for PCR (38 cycles: 94 °C, 30 s; 59 °C, 20 s; 68 °C, 60 s); vector-specific primers were pFB-F (CCTAGAACCTCGCTGGAAAGGACCTTACAC) and pFB-R (AGAGTCCCGCTCAGAAGAACTCGGATCG). PCR products were cloned into pGEM-T Easy vector (Promega, WI) and sequenced using M13 primers. The same procedure was used for RT-PCR with pFB-F and gene-specific primers: B6-R, 5′-GGGACTTACAAATGGGCCAAAGACAC-3′.
Clonogenic Assay
103 cells were plated in a 60-mm plate and allowed to recover overnight. The media were then replaced with 4OHTAM-containing media; control plates were replaced with vehicle-containing media. Treatment with 7.5 or 10 μm of TAM lasted for 3 days, although treatment with 5 μm TAM continued for 14 days with media replacement every 2 days; cells were allowed to recover for 2 weeks, fixed with alcohol, and stained with crystal violet (2% w/v) or methylene blue (1% in 50% ethanol). Colonies were counted. Experiments were done in triplicate.
Propidium Iodide Measurement of Plasma Membrane Integrity
105 cells were plated per 12-well culture dish, treated with 4OHTAM for specified periods of time, trypsinized, combined with floaters, resuspended in ice-cold 100 μm PBS, stained with 10 μm PI/RNase buffer (BD Biosciences) for 15 min in the dark at room temperature, and analyzed by flow cytometry.
Quantitative Analysis of Mitochondria and Acidic Autophagic Vacuoles
MitoTracker Red CMXRos (mitochondrial membrane potential) and Mitofluor 589 (mitochondrial mass detection) were added to cells (250 nm final concentration) for 25 min at 37 °C in the CO2 incubator. LysoTracker Blue DND-22 (lysosome/vacuole compartment) was added to cells (800 nm final concentration) for 1.5 h at 37 °C in a CO2 incubator. Cells were harvested by trypsinization and then analyzed by Flow Cytometry using a Beckman Coulter Epics XL-MCL (Beckman Coulter, Miami, FL) with System II version 3.0 software and CYAN (DakoCytomation, CO) and Summit version 3.3 software.
MDC Sequestration Assay and PicoGreen Staining of Cellular DNA
The cells were plated in 60-mm dishes (2 × 105 cells/dish) and treated with 4OHTAM for 2 days. The cells were labeled with MDC (0.05 mm in PBS) for 30 min, and the total cells (monolayer or detached) were collected in 300 μl of lysis buffer (10 mm Tris, pH 8.0, 0.1% Triton X-100) and incubated on ice with light agitation for 20 min. Following incubation, the cell suspension was transferred to a black-colored 96-well plate, and fluorescence was determined using a BioTek Mx microplate reader with an excitation of 360 nm and an emission of 520 nm. The cells then underwent three rounds of standard freeze and thaw followed staining with PicoGreen (1:200 dilution in TE buffer, pH 8.0). The fluorescence intensity of PicoGreen was determined using a BioTek Mx microplate reader with an excitation of 480 nm and an emission of 520 nm. The cell number in each well was calculated with a standard cell titration curve of PicoGreen-stained cell lysates, and the relative values of the MDC fluorescence were calculated.
Quantitative Analysis of Proteolysis of BSA by Lysosomes
The cells were treated with DQ-Red-BSA (10 μg/ml) for 1 h and then incubated at 37 °C for 3 h. The cells were then trypsinized and resuspended in FACS buffer (10% FBS/PBS). A three laser (405, 633, and 488 nm) LSR II flow cytometer (BD Biosciences) and FACSDiva version 6.1.2 software were used to acquire and analyze the samples. The DQ-Red signal was excited by the 633-nm red laser and collected using a 610/20 bandpass filter.
Analysis of Cell Viability by MTT Assay
Cells grown in 96-well plates were incubated in phenol red-free culture medium containing 1.2 mm MTT (100 μl/well) at 37 °C for 3 h. The cells were then lysed by addition of 100 μl of 10% SDS, 0.01 m HCl to each well and incubation at 37 °C for 12 h. The plates were read for absorbance at 570 nm by a microplate reader.
Confocal Microscopy and Quantification of Auto-phagosomes
MCF7.beclin cells on glass coverslips were infected with pMSCV-EGFR-LC3 for 2 days and were starved in Hanks' balanced salt solution for 6 h. The cells were mounted on the UptraCruzTM mounting medium and then analyzed by a Zeiss LSM510 confocal microscope for autophagosomes visualized as EGFR-LC3 dotted structures. The images were converted to binary images, and the numbers of autophagosomes and cell areas were measured using the ImageJ program (National Institutes of Health).
Immunoblot
Cells were lysed in RIPA buffer followed with SDS-PAGE and immunoblotting carried out as described earlier (21). The images were recorded by a BioSpectrum 500 imaging system with LM-26 and Biochemi 500 Camera 1/1.2 (UVP, Upland, CA).
Determining the Pattern of Kallikrein Production by PRCP in MCF7 and Its TAM-resistant Derivatives
To determine PRCP activity, the activation of PK to kallikrein by PRCP on cell-bound high molecular weight kininogen was assessed as described previously (22). Cells (2.5 × 104 cells/well) were grown overnight in 96-well plates. The 4OHTAM-resistant cells (described below) were maintained in medium containing 1 μm 4OHTAM. Confluent monolayers of cells were washed with HEPES-carbonated buffer (137 mm NaCl, 3 mm KCl, 12 mm NaHCO3, 14.7 mm HEPES, 5.5 mm glucose, 0.1% gelatin, 2 mm CaCl2, 1 mm MgCl2, pH 7.1, 37 °C) and blocked with 1% gelatin at 37 °C for 1 h. The substrate HD-Pro-Phe-Arg-paranitroaniline (S2302, 0.8 mm) was used to measure kallikrein production by an increase in absorbance at 405 nm after 1 h of incubation at 37 °C. To determine the effect of the anti-PRCP antibodies on PRCP-dependent PK activation, anti-PRCP antibody was titrated, and the kallikrein activity was measured after addition of the chromogenic substrate (S2302, 0.8 mm) to MCF7 cells and the 4OHTAM-resistant cells. They were then lysed by standard freeze and thaw and stained with PicoGreen for cellular DNA. The relative absorbance was normalized to cellular DNA (ng/well). Data represent the mean ± S.E. of three experiments in triplicate.
Statistical Analysis
The statistical analysis for all the experiments was done by one-way ANOVA followed by a two-tailed t test.
RESULTS
Genes Inducing Resistance to Tamoxifen Affect the Growth Characteristics and Increase Viability of MCF7 Cells
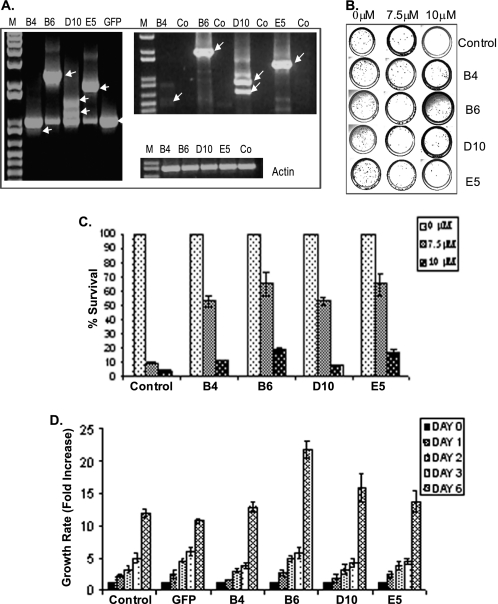
ViraPort® human fetal brain full-length cDNA library (Stratagene) in pFB vector was used to screen for genes that induced resistance to tamoxifen in MCF7 cells. Four clones resistant to a cytotoxic dose of 4OHTAM (supplemental Fig. 1A) were identified and designated B4, B6, D10, and E5 (supplemental Fig. 1B). Library inserts from these clones were isolated, re-cloned into pFB vector, and individually transduced into naive MCF7 cells (Fig. 1A). Induction of resistance was confirmed by the response of corresponding populations to 4OHTAM as shown by clonogenic assay (Fig. 1, B and C). Isolated library inserts were designated as resistance-inducing genes (RIGs).
FIGURE 1.
Re-transduction of identified cDNA inserts made MCF7 cells resistant to 4OHTAM. A, integration of corresponding cDNA inserts confirmed by PCR using genomic DNA from re-infected populations (left panel). Expression of delivered cDNA inserts confirmed by RT-PCR using total RNA from the same populations (right panel). RT-PCR of actin was used as a control. Source of the genomic DNA or total RNA is indicated on top. Co-control reactions were assembled with cDNA from parental MCF7 cells. B, clonogenic assay was performed in triplicate in p60-mm dishes, treated with vehicle or 4OHTAM for 3 days, and then recovered in drug-free media for 2 weeks. Colonies were stained with methylene blue. C, colonies were quantified and normalized to vehicle-treated controls. The average relative numbers of colonies were presented with standard deviation shown (representative graph of three independent experiments). D, indicated cell lines were cultured in drug-free media and were harvested at the indicated time points and stained with Hoechst 33342 for cellular DNA. The fluorescence values were normalized to the cells harvested on day 1.
To gain a better understanding of changes induced by the RIGs, we evaluated cell growth of RIG-expressing populations (Fig. 1D). In drug-free media, proliferation rates of B4 RIG-expressing cells were similar to parental MCF7 cells or EGFP-expressing control. However, proliferation rates for cells expressing B6, D10, and E5 RIGs were higher than the controls (Fig. 1D).
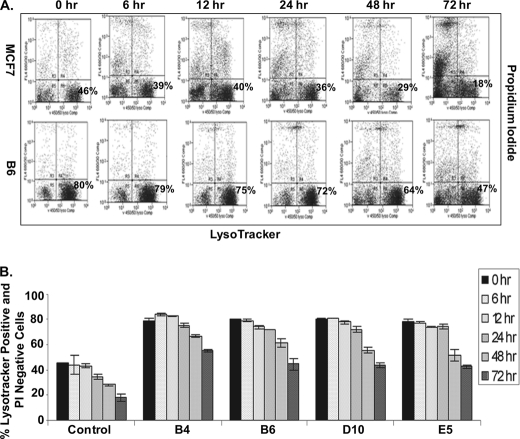
In recent studies autophagy was identified as a major response to a cytotoxic dose of 4OHTAM in MCF7 cells (7). To determine whether autophagy was affected by the RIGs, we tested the formation of acidic vesicular organelles (AVO), also called acid autophagic vacuoles, one of the morphologic features of autophagy (23, 24). AVO can be stained with the acidic dye LysoTracker Blue DND-22. To correlate AVO formation with cell viability, MCF7 cells and the RIGs-expressing cells were treated with 10 μm 4OHTAM and then co-stained with LysoTracker Blue DND-22 for AVO and PI for damaged cells with increased permeability of plasma membrane. An increase in AVO-positive cells and a decrease in PI staining for the RIG-expressing cells compared with control MCF7 cells (Fig. 2, A and B) suggested that development of autophagic vacuoles correlated with improved survival of the RIG-expressing cells.
FIGURE 2.
RIGs-expressing cells have increased AVO formation in response to 4OHTAM, which is correlated with increased plasma membrane integrity. A, MCF7, B4, B6, D10, and E5 cell lines were treated with 10 μm 4OHTAM for a time course, co-stained with propidium iodide and LysoTracker Blue DND-22, and analyzed by FACS. The Blue DND-positive and PI-negative cells were gated at the lower right corner (representative picture of three independent experiments in MCF7 cells and B6 cells). B, average percentage of Blue DND-positive and PI-negative cells in all of the cell lines in triplicate was presented as a graph with standard deviation indicated (representative graph of three independent experiments).
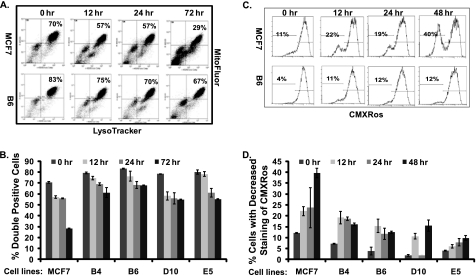
Considering that the autophagic process affected mitochondria (25), both the RIG-expressing and parental cells were co-stained with Blue DND-22 and Mitofluor 589 for the evaluation of mitochondrial mass. All of the RIG-expressing cells showed higher fluorescence intensity of Mitofluor 589 correlated with AVO positivity (Fig. 3, A and B), suggesting that the RIG-expressing cells contained more AVO and mitochondria. When the cells were also stained with MitoTracker Red CMXRos to measure mitochondrial activity, the parental MCF7 cells showed a significantly higher proportion of cells with reduced CMXRos staining during 4OHTAM treatment compared with the RIG-expressing cells (Fig. 3, C and D). This finding confirmed increased viability of the RIG-expressing cells in response to a toxic dose of 4OHTAM.
FIGURE 3.
Increased AVO formation in RIGs-expressing cells is correlated with increased mitochondrial mass and a stabilized mitochondrial transmembrane potential. A, MCF7, B4, B6, D10, and E5 cell lines were treated with 10 μm 4OHTAM and co-stained with Mitofluor and LysoTracker Blue DND-22 for a time course, and their fluorescence was measured by FACS. The Blue DND and Mitofluor double-stained cells were gated at the lower right corner (representative picture of three independent experiments in MCF7 cells and B6 cells). B, average percentage of Blue DND- and Mitofluor-positive cells from triplicate were presented as a graph with standard deviation indicated (representative graph of three independent experiments). C, MCF7, B4, B6, D10, and E5 cell lines were treated with 10 μm 4OHTAM and stained with CMXRos for a time course, and their fluorescence was measured by FACS at the indicated time points. The cells with decreased staining of CMXRos were gated on the left (representative picture of three independent experiments in MCF7 cells and B6 cells). D, average percentage of cells with decreased staining of CMXRos (in triplicate) was presented as a graph with standard deviation indicated (representative graph of three independent experiments).
Overexpression of PRCP Increases ER-dependent Cell Proliferation and Cell Resistance to 4OHTAM
Knockdown of PRCP Decreases Cell Proliferation and Sensitizes Cells to 4OHTAM-induced Cytotoxicity
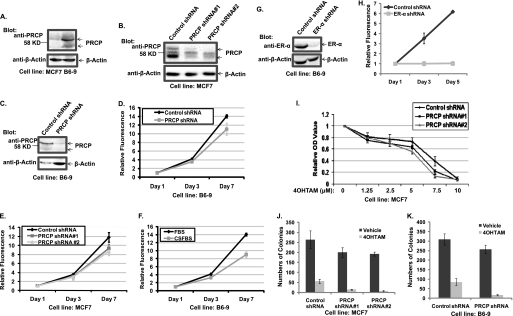
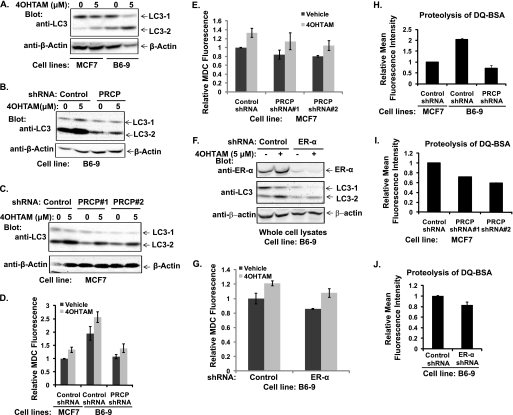
Increased AVO in the RIG-expressing cells suggested that autophagy was enhanced by the RIGs expression, so we characterized molecular events linked to one of the genes (B6) that encoded isoform A of the PRCP. Experiments were done with a cell line (B6-9) isolated by single-cell cloning from a population of B6-transduced cells with more than a 10-fold increase in PRCP mRNA as detected by quantitative PCR (data not shown). Two bands at ∼58 kDa were detected with the goat anti-PRCP antibody in parental MCF7; the signals of these bands were significantly stronger in B6-9 cells (Fig. 4A), indicating a detectable overexpression of PRCP protein. When short hairpin RNA constructs (shRNA) were introduced into parental MCF7 cells by transduction, a lower signal was observed in populations transduced with PRCP shRNA compared with the control population transduced with scrambled shRNA (Fig. 4B). Similarly, transduction of anti-PRCP shRNA2 (but not scrambled shRNA) into B6-9 cells induced a significant reduction in proteins recognized by anti-PRCP antibodies (Fig. 4C). The mouse anti-PRCP antibody (Abcam) detects three bands. Only the upper and lower bands are detected by the goat anti-PRCP antibody and are decreased in PRCP shRNA-expressing MCF7 and B6-9 cells. Based on these results, we concluded that the two increased bands detected in the B6-9 cell line were the overexpressed PRCP gene products.
FIGURE 4.
PRCP regulates cell proliferations, which are dependent on ER-α. Overexpression of PRCP decreases and knockdown of PRCP increases 4OHTAM-induced cytotoxicity. A, whole cell lysates of MCF7 and B6-9 were probed with the goat anti-PRCP antibody. Note that two bands around the 58-kDa marker are detected by the goat anti-PRCP antibody. B, whole cell lysates of MCF7-control shRNA, MCF7-PRCP shRNA1 and -2, were probed with the mouse anti-PRCP antibody (Abcam). Note that the mouse anti-PRCP antibody detects three bands, and only the signal of the upper and lower bands is decreased in PRCP shRNA-expressing cells. C, whole cell lysates of B6-9-control shRNA and B6-9-PRCP shRNA were probed with the goat anti-PRCP antibody. D and E, cells were grown in 96-well plates (2.5 × 103 cells/well) in octuplicate for the time indicated, stained with PicoGreen, and the average fluorescence intensity evaluated. Values at days 3 and 7 were normalized to the basal (day 1) value and presented as graphs with standard deviation shown (representative graph of two independent experiments). F, B6-9-control shRNA cells were grown in medium containing regular FBS or charcoal-striped FBS (CSFBS) were also compared for their proliferation by PicoGreen staining (representative graph of two independent experiments). G, whole cell lysates of B6-9-control shRNA and B6-9-ER-α shRNA were probed with anti-ER-α antibodies. H, B6-9-control shRNA cells and B6-9 ERα shRNA cells were grown in medium containing regular FBS, and their proliferation was compared by PicoGreen staining (representative graph of two independent experiments). I, MCF7-control shRNA and MCF7-PRCP shRNA1 and -2 cells in 96-well plates were treated with different doses of 4OHTAM for 3 days. The cells were analyzed with the MTT assay, and the average OD values from octuplicate were normalized to the vehicle-treated cells and presented as a graph with standard deviation indicated (representative graph of three independent experiments). There are significant differences between control shRNA and PRCP shRNA1 cells treated with 5 μm (p = 0.006) and 7.5 μm 4OHTAM (p = 6 × 10−5) and PRCP2 cells treated with 5 μm (p = 0.002) and 7.5 μm 4OHTAM (p = 2 × 10−5). There are no significant differences between control shRNA and PRCP shRNA1 (p = 0.08, 0.06, and 0.12) and PRCP2 (p = 0.06, 0.11, and 0.23) treated with 1.25, 2. 5, and 10 μm of 4OHTAM. J, clonogenic assay was performed in control shRNA and PRCP shRNA1 and -2 expressing MCF7 cells treated with vehicle or 5 μm 4OHTAM in triplicate. The average colony numbers were presented as a graph with standard deviation indicated (representative graph of two independent experiments). There are no significant differences between vehicle-treated control shRNA cells and PRCP shRNA1 (p = 0.21) and PRCP2 cells (p = 0.09). There are significant differences between TAM-treated control shRNA cells and PRCP shRNA1 cells (p = 0.03) and PRCP shRNA2 (p = 0.03) cells. K, clonogenic assay was performed in B6-9 control shRNA and B6-9-PRCP shRNA cells treated with vehicle or 5 μm 4OHTAM in triplicate. The average colony numbers were presented as a graph with standard deviation indicated (representative graph of two independent experiments). There is no significant difference between vehicle-treated B6-9-control shRNA cells and B6-9-PRCP shRNA cells (p = 0.2). There is significant difference between 4OHTAM-treated B6-9-control shRNA cells and B6-9-PRCP shRNA cells (p = 0.02).
Next we tested the effects of PRCP shRNA on proliferation of the B6-9 and MCF7 cells. Consistent with enhancement of growth in B6 cells compared with parental MCF7 (Fig. 1D), B6-9 with PRCP shRNA grew slower compared with B6-9 with control shRNA (Fig. 4D). Interestingly, the same effect was noticed for the parental cell, i.e. PRCP shRNAs also decreased proliferation of MCF7 when compared with the cells with control shRNA (Fig. 4E). These results indicated that PRCP influenced proliferation not only in the tamoxifen-resistant derivative but in the parental MCF7 cells as well. The proliferation of B6-9 cells was still dependent on ER-α function as growth of B6-9 cells in medium containing charcoal-stripped FBS was significantly slower than in medium containing regular FBS (Fig. 4F), and knockdown of ER-α (Fig. 4G) led to growth-arrest of B6-9 cells in medium containing regular FBS (Fig. 4H).
The MTT assay and clonogenic assay were used to assess response to 4OHTAM in MCF7 and B6-9 cells transduced with the control shRNA and PRCP shRNA. MTT conversion was measured after 3 days of exposure to different concentrations of 4OHTAM. The most significant decrease in MTT OD value was observed for PRCP shRNA-expressing MCF7 cells treated with 5 and 7.5 μm of 4OHTAM (Fig. 4I) compared with control shRNA-expressing MCF7 cells. Fourteen days of treatment with one concentration (5 μm) of 4OHTAM was used for the clonogenic assay, and colonies were stained with methylene blue and quantified. Knockdown of PRCP in MCF7 cells and B6-9 cells significantly reduced colony formation in response to 4OHTAM (Fig. 4, J and K). Results of this experiment also showed reduced colony formation in PRCP shRNA-expressing cells treated with vehicle (Fig. 4J). Similar results were obtained for B6-9 cells (Fig. 4K), implying that basal cell survival and proliferation in drug-free medium are reduced by knockdown of PRCP.
Overexpression of PRCP Enhances Autophagosome Formation and Lysosomal Function That Are Reduced by Knockdown of ER-α
Knockdown of PRCP Decreases Autophagosome Formation and Impairs Lysosomal Function
Increased AVO in RIGs-expressing cells implied that autophagy played a role in RIGs-mediated resistance to 4OHTAM. To test that, MCF7 and B6-9 cells were treated with 4OHTAM for 3 days, and expression of autophagosome marker LC3 was assessed. B6-9 cells had much higher basal levels of LC3-2 compared with MCF7 cells (Fig. 5A), suggesting that the basal autophagy was more active in B6-9. Treatment with 4OHTAM induced expression of LC3-2 in both cell lines, indicating that autophagy activity increased in response to 4OHTAM. When the same experiment was done using B6-9 cells transduced with control shRNA or PRCP shRNA, the basal LC3-2 level was reduced in the cells with knockdown of PRCP (Fig. 5B). We also determined expression of LC3 in parental MCF7 cells transduced with PRCP shRNA. Both of the PRCP shRNA-expressing cell lines showed decreased basal LC3-2 (Fig. 5C). There was still increased expression of LC3-2 in response to 4OHTAM in PRCP knockdown cells (Fig. 5, B and C), indicating that 4OHTAM-induced autophagy involves other regulatory mechanisms. MDC sequestration was measured to test the effects of PRCP on autophagosome formation and maturation (Fig. 5, D and E). The results showed that sequestration was significantly higher in B6-9-control shRNA cells compared with MCF7-control shRNA cells and B6-9-PRCP shRNA cells (Fig. 5D). Conversely, MDC sequestration is lower in MCF7 cells expressing PRCP shRNA (Fig. 5E). 4OHTAM treatment did not affect the difference (Fig. 5, D and E). LC3-2 and MDC sequestration were increased in 4OHTAM-treated cells, confirming that PRCP regulates basal autophagy activity independent of 4OHTAM.
FIGURE 5.
Overexpression of PRCP up-regulates autophagy and knockdown of PRCP down-regulates autophagy. A, MCF7 and B6-9 cells were treated with vehicle or 5 μm 4OHTAM for 2 days. Whole cell lysates were immunoblotted for LC3 and β-actin (representative picture of three independent experiments). B, B6-9-control shRNA and B6-9-PRCP shRNA cells were treated with vehicle or 5 μm 4OHTAM for 2 days, and whole cell lysates were immunoblotted for LC3 and β-actin (representative picture of two independent experiments). C, MCF7-control shRNA and MCF7-PRCP shRNA1 and -2 cells were treated with vehicle or 5 μm 4OHTAM for 2 days. Whole cell lysates were immunoblotted for LC3 and β-actin (representative pictures of two independent experiments). D, MCF7-control shRNA, B6-9-control shRNA, and B6-9-PRCP shRNA cells were treated with vehicle or 5 μm 4OHTAM for 2 days, and MDC sequestration was analyzed. The average MDC fluorescence (in triplicate) was normalized to the vehicle-treated B6-9-control shRNA cells and presented as a graph with standard deviation indicated (representative graph of three independent experiments). There are significant differences between B6-9-control shRNA and B6-9-PRCP shRNA cells treated with either vehicle (p = 0.03) or 5 μm 4OHTAM (p = 0.03). E, MCF7-control shRNA and MCF7-PRCP shRNA1 and -2 cells were treated with vehicle or 5 μm 4OHTAM for 2 days, and MDC sequestration assay was performed. The average MDC fluorescence in triplicate was normalized to the vehicle treated MCF7-control shRNA cells and presented as a graph with standard deviation indicated (representative graph of three independent experiments). There are significant differences between vehicle-treated MCF7-control shRNA and PRCP shRNA cells (p = 0.03 for shRNA1 and -2). F and G, B6-9-control shRNA and B6-9-ER-α shRNA cells were treated with vehicle or 5 μm 4OHTAM for 2 days. Whole cell lysates were immunoblotted for ER-α, LC3, and β-actin (F). MDC sequestration assay was performed, and the average MDC fluorescence in triplicate was normalized to the vehicle-treated B6-9-control shRNA cells and presented as a graph with standard deviation indicated (representative graph of two independent experiments). H–J, FACS analysis of proteolysis of DQ-Red-BSA in cells. H, MCF7-control shRNA, B6-9-control shRNA, and B6-9-PRCP shRNA cells were assessed, and the average mean fluorescence intensity in triplicate was normalized to the MCF7-control shRNA cells and presented as a graph with standard deviation indicated. There are significant differences between MCF7-control shRNA cells and B6-9-control shRNA cells (p = 0.007), and between B6-9-control shRNA cells and B6-9-PRCP shRNA cells (p = 0.002). I, MCF7-control shRNA and MCF7-PRCP shRNA1 and -2 cells were assessed, and the average mean fluorescence intensity in triplicate was normalized to MCF7-control shRNA cells and presented as a graph with standard deviation indicated. There are significant differences between MCF7-control shRNA cells and MCF7-PRCP-shRNA1 cells (p = 0.05) and MCF7-PRCP shRNA2 cells (p = 0.001). J, B6-9-control shRNA and B6-9-ER-α shRNA were assessed and the average mean fluorescence intensity in triplicate was normalized to B6-9-control shRNA cells and presented as a graph with standard deviation indicated (representative graphs of two independent experiments).
Because proliferation of B6-9 cells was dependent on ER-α, we tested whether knockdown of ER-α affects PRCP-mediated autophagy. B6-9 cells expressing ER-α shRNA showed decreased basal LC3-1 and LC3-2 expression (Fig. 5F) and MDC sequestration compared with the control shRNA expressing B6-9 cells (Fig. 5G). However, 4OHTAM-induced increase in LC3-2 expression and MDC sequestration was not significantly affected by knockdown of ER-α (Fig. 5G).
Next, we tested functionality of PRCP-dependent formation of autolysosomes. The cells were loaded with self-quenched DQ-Red-BSA, which could produce fluorescent derivatives upon cleavage by activated lysosomal proteases. The fluorescence of loaded cells was analyzed directly without fixation by FACS. B6-9-control shRNA cells showed increased fluorescence intensity compared with B6-9-PRCP shRNA cells and MCF7-control shRNA cells (Fig. 5H), whereas MCF7-PRCP shRNA1 and -2 cells showed significantly decreased fluorescence intensity compared with MCF7-control shRNA cells (Fig. 5I), suggesting that autolysosomal functions were also enhanced by PRCP. Knockdown of ER-α also decreased proteolysis of BSA (Fig. 5J) in B6-9 cells which together with the reduced LC3 activation and MDC sequestration suggested a function for ER-α in basal autophagy activity in B6-9 cells.
Finally, we tested whether nutrient starvation-mediated autophagy is affected by PRCP. MCF7.beclin cells stably expressing control shRNA and PRCP shRNA (supplemental Fig. 2A) were grown in the absence of doxycycline, which induced expression of Beclin 1 (supplemental Fig. 2B). Whole cell lysates were immunoblotted for LC3 and β-actin (supplemental Fig. 2C). A slight increase in LC3-1 and LC3-2 levels was noted in cells expressing control shRNA in response to nutrient starvation (supplemental Fig. 2C). The PRCP shRNA-expressing cells consistently showed decreased basal LC3-1 compared with the cells expressing control shRNA (supplemental Fig. 2C). There was also a slight increase in the LC3-1 level upon nutrient deprivation in the PRCP shRNA-expressing cells (supplemental Fig. 2C). To quantify autophagosomes, both cell lines were infected with pMSCV-EGFP-LC3. The cells were then grown in regular medium or Hanks' balanced salt solution for 6 h. The cells were fixed and analyzed with a confocal microscope. The dotted structures of EGFP-LC3 were quantified as autophagosomes (supplemental Fig. 2D). The results showed that nutrient starvation caused an increase in autophagosomes in both the control shRNA and PRCP shRNA cells. However, the amount of autophagosomes was lower in PRCP shRNA cells with or without starvation (supplemental Fig. 2E). These results suggest that PRCP mediates basal autophagy activity and does not significantly affect nutrient starvation-induced increase in autophagy.
Acquired Resistance to 4OHTAM Is Accompanied by Increased PRCP Activity, although Inhibition of PRCP Down-regulates Autophagy, Enhances 4OHTAM-induced Cytotoxicity, and Prevents and Reverses Acquired 4OHTAM Resistance in MCF7 Cells
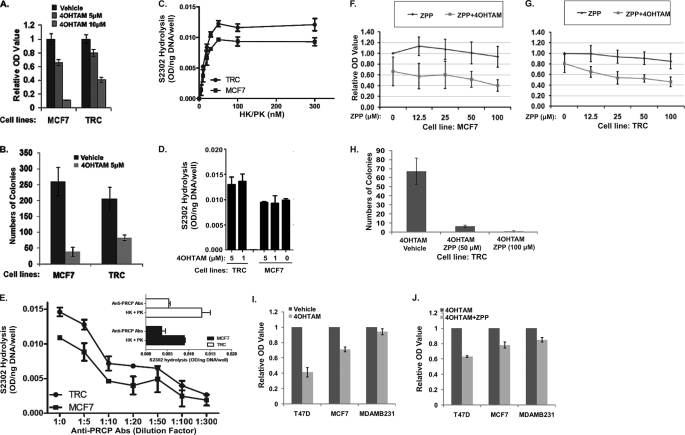
Cytoprotective effect of PRCP against 4OHTAM suggested that it might be involved in fortuitously acquired resistance to 4OHTAM. To test this possibility, we developed a 4OHTAM-resistant cell line by culturing MCF7 cells in the presence of 5 μm 4OHTAM for 3 months. Resistant cells grew slower than MCF7 cells (supplemental Fig. 3, A–C); their proliferation was further decreased in estrogen-free media but could be enhanced by β-estradiol (supplemental Fig. 3B), suggesting that these cells had functional ER. Lower doses of 4OHTAM (1 μm) had no effect on their proliferation (supplemental Fig. 3, B and C), and they also showed increased cell viability in response to cytotoxic doses of 4OHTAM (Fig. 6A). In a clonogenic assay, these cells produced more colonies after treatment with 4OHTAM compared with parental MCF7 cells (Fig. 6B) and could be considered resistant to 4OHTAM. We referred to these cells as TAM-resistant cells (TRC cells).
FIGURE 6.
MCF7 cells that acquired resistance to 4OHTAM show increased PRCP activity. Inhibition of PRCP enhances TAM-induced cytotoxicity in 4OHTAM-sensitive and -resistant MCF7 cells and shows more effect in ER-positive T47D and MCF7 cells than ER-negative MDAMB231 cells. A, MCF7 and TRC cells were treated with vehicle or 4OHTAM (5 and 10 μm) for 3 days. The cell viability was assessed by the MTT assay. The average OD values of octuplicate were normalized to the vehicle-treated cells and presented as a graph with standard deviation indicated (representative experiments of three independent experiments). B, MCF7 cells and TRC cells were treated with vehicle or 5 μm 4OHTAM for 14 days and recovered for 14 days. The numbers of cell colonies were presented as a graph with standard deviation indicated (representative graph of two independent experiments). C, activation of the high molecular weight kininogen-PK complex on human breast carcinoma cell lines. Confluent monolayers of MCF7 (■) or TRC (●) cell cultures were incubated with the various concentrations of the complex of high molecular weight kininogen -PK and the generation of kallikrein by PRCP measured after 1 h. The substrate HD-Pro-Phe-Arg-paranitroaniline (S2302, 0.8 mm) was used to determine kallikrein activity. The absorbance of hydrolyzed S2302 was normalized to cellular DNA (ng) measured by PicoGreen. Data represent the mean ± S.E. of three experiments in triplicate. The absence of standard error bars indicates that the variation was too little to be visualized. D, effect of 4-hydroxytamoxifen on PRCP-dependent PK activation in MCF7 and TRC cells. Cells were treated with 1 or 5 μm 4OHTAM and analyzed for kallikrein activity. The absorbance of hydrolyzed S2302 was normalized to cellular DNA (ng) measured by PicoGreen. Data represent the mean ± S.E. of three experiments in triplicate. E, effect of anti-human PRCP IgG on the activation of PK on the cell-bound high molecular weight kininogen. Confluent monolayers of MCF7 (■) or TRC (●) cells were treated with high molecular weight kininogen (20 nm) and incubated for 1 h at 37 °C. Cells were subsequently incubated with the increasing amount of anti-human PRCP IgG in the presence of PK (20 nm) and analyzed for kallikrein activity. S2302 (0.8 mm) was used to determine kallikrein activity. The inset shows the inhibition of the PRCP-PK activation by anti-human PRCP IgG (1:100 dilution) on MCF7 and TRC cells. The absorbance of hydrolyzed S2302 was normalized to cellular DNA (ng) measured by PicoGreen. Data represent the mean ± S.E. of three experiments in triplicate. F and G, MCF7 cells (F) and the TRC cells (G) (in triplicate) were treated with different doses of ZPP and/or 5 μm 4OHTAM for 3 days and analyzed by the MTT assay. The average OD values were normalized to the vehicle-treated cells and presented as graphs with standard deviation indicated (representative graph of three independent experiments). There is no significant difference between the vehicle-treated and ZPP-treated MCF7 cells (one-way ANOVA: F = 2.29, p = 0.06). There is significant difference between TAM/vehicle-treated and TAM/ZPP-treated MCF7 cells (one-way ANOVA: F = 2.75, p = 0.04). There is no significant difference between the vehicle-treated and ZPP-treated TRC cells (one-way ANOVA: F = 1.88, p = 0.14). There is significant difference between 4OHTAM/vehicle-treated and 4OHTAM/ZPP-treated TRC cells (one-way ANOVA: F = 13.9, p = 0). H, clonogenic assay of the TRC cells treated with vehicle or 5 μm 4OHTAM in combination with vehicle or 50 and 100 μm of ZPP. The average numbers of colonies in triplicate were normalized to vehicle-treated cells and presented as a graph with standard deviation indicated (representative graph of two independent experiments). There are significant differences between 4OHTAM/vehicle-treated TRC cells and 4OHTAM/ZPP-treated cells (p = 0.02 and 0.001). I and J, T47D, MCF7, and MDAMB231 cells were treated with vehicle or 5 μm 4OHTAM in the presence of vehicle or 100 μm ZPP for 3 days. The cell viability was assessed by the MTT assay. The average OD values of octuplicate were normalized to the vehicle-treated (I) or 4OHTAM-treated (J) cells and presented as a graph with standard deviation indicated (representative experiments of three independent experiments).
The enzymatic activity of PRCP was examined in MCF7 and TRC cells by activation of PK to kallikrein (22). The TRC cells showed increased PK activation compared with MCF7 cells (Fig. 6C). 4OHTAM did not significantly affect PK activation in both MCF7 and the TRC cells (Fig. 6D) and the activity of recombinant PRCP protein (data not shown). The PK activation in both MCF7 and TRC cells was blocked by anti-PRCP antibodies (Fig. 6E) indicating that PK activation was mediated by PRCP in MCF7 cells similar to previous results in endothelial cells (22). These results suggested that in vitro acquired resistance to 4OHTAM was accompanied by increased activity of PRCP in MCF7 cells.
A specific inhibitor of PRCP (ZPP) (17) was used to test whether the enzymatic activity of PRCP was responsible for the observed effects. MCF7 cells were treated with 5 μm 4OHTAM for 3 days, and different concentrations of ZPP were included; cell viability was then assessed by MTT assay. Treatment with ZPP alone did not show significant adverse effects up to 100 μm concentration (Fig. 6F). However, when combined with 5 μm 4OHTAM, ZPP decreased cell viability in a dose-dependent manner (Fig. 6F), suggesting that inhibition of PRCP enzymatic activity enhanced 4OHTAM-induced cytotoxicity. When the same experiment was repeated with the TRC cells, we did not observe a significant effect of ZPP on cell viability, whereas a combination of ZPP with 5 μm 4OHTAM reduced viability of TRC in a dose-dependent manner (Fig. 6G) similar to effects observed with parental MCF7 cells (Fig. 6G). Notably, the attempt to develop TRC cells in the presence of ZPP was unsuccessful, suggesting that ZPP blocked molecular changes required for resistance. No significant difference in PRCP mRNA level and protein level was detected in MCF7 and TRC cells in the presence or absence of 4OHTAM (data not shown).
The effect of ZPP on the 4OHTAM-induced cytotoxicity in the TRC cells was further assessed by the clonogenic assay. The TRC cells were cultured in 5 μm 4OHTAM with ZPP (50 or 100 μm), allowed to recover, and form colonies, which were then stained with methylene blue and quantified. The colony formation was significantly reduced by 50 μm ZPP and no colonies formed in 100 μm ZPP (Fig. 6H), suggesting that resistance of TRC could be reversed by ZPP. Similar results were seen in B6-9 cells treated with 4OHTAM and ZPP (data not shown).
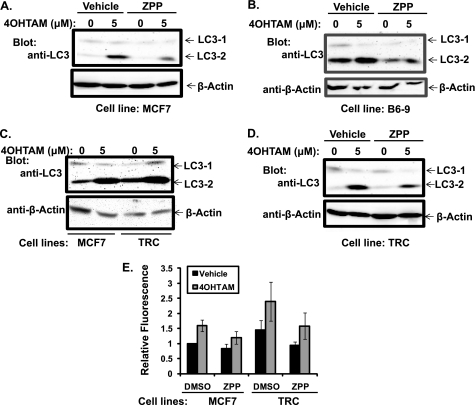
To test whether the effect of PRCP inhibition on 4OHTAM-induced cytotoxicity is specific for ER-positive cells, we examined the response of ER-positive T47D cells and ER-negative MDAMB-231 cells to 4OHTAM and 4OHTAM plus ZPP. Compared with MCF7 cells, T47D cells showed a decrease, and MDAMB231 showed an increase in cell viability in response to 5 μm 4OHTAM (Fig. 6I). Addition of ZPP enhanced 4OHTAM-induced cytotoxicity in all of the cell lines, although the effect was more pronounced in ER-positive T47D cells and MCF7 cells than MDAMB231 cells (Fig. 6J). Inhibition of PRCP enzymatic activity by ZPP reduced accumulation of LC3-2 in both MCF7 cells (Fig. 7A) and B6-9 cells (Fig. 7B) treated with vehicle and 4OHTAM, suggesting that PRCP activity and autophagy were directly connected. Furthermore, this result confirmed the links between enzymatic activity of PRCP, resistance to 4OHTAM, and increased autophagic activity in B6-9 cells. Similar effects of ZPP in parental MCF7 cells and their resistant derivative suggested that the increased autophagic activity in B6-9 cells was due to overexpression of PRCP.
FIGURE 7.
Autophagy is inhibited by ZPP in MCF7s, B6-9, and the TRC cells, which show higher autophagy activity. A and B, MCF7 (A) and B6-9 (B) cells were treated with 4OHTAM/vehicle and 4OHTAM/ZPP for 2 days. Whole cell lysates were immunoblotted for LC3 and β-actin (representative picture of three independent experiments). C, MCF7 cell and the TRC cells were treated with 4OHTAM for 2 days. Whole cell lysates were immunoblotted for LC3 and β-actin (representative picture of three independent experiments). D, TRC cells were treated with 4OHTAM/vehicle and 4OHTAM/ZPP for 2 days. Whole cell lysates were immunoblotted for LC3 and β-actin (representative picture of three independent experiments). E, MCF7 and TRC cells were treated with 4OHTAM/vehicle and 4OHTAM/ZPP for 2 days. MDC sequestration assay was performed. The average MDC fluorescence in triplicate was normalized to the vehicle treated MCF7 cells and presented as a graph with standard deviation as error bars (representative graph of three independent experiments).
Increased autophagy was shown to be a mechanism for acquired resistance to 4OHTAM in MCF7 cells (9). Consistent with this finding, the developed TRC cells also showed increased expression of LC3-1 and LC3-2 compared with MCF7 cells (Fig. 7C). In the presence of ZPP, the LC3-2 levels were also decreased in the TRC cells (Fig. 7D). When MDC sequestration was analyzed in MCF7 and TRC cells, the results showed that there was a significantly increased MDC fluorescence in the TRC cells compared with MCF7 cells (Fig. 7E). The MDC sequestration was also reduced by ZPP in both MCF7 and TRC cells (Fig. 7E). These results further supported a role for the enzymatic activity of PRCP in the regulation of autophagy in MCF7 and TRC cells.
To test whether PRCP also changes cell response to other therapeutic drugs, we treated MCF7 and B6-9 cells with different chemotherapeutic drugs and calculated the IC50 concentration of each drug. The results showed that B6-9 cells have increased resistance to mafosfamide, melphalan, and raloxifen and increased sensitivity to fluorouracil (supplemental Table 1). These results suggested that PRCP-mediated resistance is not restricted to 4OHTAM.
DISCUSSION
Using a functional genomic approach, we have isolated genes that convey resistance to the cytotoxic action of 4OHTAM. Two important paradigms have been considered in screening for drug resistance genes using the MCF7/4OHTAM cell culture model for these experiments. First, parental cells used in the study (MCF7) express estrogen receptor and respond to estrogen in many different ways (26). In clinical practice, a certain level of estrogen is present even in postmenopausal women (27), and TAM treatment of breast cancer patients and concomitant emergence of resistance to the drug take place in the presence of this hormone. It seemed logical to preserve regular cell culture conditions that approximate a clinically relevant situation, so we decided against the removal of estrogen from tissue culture media during selection. It is quite possible that a similar selection done in phenol red-free media with estrogen-depleted serum would have produced different results. Second, the selection screen described here targets overexpressed genes; it cannot detect genes that have to be repressed to make cells resistant, as well as genes that do not change their expression level while their proteins undergo modification (e.g. phosphorylation, etc). This limitation may prevent identification of the complete pathway involved in resistance to a specific drug, although even partial description of a pathway can point out potential drug targets or resistance biomarkers.
Resistance-inducing genes (Fig. 1) recovered after selection contain complete (B4, B6, and D10) or substantial parts (E5) of the corresponding protein-coding regions. To confirm protective effects of selected RIGs, we recovered them from surviving cellular clones, re-cloned them into the initial pFB vector, and used the recombinant retroviruses to introduce individual RIGs into naive populations of MCF7 cells. This step allowed us to avoid potential interference from ill-defined genomic mutations induced in surviving cellular clones by drug exposure. To avoid effects of clonal variability, we used populations of infected cells rather than single cell clones for some of the downstream testing.
The general characterization of all the four RIGs suggest that they regulate autophagy activity upon 4OHTAM treatment because one of the common features shared by the cells with the RIGs is increased AVO (Fig. 2). This increase is accompanied by increased cell viability demonstrated by the increases in colony formation and plasma membrane integrity and by the retaining of mitochondrial transmembrane potential in the RIG-expressing cells treated with a cytotoxic dose of 4OHTAM (Fig. 3). This feature was further investigated by a detailed characterization of one of the four genes, PRCP by overexpression, knockdown, and inhibition of enzymatic activity.
Compared with MCF7 cells, the PRCP-overexpressing B6-9 cell line showed an increase in proliferation and autophagy activity indicated by significantly increased expression of LC3-2, increased MDC sequestration, and enhanced proteolysis of BSA in drug-free medium. These effects were reversed by the knockdown of the overexpressed PRCP in B6-9 cells. Knockdown of endogenous PRCP in MCF7 cells resulted in decreases in proliferation, expression of LC3-2, MDC sequestration, and proteolysis of BSA (Fig. 4). Changes induced by PRCP overexpression or knockdown in LC3 expression, MDC sequestration, and proteolysis of BSA are independent of 4OHTAM treatment, suggesting that PRCP regulates basal autophagy activity in MCF7 cells. Increase in LC3-2 level and MDC sequestration induced by 4OHTAM was less significantly affected by PRCP (Fig. 5, A–E), implying that 4OHTAM-induced autophagy is mechanistically different from that mediated by PRCP.
Recent studies in mammalian cells indicate that autophagy promotes cell survival by degrading cytoplasm, specific long lived proteins, and mitochondria in response to nutrient deprivation and cellular stress (11–14). Autophagy is a multistage process that includes formation of autophagosomes at the early stage, which are then fused with lysosomes to form autolysosomes (10). 4OHTAM-induced autophagy requires beclin 1 for the formation of autophagosomes (9). Overexpression of beclin 1 protects MCF7 cells from 4OHTAM (28), whereas knockdown of beclin 1 sensitizes MCF7 cells to 4OHTAM (9, 29). Acquired 4OHTAM resistance developed in vitro in MCF7 cells is accompanied by increased autophagy activity and can be reversed by the blockade of autophagy (9, 29). These results have redefined autophagy as a mechanism for 4OHTAM resistance in ER-positive breast cancer cells. Consistent with these findings, our results show that treatment of the MCF7 cells with 5 μm 4OHTAM activates autophagy, indicated by the increases in LC3-2 expression and MDC sequestration (Fig. 5, A–E), suggesting that autophagy is induced by 4OHTAM in our experimental system. Qadir et al. (29) show that microautophagy protects MCF7 cells from 4OHTAM-induced mitochondrial depolarization. Our results showed that the RIG-expressing cells have increased AVO formation, which is correlated with the maintenance of mitochondrial transmembrane potential in response to 4OHTAM (Fig. 3). These results suggest that autophagy attenuates 4OHTAM-induced damage of mitochondria and cell death.
The cytoprotective effect of PRCP is well correlated with its function in the up-regulation of autophagy. B6-9 cells showed basally increased cell viability compared with MCF7 cells in drug-free medium. The PRCP-mediated autophagy appears additive to 4OHTAM-induced autophagy and is correlated with increased cell survival as demonstrated by increased colony formation in 4OHTAM-treated B6-9 cells and decreased cell viability and colony formation in the PRCP knockdown cells treated with 4OHTAM. Our results also show that in vitro acquired resistance to 4OHTAM is associated with increased PRCP activity and autophagy. The TRC cells derived from MCF7 cells constantly treated with 5 μm 4OHTAM for 3 months lose response to 1 μm 4OHTAM and are resistant to 5 μm 4OHTAM. The TRC cells have increased LC3-2 expression and MDC sequestration, which is consistent with the findings of Samaddar et al. (9) that autophagy plays a role in acquired 4OHTAM resistance. Both the B6-9 and TRC cells showed decreased proliferation in medium containing charcoal-stripped FBS, suggesting that ER is still functional in the 4OHTAM-resistant derivatives. Their response to estrogen deprivation implies that altered ER signaling rather than loss of ER function accounts for the resistance to 4OHTAM in these cells. Depletion of ER-α in B6-9 cells induced growth arrest and decreased basal autophagy activity in B6-9 cells, implying that ER-α plays a role in cell proliferation and in basal autophagy mediated by PRCP. These results are consistent with the clinical findings that TAM-insensitive ER-positive breast cancers respond to aromatase inhibitors that block synthesis of estrogen (30, 31) and that altered ER signaling contributes to TAM resistance (28, 32, 33). PRCP is a serine protease that belongs to the prolyl peptidase family (34). PRCP selectively cleaves the peptide bond after the C terminus of penultimate proline in oligopeptides (35). Inhibition of PRCP by the specific inhibitor ZPP reduces LC3-2 expression and MDC sequestration in drug-free medium, suggesting that enzymatic activity of PRCP is responsible for the regulation of autophagy. Interestingly, although ZPP is not cytotoxic by itself, it shows a dose-dependent enhancement in 4OHTAM-induced cytotoxicity in MCF7 cells (Fig. 6C), suggesting that PRCP-mediated autophagy depends on its enzymatic activity that is protective against 4OHTAM-induced cytotoxicity. ZPP prevented the development of acquired 4OHTAM resistance in MCF7 cells and enhanced 4OHTAM-induced cytotoxicity in the 4OHTAM resistance cells (Fig. 6D). The regulatory mechanism underlying the enzymatic activity of PRCP is currently unclear. We found increased activity of PRCP (Fig. 6) without alteration in PRCP expression (data not shown) in the MCF7 cells that acquired resistance to 4OHTAM, which suggest that PRCP activity can be regulated via a post-translational modification mechanism. Future investigation will elucidate the mechanism by which PRCP activity is regulated in the TRC cells.
The mechanism underlying PRCP-mediated autophagy needs to be further elucidated. There is no documented function for the known PRCP substrates such as angiotensin II, angiotensin III, and des-Arg9-bradykinin (36) in the regulation of autophagy. One member of the prolyl peptidase family, prolyl oligopeptidase, is found to play a role in autophagy by down-regulation of inositol 1,4,5-trisphosphate (37). Whether PRCP also regulates autophagy similarly through regulation of inositol 1,4,5-trisphosphate has to be investigated. It is noteworthy that ZPP also inhibits prolyl oligopeptidase that targets the PRCP substrates in vitro (38, 39). It is possible that the general inhibition of prolyl oligopeptidase and other prolyl peptidase family members by ZPP in MCF7 cells may contribute to the enhanced cytotoxicity by 4OHTAM. Consistent with the role of PRCP in the promotion of cell survival, another prolyl peptidase family member dipeptidyl peptidase 7 that belongs to the same subfamily of PRCP, is crucial for cell survival in quiescent lymphocytes and fibroblasts (40). Whether DPP7, prolyl oligopeptidase, and the other prolyl peptidases also regulate autophagy, cell survival and 4OHTAM resistance in MCF7 cells require further investigation. Previous studies have identified complex mechanisms that involve signal transduction pathways and gene transcription regulation in TAM resistance (33, 41–44). Altered ER functions and the cross-talk between ER and other growth factor signaling pathways represent a major resistance mechanism for TAM as well as other anti-estrogen therapeutic drugs (4, 28, 32, 33, 45). In this study, PRCP seems to regulate proliferation and autophagy via an ER-α-dependent mechanism. Whether and how PRCP modulates ER-α signaling pathways need to be elucidated in future studies.
In conclusion, we have for the first time identified PRCP as a regulator of proliferation and autophagy and a molecular regulator of 4OHTAM resistance. Further study on the elucidation of the role for PRCP in anti-estrogen resistance in vivo in ER-positive breast cancer patients and the mechanisms underlying proliferation and autophagy mediated by PRCP and possibly other prolyl peptidases may help understand the resistance to TAM and potentially to other therapeutic drugs, thus identifying possible therapeutic targets.
Supplementary Material
Acknowledgments
We thank the RUSH University flow cytometry facility supported by the James B. Pendleton Charitable Trust. We are also indebted to Jeffrey Martinson (Rush University Flow Cytometry Facility) and Mary Paniagua and Jeffrey Nelson (Northwestern University Flow Cytometry Facility) for their help with FACS experiments. We thank Dr. Lothar A. Blatter and Dr. Christoph Littwitz for help with the confocal microscope. We are grateful to Dr. Beth Levine (University of Texas Southwestern Medical Center) for providing MCF7.beclin cells. We also thank Dr. Trono Didier (École Polytechnique Fédérale de Lausanne (EPFL) School of Life Sciences) for providing the lentiviral packaging and enveloping vectors psPAX2 and pMD2.G.
This work was supported, in whole or in part, by National Institutes of Health Grant P20RR021929 from NCRR (to Z. S.-M.). This work was also supported by Susan G. Komen Breast Cancer Foundation Grants BCTR 0100916 and BCTR 0504311 (to V. V. L.) and Department of Defense Grant W81XWH-07-1-0505 (to V. V. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- TAM
- tamoxifen
- RIG
- resistance-inducing gene
- 4OHTAM
- 4-hydroxytamoxifen
- ER
- estrogen receptor
- PRCP
- prolylcarboxypeptidase
- AVO
- acidic vesicular organelles
- ZPP
- N-benzyloxycarbonylprolylprolinal
- MDC
- monodansylcadaverine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- EGFP
- enhanced GFP
- PI
- propidium iodide
- PK
- prekallikrein
- ANOVA
- analysis of variance.
REFERENCES
- 1. Jordan V. C. (2004) Cancer Cell 5, 207–213 [DOI] [PubMed] [Google Scholar]
- 2. Arafah B. M., Griffin P., Gordon N. H., Pearson O. H. (1986) Cancer Res. 46, 3268–3272 [PubMed] [Google Scholar]
- 3. Reddel R. R., Murphy L. C., Hall R. E., Sutherland R. L. (1985) Cancer Res. 45, 1525–1531 [PubMed] [Google Scholar]
- 4. Clarke R., Liu M. C., Bouker K. B., Gu Z., Lee R. Y., Zhu Y., Skaar T. C., Gomez B., O'Brien K., Wang Y., Hilakivi-Clarke L. A. (2003) Oncogene 22, 7316–7339 [DOI] [PubMed] [Google Scholar]
- 5. Poole R., Paridaens R. (2007) Curr. Opin. Oncol. 19, 564–572 [DOI] [PubMed] [Google Scholar]
- 6. Brodie A., Macedo L., Sabnis G. (2010) J. Steroid Biochem. Mol. Biol. 118, 283–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bursch W., Ellinger A., Kienzl H., Török L., Pandey S., Sikorska M., Walker R., Hermann R. S. (1996) Carcinogenesis 17, 1595–1607 [DOI] [PubMed] [Google Scholar]
- 8. Schoenlein P. V., Periyasamy-Thandavan S., Samaddar J. S., Jackson W. H., Barrett J. T. (2009) Autophagy 5, 400–403 [DOI] [PubMed] [Google Scholar]
- 9. Samaddar J. S., Gaddy V. T., Duplantier J., Thandavan S. P., Shah M., Smith M. J., Browning D., Rawson J., Smith S. B., Barrett J. T., Schoenlein P. V. (2008) Mol. Cancer Ther. 7, 2977–2987 [DOI] [PubMed] [Google Scholar]
- 10. Eskelinen E. L. (2005) Autophagy 1, 1–10 [DOI] [PubMed] [Google Scholar]
- 11. Livesey K. M., Tang D., Zeh H. J., Lotze M. T. (2009) Curr. Opin. Investig. Drugs 10, 1269–1279 [PubMed] [Google Scholar]
- 12. Cecconi F., Levine B. (2008) Dev. Cell 15, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Azad M. B., Chen Y., Gibson S. B. (2009) Antioxid. Redox. Signal. 11, 777–790 [DOI] [PubMed] [Google Scholar]
- 14. Wang J., Whiteman M. W., Lian H., Wang G., Singh A., Huang D., Denmark T. (2009) J. Biol. Chem. 284, 21412–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lien E. A., Solheim E., Lea O. A., Lundgren S., Kvinnsland S., Ueland P. M. (1989) Cancer Res. 49, 2175–2183 [PubMed] [Google Scholar]
- 16. Clarke R., Leonessa F., Welch J. N., Skaar T. C. (2001) Pharmacol. Rev. 53, 25–71 [PubMed] [Google Scholar]
- 17. Shariat-Madar Z., Rahimy E., Mahdi F., Schmaier A. H. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H2697–H2703 [DOI] [PubMed] [Google Scholar]
- 18. Liang X. H., Yu J., Brown K., Levine B. (2001) Cancer Res. 61, 3443–3449 [PubMed] [Google Scholar]
- 19. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 20. Kim S. K., Yang J. W., Kim M. R., Roh S. H., Kim H. G., Lee K. Y., Jeong H. G., Kang K. W. (2008) Free Radic. Biol. Med. 45, 537–546 [DOI] [PubMed] [Google Scholar]
- 21. Duan L., Chen G., Virmani S., Ying G., Raja S. M., Chung B. M., Rainey M. A., Dimri M., Ortega-Cava C. F., Zhao X., Clubb R. J., Tu C., Reddi A. L., Naramura M., Band V., Band H. (2010) J. Biol. Chem. 285, 1555–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shariat-Madar Z., Mahdi F., Schmaier A. H. (2002) J. Biol. Chem. 277, 17962–17969 [DOI] [PubMed] [Google Scholar]
- 23. Bursch W., Ellinger A., Gerner C., Frohwein U., Schulte-Hermann R. (2000) Ann. N.Y. Acad. Sci. 926, 1–12 [DOI] [PubMed] [Google Scholar]
- 24. Scarlatti F., Bauvy C., Ventruti A., Sala G., Cluzeaud F., Vandewalle A., Ghidoni R., Codogno P. (2004) J. Biol. Chem. 279, 18384–18391 [DOI] [PubMed] [Google Scholar]
- 25. Goldman S. J., Taylor R., Zhang Y., Jin S. (2010) Mitochondrion 10, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Levenson A. S., Jordan V. C. (1997) Cancer Res. 57, 3071–3078 [PubMed] [Google Scholar]
- 27. Purohit A., Reed M. J. (2002) Steroids 67, 979–983 [DOI] [PubMed] [Google Scholar]
- 28. Gutierrez M. C., Detre S., Johnston S., Mohsin S. K., Shou J., Allred D. C., Schiff R., Osborne C. K., Dowsett M. (2005) J. Clin. Oncol. 23, 2469–2476 [DOI] [PubMed] [Google Scholar]
- 29. Qadir M. A., Kwok B., Dragowska W. H., To K. H., Le D., Bally M. B., Gorski S. M. (2008) Breast Cancer Res. Treat. 112, 389–403 [DOI] [PubMed] [Google Scholar]
- 30. Adamo V., Iorfida M., Montalto E., Festa V., Garipoli C., Scimone A., Zanghi M., Caristi N. (2007) Ann. Oncol. 18, Suppl. 6, 53–57 [DOI] [PubMed] [Google Scholar]
- 31. Miyoshi Y., Murase K., Saito M., Oh K. (2010) Breast Cancer 17, 86–91 [DOI] [PubMed] [Google Scholar]
- 32. Massarweh S., Schiff R. (2006) Endocr. Relat. Cancer 13, S15–S24 [DOI] [PubMed] [Google Scholar]
- 33. Normanno N., Di Maio M., De Maio E., De Luca A., de Matteis A., Giordano A., Perrone F. (2005) Endocr. Relat. Cancer 12, 721–747 [DOI] [PubMed] [Google Scholar]
- 34. Rosenblum J. S., Kozarich J. W. (2003) Curr. Opin. Chem. Biol. 7, 496–504 [DOI] [PubMed] [Google Scholar]
- 35. Skidgel R. A., Erdös E. G. (1998) Immunol. Rev. 161, 129–141 [DOI] [PubMed] [Google Scholar]
- 36. Odya C. E., Marinkovic D. V., Hammon K. J., Stewart T. A., Erdös E. G. (1978) J. Biol. Chem. 253, 5927–5931 [PubMed] [Google Scholar]
- 37. Williams R. S., Eames M., Ryves W. J., Viggars J., Harwood A. J. (1999) EMBO J. 18, 2734–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gass J., Khosla C. (2007) Cell. Mol. Life Sci. 64, 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. García-Horsman J. A., Männistö P. T., Venäläinen J. I. (2007) Neuropeptides 41, 1–24 [DOI] [PubMed] [Google Scholar]
- 40. Mele D. A., Bista P., Baez D. V., Huber B. T. (2009) Cell Cycle 8, 2425–2434 [DOI] [PubMed] [Google Scholar]
- 41. Choi J. Y., Barlow W. E., Albain K. S., Hong C. C., Blanco J. G., Livingston R. B., Davis W., Rae J. M., Yeh I. T., Hutchins L. F., Ravdin P. M., Martino S., Lyss A. P., Osborne C. K., Abeloff M. D., Hayes D. F., Ambrosone C. B. (2009) Clin. Cancer Res. 15, 5258–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gururaj A. E., Rayala S. K., Vadlamudi R. K., Kumar R. (2006) Clin. Cancer Res. 12, 1001s–1007s [DOI] [PubMed] [Google Scholar]
- 43. Musgrove E. A., Sutherland R. L. (2009) Nat. Rev. Cancer 9, 631–643 [DOI] [PubMed] [Google Scholar]
- 44. Riggins R. B., Schrecengost R. S., Guerrero M. S., Bouton A. H. (2007) Cancer Lett. 256, 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arpino G., De Angelis C., Giuliano M., Giordano A., Falato C., De Laurentiis M., De Placido S. (2009) Oncology 77, Suppl. 1, 23–37 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.