Abstract
The mitochondrial inner membrane (IM) serves as the site for ATP production by hosting the oxidative phosphorylation complex machinery most notably on the crista membranes. Disruption of the crista structure has been implicated in a variety of cardiovascular and neurodegenerative diseases. Here, we characterize ChChd3, a previously identified PKA substrate of unknown function (Schauble, S., King, C. C., Darshi, M., Koller, A., Shah, K., and Taylor, S. S. (2007) J. Biol. Chem. 282, 14952–14959), and show that it is essential for maintaining crista integrity and mitochondrial function. In the mitochondria, ChChd3 is a peripheral protein of the IM facing the intermembrane space. RNAi knockdown of ChChd3 in HeLa cells resulted in fragmented mitochondria, reduced OPA1 protein levels and impaired fusion, and clustering of the mitochondria around the nucleus along with reduced growth rate. Both the oxygen consumption and glycolytic rates were severely restricted. Ultrastructural analysis of these cells revealed aberrant mitochondrial IM structures with fragmented and tubular cristae or loss of cristae, and reduced crista membrane. Additionally, the crista junction opening diameter was reduced to 50% suggesting remodeling of cristae in the absence of ChChd3. Analysis of the ChChd3-binding proteins revealed that ChChd3 interacts with the IM proteins mitofilin and OPA1, which regulate crista morphology, and the outer membrane protein Sam50, which regulates import and assembly of β-barrel proteins on the outer membrane. Knockdown of ChChd3 led to almost complete loss of both mitofilin and Sam50 proteins and alterations in several mitochondrial proteins, suggesting that ChChd3 is a scaffolding protein that stabilizes protein complexes involved in maintaining crista architecture and protein import and is thus essential for maintaining mitochondrial structure and function.
Keywords: Bioenergetics, Confocal Microscopy, Electron Microscopy (EM), Mitochondria, Protein-Protein Interactions, ChChd3, Crista Junctions, Crista Morphology, Mitochondrial Dynamics, Protein Import
Introduction
Mitochondria are ubiquitous and dynamic organelles that undergo frequent fission and fusion and are actively transported to specific cellular locations based on energy requirements. The highly organized structure of mitochondria and the nature of mitochondrial protein complexes enable them to sense the cellular environment and regulate ATP synthesis, metabolism, and other key physiological processes according to the energy requirements. In the past decade, several crucial regulators of mitochondrial structure, dynamics and function, have been identified and characterized in fungi, Drosophila, and mammals (1, 2). Evolutionarily conserved large GTPases, mitofusin1 and 2 (Mfn1 and Mfn2) located on the outer membrane (OM)3 (3) and optic atrophy 1 (OPA1) located on the inner membrane (IM) and intermembrane space (IMS), have been identified as key modulators for mitochondrial fusion (4). On the other hand, the large GTPase dynamin-related protein 1 (DRP1/DLP1/DNM1) (5) along with fission protein 1 homolog (FIS1) (6) on the OM have been well characterized as the proteins responsible for mitochondrial fission. Mutations in some of these proteins have been associated with debilitating conditions such as neurodegenerative diseases, cancer, and type II diabetes (7, 8).
Although significant advances have been made in identifying the proteins involved in mitochondrial morphology and dynamics, very little is known about the protein complexes that control the biogenesis of cristae and crista junctions (CJs). Recent studies have suggested the involvement of IM proteins F1,F0-ATP synthase (9), OPA1 (10, 11), mitofilin (12), and MICS1 (13) in regulating the crista morphology. Furthermore, OPA1 and mitofilin are suggested to be at the CJs and involved in CJ formation and control the CJ opening (10, 12, 14).
Coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3/FLJ20420/LOC54927/RIKEn cDNA 0610041L09) was previously identified in our laboratory as a cAMP-dependent protein kinase A (PKA) substrate in mitochondria (15). Proteomic studies and in situ hybridization experiments identified ChChd3 as a highly abundant protein at synaptic membranes and in neurons of rat brain throughout the gray matter, dorsal root ganglion, and spinal cord (16). Furthermore, ChChd3 was found to be significantly down-regulated in mitochondrial proteomic analysis of a cell line model of familial amyotrophic lateral sclerosis expressing SOD1 mutant G93A compared with the control cells expressing WT SOD1 (17), suggesting its possible role in the pathophysiology of human disease.
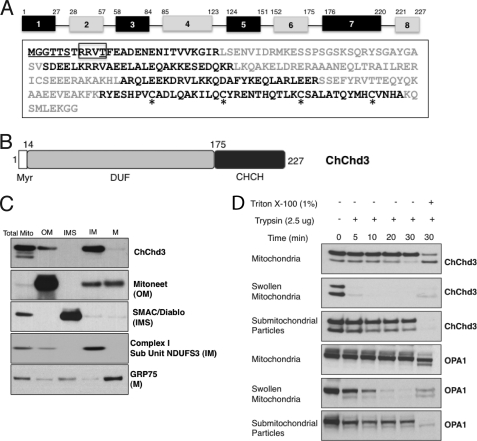
ChChd3 is highly conserved in mammals with human and mouse protein sharing ∼92% sequence similarity. Orthologs of the protein are present throughout the metazoans, whereas no homologs are so far seen in fungi or plants. In humans, the ChChd3 gene maps on chromosome 7 (7q32.3-q33), and the coding sequence of the protein has 8 exons that can potentially generate nine splice variants. Full-length ChChd3 has 227 amino acids with an N-terminal myristoylation motif followed by a DUF737 domain (domain of unknown function), and a coiled-coil helix-coiled-coil helix (chch) domain (Fig. 1, A and B). DUF domain belongs to the large set of protein families in the data base whose function is not known. The C-terminal chch domain, on the other hand, is primarily seen in mitochondrial proteins and was known to be involved in the protein import and metal binding in the IMS (18–21). Each helix in the chch domain contains two of the four conserved cysteines arranged in CX9C or CX3C manner, and in the functional mature protein the cysteines are known to be oxidized forming two intramolecular disulfide bonds, essential for the proper folding and structural stabilization of the protein. The mitochondrial disulfide relay system mediated by Mia40 and Erv1 has been proposed as the principal mechanism for the transport and subsequent folding of the cysteine-containing proteins in the IMS (22).
FIGURE 1.
Domain organization and mitochondrial localization of ChChd3. A, amino acid sequence and exon organization of ChChd3. Different exons are shown in alternate black and gray colors. The myristoylation motif at the N terminus is underlined. The consensus site for the PKA phosphorylation is shown in the box, and the previously identified PKA phosphorylation site is underlined. The CX9C-CX9C motif cysteines in the chch domain are highlighted with asterisk. B, schematic diagram of the ChChd3 protein. DUF, domain of unknown function, CHCH, coiled-coil helix-coiled-coil helix domain. C and D, ChChd3 in mitochondria is primarily localized to the IM facing toward the IMS. Mouse liver mitochondrial subfractions were separated by SDS-PAGE and analyzed by immunoblotting by using antibodies against ChChd3 and known mitochondrial marker proteins. ChChd3 is enriched in the IM fraction similar to that of the IM marker protein, NDUFS3. M, matrix (C). SDS-PAGE and immunoblot analysis of trypsin-treated samples of mitochondria, swollen mitochondria, and submitochondrial particles (D).
Although there is some evidence that ChChd3 is an abundant protein and localized to the mitochondria (15, 23), the role of the protein has remained poorly understood with no systematic study describing its cellular localization and/or biological function. In this comprehensive study, we describe a potential role of this protein in regulating mitochondrial structure and function. We show that ChChd3 is localized to the IM facing the IMS and exists in a complex with the IM proteins mitofilin and OPA1 and the OM protein Sam50. Furthermore, we analyzed the consequence of down-regulation of ChChd3 in HeLa cells by using the RNAi method and demonstrate that ChChd3 plays an essential role in maintaining CJ architecture, crista morphology, and mitochondrial function.
EXPERIMENTAL PROCEDURES
Antibodies and Plasmids
Polyclonal antibodies against internal peptide sequence corresponding to ChChd3 mouse (QAKKESEHQRRLKQARDLERER) and human (QAKKESEDQKRLKQAKELDRER) protein sequence were generated in rabbits (Invitrogen). The other commercial antibodies used are described in supplemental material.
The mouse cDNA clone for ChChd3 (BC021941) in pCMV-SP6 vector was procured from Invitrogen (catalog no. 5124504) and subsequently subcloned into a modified C-terminally FLAG-tagged pCMV-SP1 vector by using EcoRI and SalI restriction sites. Point mutations G2A ChChd3 and K38A Drp1GFP were made by QuikChange® site-directed mutagenesis (Stratagene). ΔCT and ΔNT mutants were made by PCR amplification and subcloning by standard protocols. The cDNA clones for mitofilin (catalog no. SC320269, reference sequence NM_006839) in pCMV6-AC vector and Sam50 (catalog no. SC108289, reference sequence NM_015380) in pCVM6-XL5 vector were purchased from OriGene and subcloned into C-terminally FLAG tagged pCMV-SP1 vector. The cDNA clones for Drp1 GFP and mito-RFP were received as gifts from Dr. Stefan Strack at the University of Iowa and Dr. Luca Scorrano at Venetian Institute of Molecular Medicine, respectively.
Isolation and Fractionation of Mitochondria
Mitochondria from mouse liver were isolated, and Histodenz gradient purified as described previously (24). Mitochondrial subfractions were obtained by swell-shrink procedure and purified by using sucrose gradient ultracentrifugation method (24). Details can be found in supplemental material.
Trypsin Digestion Assay
For trypsin digestion experiments, mitochondria were isolated without protease inhibitors throughout the preparation. 100 μg of freshly isolated mitochondria, swollen mitochondria, and submitochondrial particles were incubated with 2.5 μg of trypsin in 0.125 m sucrose and 10 mm Tris-HCl, pH 8, for 5–30 min with or without 1% Triton X-100 at room temperature. 200 μg of soybean trypsin inhibitor was added to quench the reaction, and the samples were immediately boiled in the presence of 2× Laemmli sample buffer. 10 μg of the sample was separated on SDS-PAGE and analyzed by immunoblot.
RNA Interference (RNAi)
A 29-mer short hairpin RNA (shRNA), 5′-TATCAGAAAGCTGCTGAGAGGTGGAAGC-3′ corresponding to the nucleotide sequence 489–516 of human ChChd3 in a pRS vector (OriGene) with U6 promoter and puromycin-resistant gene, was used to generate stable knockdown of ChChd3 in HeLa cells. Control stable cells were generated by using a scrambled sequence 5′-GCACTACCAGAGCTAACTCAGATAGTACT-3′ in pRS vector. HeLa cells maintained in DMEM with 10% FBS and 1% GlutaMAX grown to 40% confluency in 35 mm plates were transfected with ChChd3-shRNA or scrambled-control using Turbofectin (OriGene) according to the manufacturer's protocol, and stable cell lines were selected in 0.5 μg/ml puromycin (Sigma).
Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR) Measurements
An XF24 extracellular flux analyzer (Seahorse Biosciences) was used to measure the rates of oxygen consumption and extracellular acidification in 24-well plates (Seahorse Biosciences) as described previously (25) with minor modifications. Briefly, cells were plated in growth medium at 3 × 104 cells/well 16 h prior to experimentation. The assay medium was unbuffered DMEM (Sigma) supplemented with 10 mm glucose, 10 mm pyruvate, and 2 mm glutamine, pH 7.4, to which 10× additions were made from the cartridge injection ports. Assay cycles included 3 min of mixing and a 2 min waiting period, followed by 3 min of measurement. An updated algorithm that more accurately accounts for oxygen diffusion through the plastic plate was used for data analysis (26).
ATP Measurements
Cellular ATP levels were measured by using a CellTiter-Glo® luminescent cell viability assay (Promega). Briefly, control and ChChd3 knockdown cells were seeded in a 96-well plate with 35,000 cells per well in a 100 μl volume. After 6 h, 100 μl of CellTiter-Glo® reagent was added to each well and mixed for 2 min. After 10 min of incubation at room temperature, luminescence was measured in a GENios microplate reader (Tecan). ATP levels were normalized to protein levels.
Transmission Electron Microscopy
Control and ChChd3 knockdown HeLa cells were grown to 60% confluency on poly-d-lysine-coated glass dishes (MatTek) and fixed with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.4, for 5 min at room temperature, followed by 30 min on ice for optimal cellular ultrastructural preservation. The cells were washed five times, 2 min each in ice-cold 0.1 m sodium cacodylate buffer, pH 7.4, with 3 mm calcium chloride. The cultured cells were post-fixed in ice-cold 1% osmium tetroxide (Electron Microscopy Sciences) with 0.8% potassium ferrocyanide (Sigma) and 3 mm calcium chloride (Sigma). After 30 min, the cells were washed five times in double-distilled water for 2 min each and stained en block in ice-cold 2% uranyl acetate overnight to increase membrane contrast. The cultured cells were dehydrated in an ethanol series for 3 min each with 20, 50, 70, and 90% ethanol on ice followed by four changes of 100% ethanol at room temperature. The dehydrated cells were infiltrated in a 1:1 ratio of absolute ethanol to Durcupan ACM epoxy resin (Electron Microscopy Sciences) for 30 min followed by three changes of Durcupan ACM epoxy resin for 1 h each. The dishes with epoxy resin infiltrated culture cells were then placed in a vacuum oven at 60 °C for 48–72 h for the resin polymerization. After polymerization, the coverslips were removed from the dishes, and plastic blocks were sawed out and glued on to dummy blocks. 80 nm ultrathin serial sections were prepared using a Reichert-Jung Ultra-cut E ultramicrotome and Diatome diamond knife. The sections were supported on copper grids/75 mesh (Ted Pella). The 80-nm sections were post-stained in Sato lead for 1 min, and the stained sections were imaged onto negatives using a JEM-1200EX I electron microscope operated at 80 kV. The negatives were digitized at 1800 dpi using a Nikon CoolScan system, giving an image size of 4033 × 6010 pixel array and a pixel resolution of 1.77 nm.
Electron Tomography
For electron tomography, sections were cut from the same blocks used for conventional transmission electron microscopy at a nominal thickness of 0.5 μm and collected on 100:100 clamshell grids. These sections were poststained for 15 min in a 2% uranyl acetate solution followed by 15 min in Sato lead solution. Two sizes of colloidal gold particles, 15 and 20 nm in diameter, were deposited on opposite sides of the section to serve as fiducial cues. For stability in the beam, the section was coated with carbon. For each reconstruction, a single series of images was collected with a JEOL 4000EX intermediate high voltage electron microscope operated at 400 kV. The specimens were irradiated for about 30 min before initiating a tilt series to limit anisotropic specimen thinning during image collection. During data collection, the illumination was held to near parallel beam conditions. Single-tilt series were recorded using a 4 × 4k slow scan CCD camera controlled by serial EM at 12,000 magnification. Angular increments of 2° usually from −60 to +60° about an axis perpendicular to the optical axis of the microscope were achieved using a computer-controlled goniometer to increment accurately the angular steps. The pixel resolution was 1.2 nm. To improve the signal-to-noise ratio, each image was binned down two times by averaging adjacent pixels so that the final pixel resolution was 2.4 nm. The IMOD software package (27) was used for the complete image processing unless there were problems, and then TxBR software was used to refine the alignment and reconstruction (28). Six reconstructed volumes were analyzed (three control and three ChChd3 knockdown). Volume segmentation was performed by manual tracing in the planes of highest resolution (X-Y) with the program Xvoxtrace (29). The reconstructions were visualized using Analyze (Mayo Foundation) and the surface-rendering graphics of Synu (National Center for Microscopy and Imaging Research) as described by Perkins et al. (29). These programs allow one to step through slices of the reconstruction in any orientation and to model and display features of interest in three dimensions. Measurements of mitochondrial outer and inner boundary and crista membrane surface areas and volumes were made within segmented volumes by the programs Synuarea and Synuvolume, respectively (National Center for Microscopy and Imaging Research, University of California San Diego). Crista junction opening sizes and crista junction densities were measured with ImageJ (National Institutes of Health) using the tomographic volumes.
Immunoprecipitation
For immunoprecipitation of transiently expressed FLAG-tagged proteins, HEK293 cells were transfected with Lipofectamine 2000 (Invitrogen) by following the manufacturer's protocols. Approximately 20 h post-transfection, cells were harvested in buffer A (50 mm Tris-HCl, 150 mm NaCl, 1 mm DTT, 2 mm EDTA, and 1% Triton X-100, pH 7.4) with a protease inhibitor mixture set (Calbiochem), and the cell lysates were incubated with anti-FLAG M2 affinity gel (Sigma) for 2 h at 4 °C. The resin was washed with buffer A (four times) and eluted with Laemmli sample buffer, and samples were analyzed on SDS-PAGE followed by immunoblot.
For immunoprecipitating ChChd3 from mouse liver mitochondria, gradient-purified mitochondria was lysed in buffer A with protease inhibitor mixture set and pre-cleared on protein A-agarose beads (Pierce) for 1 h at 4 °C to prevent any nonspecific binding. Pre-cleared samples were incubated with ChChd3 antibody or the corresponding preimmune serum at 4 °C for 5 h and immunocaptured with protein A-agarose beads. After overnight incubation, the beads were washed five times with 50 mm Tris-HCl, 150 mm NaCl, pH 7.4, with a protease inhibitor mixture, eluted with Laemmli sample buffer, and analyzed on the SDS-PAGE followed by immunoblot.
Separation of Soluble and Insoluble Fractions
Soluble/cytoplasmic and insoluble/mitochondrial fractions were isolated from the control and ChChd3 knockdown HeLa cells as described earlier (30). In brief, cells were lysed with the lysis buffer (80 mm KCl, 250 mm sucrose, 50 μg/ml digitonin, and 1 mm DTT) with a protease inhibitor mixture, and centrifuged for 5 min at 10,000 × g at 4 °C. The supernatant (cytoplasmic fraction) and the pellet containing mitochondrial fraction were used for Western blot analysis.
Statistical Analysis
Data from populations of mitochondria from EM micrographs are represented as mean ± S.E. unless otherwise mentioned. Comparisons between the two groups were made using a Student's t test unless otherwise stated. Statistical analyses were performed using Microsoft Excel and GraphPad Prism.
RESULTS
ChChd3 Is Myristoylated at the N Terminus
ChChd3 has a well conserved myristoylation site at the N-terminal Gly-2 and a PKA phosphorylation site at Thr-11 (Fig. 1A). N-Myristoylation is a co-translational modification known to promote membrane binding that is essential for proper protein localization or biological function (31). To determine whether ChChd3 is myristoylated, we immunoprecipitated transiently expressed FLAG-tagged ChChd3 from HEK293 cells and separated it by SDS-PAGE, and tryptic peptides were analyzed by mass spectrometry. Mass spectrometric analysis identified a peptide of m/z 859.5251, corresponding to the myristoylated N terminus (myrGGTASTR). Fragmentation of this peptide unequivocally showed that the N-terminal glycine is myristoylated (supplemental Fig. S1).
ChChd3 Resides on the Inner Mitochondrial Membrane Facing the Intermembrane Space
We initially identified ChChd3 in purified mouse liver mitochondrial fractions and later showed that both endogenous and transiently expressed ChChd3 is localized to mitochondria (15). To further define the submitochondrial location of ChChd3, we fractionated purified mouse liver mitochondria into OM, IMS, IM, and matrix compartments as described previously (24). Equal amounts of protein from each of these fractions were analyzed by immunoblot with antibodies against ChChd3 and known marker proteins. ChChd3 was primarily enriched in the IM fraction along with trace amounts associated with the OM (Fig. 1C). To further dissect the orientation of ChChd3 on the IM, we performed limited trypsinolysis on intact mitochondria, mitochondria swollen in hypotonic media (swollen mitochondria, with ruptured OM and intact IM), and submitochondrial particles (inside-out mitochondrial vesicles that expose the matrix face of the IM). Immunoblotting of the trypsin-digested samples revealed that ChChd3 in swollen mitochondria is susceptible to more rapid trypsinolysis compared with that of submitochondrial particles and pure mitochondria (Fig. 1D) suggesting that ChChd3 is located on the IM facing the IMS. When submitochondrial particles and mitochondrial membranes were solubilized with 1% Triton X-100, ChChd3 was digested completely. Proteolysis of ChChd3 was similar to that of OPA1 (Fig. 1D), an IM protein facing the IMS (4), thus suggesting that ChChd3 is localized on the IM facing IMS.
Loss of ChChd3 in HeLa Cells Results in Abnormal Mitochondrial Morphology and Altered Levels of Drp1 and Opa1
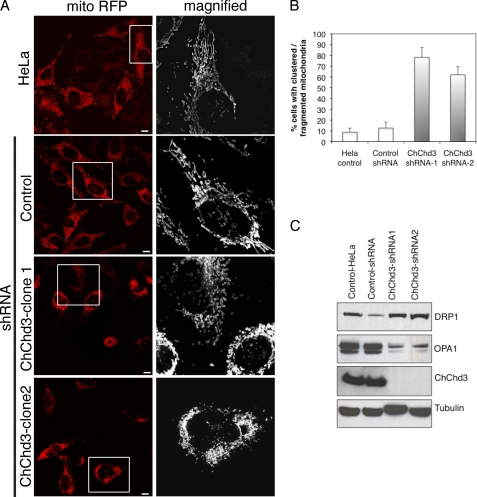
To explore the functional role of ChChd3 in mitochondria, we silenced ChChd3 gene expression using RNAi. For this purpose, we generated a panel of HeLa clonal cell lines that stably express shRNA specific for ChChd3 or a scrambled control sequence. The mitochondrial morphology in the scrambled control (control-shRNA) and from two independent clones of ChChd3 knockdown cells was assessed by using transiently expressed matrix-targeted RFP (mito-RFP). Interestingly, ∼70% of the cells lacking ChChd3 showed fragmentation of the mitochondrial network and clustering of the mitochondria around the nucleus (Fig. 2, A and B) compared with the control-shRNA and control-HeLa cells, which displayed elongated and evenly distributed mitochondria (Fig. 2A).
FIGURE 2.
Loss of ChChd3 in HeLa cells results in abnormal mitochondrial morphology. A, mitochondria in ChChd3 knockdown cells are clumpy, fragmented, and clustered around the nucleus. Representative confocal microscopic images of HeLa-control, control-shRNA, and ChChd3-shRNA HeLa cells expressing matrix-targeted RFP (mito-RFP). Two independent clones of ChChd3-shRNA (ChChd3-shRNA clone 1 and ChChd3-shRNA clone 2) were analyzed to avoid the selection artifacts. Scale bar, 10 μm. B, quantification of mitochondrial abnormalities in ChChd3 knockdown cells. 300 cells each from the control and ChChd3 knockdown cells were analyzed under confocal microscope for mitochondrial fragmentation and clumping. Mean ± S.D. from three independent experiments is shown. C, HeLa control, control-shRNA, and ChChd3-shRNA cell lysates were assessed for the changes in the protein levels of the key regulators of mitochondrial fusion and fission. ChChd3-shRNA1 and shRNA2 represent two different clones derived from a single shRNA sequence.
Because it appeared that loss of ChChd3 results in an imbalance between mitochondrial fusion and fission, we next examined these cells for changes in protein levels of the key players of mitochondrial fusion (Mfn1, Mfn2, and OPA1) and fission (Drp1) processes (1). We found a significant increase in DRP1 and a decrease in both the long and short isoforms of OPA1 protein levels in ChChd3 knockdown cells (Fig. 2C). No changes in levels of Mfn1 and Mfn2 were observed (data not shown).
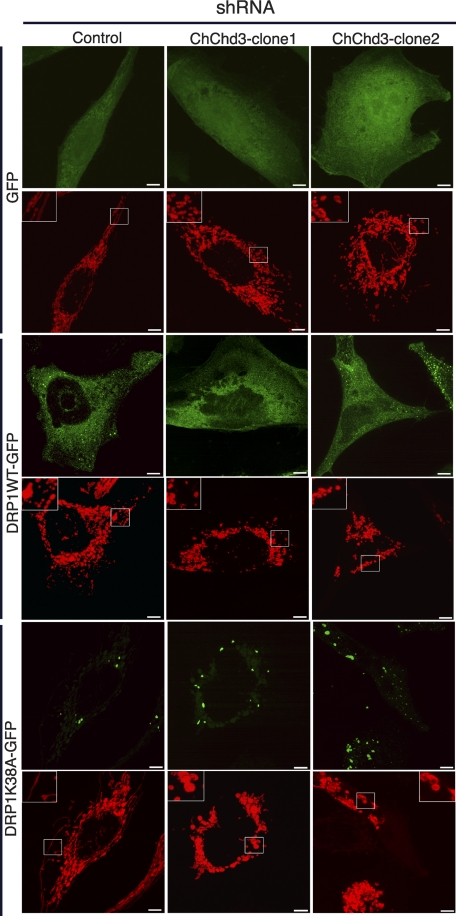
The increased fragmentation observed in ChChd3 knockdown cells could indicate either increased fission or decreased fusion in the absence of ChChd3. To distinguish these possibilities, we used the dominant negative mutant of the fission component DRP1, DRP1K38A. GFP-tagged DRP1 or DRP1K38A were co-expressed with mito-RFP in control-shRNA and ChChd3-shRNA cells, and mitochondrial morphology was analyzed by fluorescence microscopy. As reported previously (32), expression of DRP1K38A in control-shRNA cells resulted in elongated and interconnected mitochondria, visibly different from that of the cells expressing WT DRP1 or GFP alone (Fig. 3). On the other hand, expression of DRP1K38A in cells lacking ChChd3 displayed punctiform mitochondria similar to that of DRP1WT and GFP-expressing cells indicating that the organelles are unable to fuse in the absence of ChChd3 (Fig. 3).
FIGURE 3.
ChChd3 is required for mitochondrial fusion in HeLa cells. Inhibition of fission by K38A-Drp1 does not restore tubular mitochondrial network in the absence of ChChd3. Representative confocal micrographs of control-shRNA or ChChd3-shRNA HeLa cells expressing mito-RFP (red) and GFP or Drp1WTGFP or dominant negative mutant of Drp1, Drp1K38AGFP (green) are shown. Scale bars, 5 μm.
Loss of ChChd3 Expression Leads to Significant Defects in Energy Production
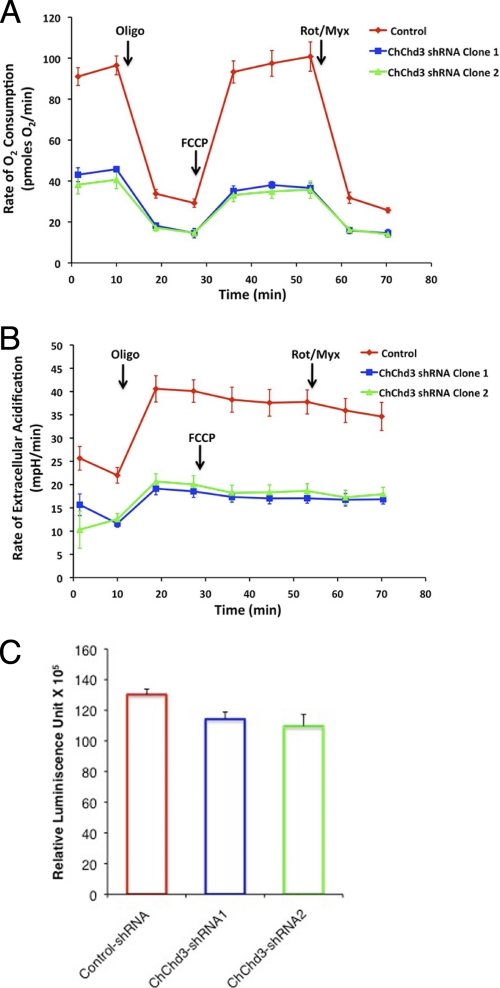
To test whether the alterations observed in the mitochondrial dynamics have affected mitochondrial function, we measured rates of endogenous, state 4, and uncoupler-stimulated mitochondrial respiration and glycolytic activity of control-shRNA and two clones of ChChd3-shRNA cells using a Seahorse extracellular flux analyzer. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) as a function of glycolytic lactate production were simultaneously measured in the presence of 10 mm glucose, 10 mm pyruvate, and 2 mm glutamine as oxidizable substrates. Strikingly, knockdown of ChChd3 produced a dramatic decrease in rates of both the oxygen consumption and lactate production indicating significantly diminished respiratory and glycolytic capacity (Fig. 4, A and B).
FIGURE 4.
ChChd3 depletion impairs cellular bioenergetics. A and B, ChChd3 knockdown cells show a drastic decrease in the OCR and ECAR. OCR and ECAR were measured simultaneously by using the Seahorse extracellular flux analyzer in real time. Rates shown are the averages of four wells measured for 3 min after every 5 min. Also shown are the OCR and ECAR values after addition of 1 μm oligomycin, 200 nm carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), and 100 nm rotenone + myxothiozol (Rot/Myx). C, cellular ATP levels in control-shRNA and ChChd3-shRNA HeLa cells were measured by luminescence-based CellTiter-Glo® cell viability assay. ATP levels were normalized to the protein levels. ChChd3-shRNA1 and shRNA2 represent two different clones derived from a single shRNA sequence, Error bars represent standard deviation from triplicate samples. p < 0.01.
We next assessed the cellular ATP levels in control-shRNA and ChChd3-shRNA cells in a luciferase-based luminescence reaction. Despite the drastically reduced capacity to generate ATP, the total cellular ATP level was reduced only 13% in the absence of ChChd3 (Fig. 4C). These data suggest that ChChd3 knockdown cells have lower energy consumption and defective cellular metabolism characterized by reduced cellular oxygen consumption and glycolysis rates.
Diminished ChChd3 Expression Alters Crista Architecture
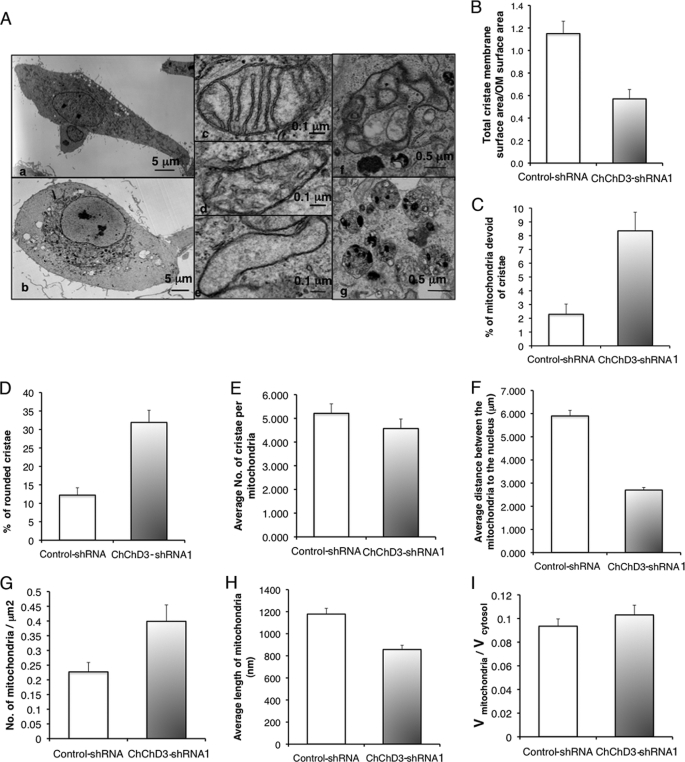
To further understand the changes in mitochondrial morphology and defective bioenergetics due to the loss of ChChd3, we analyzed the ultrastructure of mitochondria in ChChd3 knockdown cells by transmission electron microscopy. The transmission electron microscopy analysis revealed that loss of ChChd3 induced remarkable changes in the mitochondrial IM (Fig. 5). The majority of the mitochondria in ChChd3 knockdown cells have reduced crista content along with the partial remodeling of cristae from lamellar to tubular profiles (Fig. 5, A, panels c and d, and D)). Furthermore, often the mitochondria in ChChd3 knockdown cells were found to be completely devoid of cristae (Fig. 5, A, panel e, and C). Quantification of the membrane surface area of mitochondria from the control and ChChd3 knockdown cells showed that knockdown cells have a 50% reduction in the crista surface area compared with that of control cells (Fig. 5B). No significant change in the number of crista per unit volume of mitochondria between the control and ChChd3 knockdown cells was found suggesting that crista fragmentation did not occur (Fig. 5E).
FIGURE 5.
Loss of ChChd3 results in crista remodeling and perinuclear clumping and fragmentation of mitochondria. A, EM analysis of control-shRNA and ChChd3-shRNA HeLa cells showed that mitochondria in ChChd3-shRNA cells cluster around the nucleus. Altered cristae and an increase in autophagy/mitophagy were also prevalent. Panel a, control-shRNA-expressing HeLa cells have mitochondria distributed throughout the cytoplasm. Panel b, in contrast, ChChd3-shRNA cells show clustering of mitochondria around the nucleus. Panel c, mitochondria of control cells have predominantly lamellar crista, whereas the mitochondria in the ChChd3 knockdown cells have lower crista density and more tubular crista (panel d). Panel e, common in these cells is mitochondria devoid or nearly devoid of cristae. Panel f, an autophagosome in a ChChd3 knockdown cell engulfing mitochondria. Panel g, multivesicular bodies are seen high in number in ChChd3 knockdown cells. B–I, quantification of mitochondrial abnormalities in ChChd3 knockdown cells from EM analysis of control and ChChd3 knockdown cells. ChChd3 knockdown cells have significantly reduced crista surface area (B), higher percentage of mitochondria without crista (C), and altered crista (D) with no change in number of cristae per mitochondria (E). B, bar graph showing the ratio of crista membrane surface area to the OM surface area in control and ChChd3 knockdown cells. OM surface area and the crista membrane surface area from 25 mitochondria each from control and ChChd3 knockdown cells were measured. p < 0.001. C, bar graph showing the percentage of mitochondria completely devoid of cristae measured from 10 different cells as the total number of mitochondria completely devoid of cristae in a cell divided by the total number of mitochondria in the cell. A total of 414 mitochondria in control and 822 mitochondria in ChChd3 knockdown cells were counted. p = 0.0015. D, bar graph showing the percentage of mitochondria with small and rounded cristae. Measured as above in C from 100 mitochondria from control and ChChd3 knockdown cells, 10 were randomly chosen from each cell from the same 10 cells used in C. p = 0.0015. The average number of cristae per mitochondria were counted from the same 10 cells and 100 mitochondria used in C–E. F–I, mitochondria in ChChd3 knockdown cells are fragmented and clustered around the nucleus. F, statistical analysis of the closest distance between the mitochondrial outer membrane and nuclear membrane. 448 mitochondria from five control cells and 339 mitochondria from five ChChd3 knockdown cells were used for measurements. p < 0.01. G, bar graph of the average number of mitochondria per μm2 in control and ChChd3 knockdown HeLa cells. The number of mitochondria per cell was measured as the number/cytoplasm cross-sectional area for 10 cells each of control and ChChd3 knockdown. There were a total of 414 mitochondria in the control cells and 822 mitochondria in the knockdown cells. p = 0.019. H, bar graph of quantification of mitochondrial volume over total cytoplasmic volume measured from 10 cells each of control and ChChd3 knockdown. I, bar graph of average length of mitochondria measured from the same 10 cells used in H. 165 mitochondria from each control and ChChd3 knockdown were measured. p < 0.001. Means ± S.E. used throughout.
In addition to the alterations in the IM, the ChChd3 knockdown cells also showed clustering of mitochondria around the nucleus and large areas of cytoplasm devoid of mitochondria (Fig. 5A, panel b). Quantification of the closest distance between the nuclear membrane and the OM showed that in ChChd3 knockdown cells mitochondria were about twice as close to the nucleus as in control cells (Fig. 5F). Additionally, the ChChd3 knockdown cells showed reduced length and increase in number of mitochondria, with no significant change in mitochondrial volume per cell volume (Fig. 5, G–I) compared with that of the control cells, thus suggesting an increase in mitochondrial fragmentation.
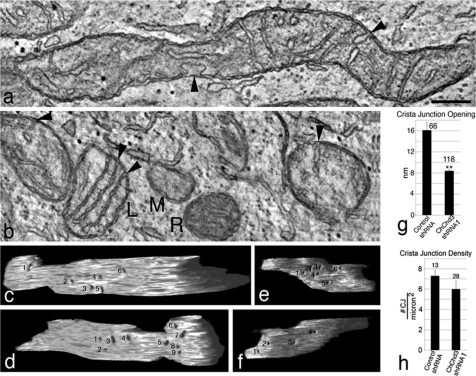
Recent studies have suggested that crista remodeling is accompanied by changes in CJ architecture (10, 14, 33). The IM proteins mitofilin and OPA1, which regulate crist morphology, have been shown to localize at the CJs (4, 12, 34). Studies by independent groups have suggested that during apoptosis disassembly of OPA1 oligomers at the CJs causes changes in the CJ diameter (10, 14). Down-regulation of mitofilin in HeLa cells resulted in complete loss of CJs (12). Because we noticed major alterations in crista structures in the absence of ChChd3, we decided to further analyze the morphology of the IM by using tomographic three-dimensional reconstructions to look into CJ opening diameter and density (Fig. 6). No changes in the number of CJs were observed in ChChd3 knockdown cells compared with that of the control-shRNA expressing cells (Fig. 6h). However, the CJ opening diameter in mitochondria in cells lacking ChChd3 is remarkably reduced to half the size (∼8 nm) compared with that of control mitochondria (∼16 nm, Fig. 6g), indicating that ChChd3 is involved in maintaining CJ architecture but not in their formation.
FIGURE 6.
Electron tomography reveals that crista junctions in ChChd3 knockdown mitochondria are smaller than those in control mitochondria. a, 2.4-nm-thick slice through the tomographic volume of a control mitochondrion. Control mitochondria are generally longer than their ChChd3 knockdown counterpart. Two crista junctions are shown (arrowheads) with the one at bottom having a very large opening. Scale bar, 200 nm and applies to all panels. b, 2.4-nm thick slice through the tomographic volume of five ChChd3 knockdown mitochondria. These mitochondria are generally smaller and often found close to each other suggesting fission has occurred. The amount of cristae present can differ remarkably (compare L, similar cristae complement to control mitochondria, with M, tiny mitochondrion devoid of cristae), and the matrix density can also differ substantially (compare M with R). Their crista junction openings are characteristically smaller (arrowheads). c and d, side views of a segmented and surface-rendered inner membrane from a control mitochondrial volume with numbered crista junctions. Six crista junctions are shown on one side and nine on the other side. e and f, side views of a segmented and surface-rendered inner membrane from a ChChd3 knockdown mitochondrial volume showing smaller crista junctions (numbered) than the control. Six crista junctions are shown on one side and four on the other side. g, crista junction openings in ChChd3 knockdown mitochondria are about half the size of control openings. The mean crista junction width at its largest opening in tomographic reconstructions is compared. The number of measurements is shown above each bar. Error bar, S.E. (**, p < 0.01). h, density of crista junctions is no different in control and ChChd3 knockdown mitochondria. The total number of crista junctions per mitochondrial volume was counted and divided by the mitochondrial surface area derived from the tomographic volume to determine the crista junction density. Error bar, S.E.
ChChd3 Interacts with Mitofilin, Sam50, OPA1, and HSP70
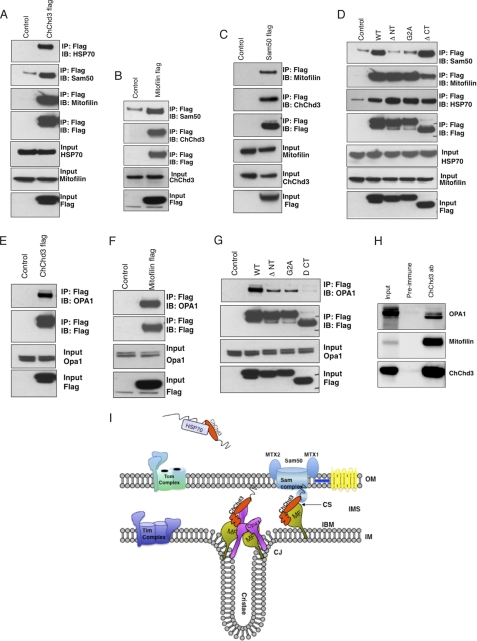
To further understand the role of ChChd3 in regulating mitochondrial structure, dynamics, and function, we sought to identify its potential binding partners. For this purpose, we immunoprecipitated the FLAG-tagged ChChd3 or vector control from HEK293 cells and analyzed the samples by MS/MS sequencing. MS/MS data identified two proteins, mitofilin and HSP70, exclusively in the ChChd3 FLAG sample (supplemental Fig. S2 and supplemental Table S1). These interactions were further confirmed by immunoblot with antibodies against mitofilin and HSP70 (Fig. 7A).
FIGURE 7.
ChChd3 interacts with Mitofilin, Sam50, HSP70, and OPA1. A–C, FLAG-tagged ChChd3 (A), mitofilin (B), and Sam50 (C) were transiently expressed in HEK293 cells and immunoprecipitated (IP) with FLAG resin. Eluted samples were analyzed on SDS-PAGE followed by immunoblot (IB) with the indicated antibodies. D, ChChd3 binds to mitofilin through the chch domain and to Sam50 through N-terminal myristoylation or myristoylated confirmation. E–H, ChChd3 and mitofilin preferentially interact with the shorter isoform of OPA1 in HEK293 cells (E–G) and in mitochondria (H). FLAG-tagged full-length WT ChChd3 (E) or FLAG-tagged mitofilin (F) or ChChd3 mutant proteins (G) were expressed in HEK293 cells and immunoprecipitated with FLAG resin. Sam50 levels are not shown in the input as the antibodies available for Sam50 failed to detect endogenous protein in total cell lysates of HEK293 cells. H, ChChd3 protein, immunoprecipitated from mouse liver mitochondria by using ChChd3 antibody, binds efficiently with mitofilin and the soluble IMS isoform of OPA1. 5% of the total input is shown throughout. I, proposed model for localization of ChChd3 in mitochondria. ChChd3 is synthesized in the cytoplasm and kept in a reduced and soluble form with the help of HSP70, before it is imported and folded in the mitochondria. In mitochondria, ChChd3 may exist at two discrete foci: 1) at CJ and (2) at contact sites (CS). At the CJs, ChChd3 would form a complex with mitofilin and OPA1, and at the contact sites it would associate with mitofilin and Sam50 thereby influencing the regulation of crista biogenesis and mitochondrial protein import, respectively. MF, mitofilin; IBM, inner boundary membrane.
Following our findings, Xie et al. (35) reported ChChd3 as a part of the 340-kDa complex immunoprecipitated with the mitofilin antibody. Sam50, metaxin 1, and metaxin 2, the components of the SAM complex on the OM involved in β-barrel protein assembly, have also been immunoprecipitated (36) in the same complex. Because the reported study was restricted to immunoprecipitation with mitofilin antibody alone, we further sought to systematically analyze the specificity of these protein-protein interactions in which ChChd3 is involved. Because of the lack of availability of antibodies for metaxin 1 and 2, we restricted our studies to Sam50 and mitofilin in this complex. Immunoprecipitation of FLAG-tagged ChChd3 from HEK293 cells showed the presence of Sam50 (Fig. 7A). In a reciprocal experiment, endogenous ChChd3 was co-purified from the immunoisolated samples of mitofilin FLAG and Sam50 FLAG (Fig. 7, B and C) further confirming these protein-protein interactions.
To characterize the binding regions of mitofilin and Sam50 on ChChd3, we mapped the regions of ChChd3 that are required for their interaction. For this, we used transiently expressed C-terminally FLAG-tagged mutants that lack the N-terminal 14 amino acids (ΔNT) or the chch domain (ΔCT) in HEK293 cells. Immunoprecipitation of these mutant proteins from HEK293 cells showed that ΔNT associates with mitofilin similar to that of the WT protein (Fig. 7D). However, interaction between ΔCT and mitofilin is significantly reduced compared with that of WT or ΔNT. In contrast, Sam50 showed efficient binding to ΔCT (Fig. 7D). On the other hand, ΔNT failed to bind Sam50 indicating that ChChd3 binds to mitofilin through the chch domain and to Sam50 through its N terminus. As we identified that ChChd3 is myristoylated at the N terminus, we hypothesized that N-myristoylation may be involved in binding to Sam50. To test this, we generated the G2A mutant of ChChd3 and assessed the ability of this mutant protein to pull down Sam50 and mitofilin. Although G2A can efficiently pull down mitofilin, it was unable to bind Sam50 (Fig. 7D), thus suggesting that ChChd3 binds to Sam50 via its myristoyl moiety or in a myristoylated conformation. It seems that in mitochondria, myristoylation recruits the N terminus of ChChd3 to the OM where the SAM complex is located, whereas the chch domain is recruited to the IM where it associates with mitofilin (Fig. 7I, model). HSP70 did not show any preference and was found to be associated with all the mutants (Fig. 7D). The chch domain proteins are synthesized in the cytosol and are imported into the mitochondria in a completely reduced form. Molecular chaperones HSP70 and HSP90 are known to bind the preproteins and prevent premature folding and to assist in transport to the mitochondria (37). Hence it is likely that HSP70 associates with ChChd3 in the reduced state in the cytoplasm before it is imported into the mitochondria (Fig. 7I, model).
Down-regulation of ChChd3 resulted in reduced OPA1 protein levels, impaired mitochondrial fusion, and crista remodeling along with narrowing of the CJ diameter. OPA1 has been shown to be involved in regulating crista remodeling independently of its role in mitochondrial fusion (10, 14). Oligomers of OPA1 at the CJs are known to control the CJ opening diameter. Because the loss of ChChd3 leads to the alterations in crista morphology along with narrowing of CJ opening diameter, we hypothesized a plausible functional connection between ChChd3 and OPA1. Additionally, mitofilin, ChChd3, and OPA1 are localized in the same compartment in mitochondria, with IM facing the IMS. Hence we assessed the possibility of the complex between these three proteins. Immunoprecipitation experiments confirmed that both the ChChd3 and mitofilin could efficiently interact with OPA1 (Fig. 7, E and F). Interestingly, the soluble IMS form of OPA1 preferentially associated with both ChChd3 and mitofilin (Fig. 7, E and F). Furthermore, ChChd3 mutants ΔNT, G2A, and ΔCT showed reduced affinity toward binding to OPA1 (Fig. 7G). Analysis of these mutant proteins by immunofluorescence revealed that although WT protein is exclusively localized to mitochondria, ΔCT failed to localize to mitochondria. On the other hand, ΔNT and G2A are diffused throughout the cell and exhibit partial mitochondrial localization.4 Hence, it is likely that the interaction between OPA1 and ChChd3 is specific to mitochondrial IMS where soluble OPA1 is generated by the cleavage of the full-length transmembrane OPA1. To further confirm this complex in mitochondria, we immunoprecipitated ChChd3 from purified mouse liver mitochondria and analyzed for OPA1 and mitofilin binding. ChChd3 showed efficient binding to mitofilin and to the soluble IMS form of OPA1 in mitochondrial samples (Fig. 7H). The interaction of ChChd3 with OPA1 together with a decrease in the protein levels of OPA1 and reduced mitochondrial fusion in ChChd3 knockdown cells (Figs. 2D and 3) suggest that ChChd3 may play an essential role in regulating OPA1 function, specifically the soluble IMS form.
ChChd3 Knockdown Cells Show a Drastic Reduction in the Protein Levels of Mitofilin and Sam50
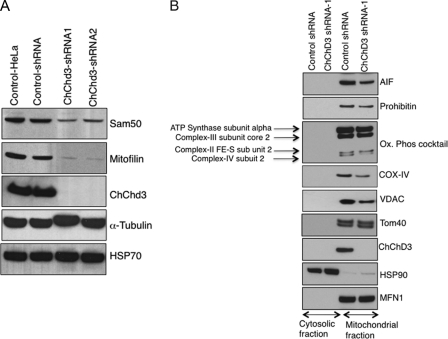
Because we found that ChChd3 interacts with mitofilin, Sam50, and OPA1 (Fig. 7), and ChChd3 knockdown cells show reduced OPA1 levels (Fig. 2C), we analyzed the protein levels of mitofilin and Sam50. Immunoblot analysis of the total cell lysates of control-shRNA and ChChd3-shRNA cells revealed a drastic reduction in both the mitofilin and Sam50 proteins in ChChd3 knockdown cells (Fig. 8A). RT-PCR revealed no changes in mRNA levels (supplemental Fig. S3), suggesting that there is no effect in the transcription of these proteins, and the decrease in protein levels is due to differences in translation or post-translational mechanisms. The levels of HSP70, another ChChd3-associated protein, did not change in the ChChd3 knockdown cells (Fig. 8A).
FIGURE 8.
Depletion of ChChd3 leads to major loss of mitofilin and Sam50 and reduction in specific mitochondrial proteins. A, equal amounts of protein from control and ChChd3 knockdown HeLa cell lysates were analyzed on the immunoblot against the antibodies indicated. Protein load was normalized against tubulin. B, ChChd3 knockdown cells show reduced levels of mitochondrial inner membrane proteins AIF, prohibitin, Cox-IV, and Cox-II and the outer membrane protein VDAC. Cytoplasmic and mitochondrial fractions from control and ChChd3 knockdown cells were separated as described under “Experimental Procedures,” and equal amounts of protein (15 μg) from each of these fractions were analyzed on SDS-PAGE followed by Western blotting. HSP90 and MFN1 were used as loading controls for cytoplasmic and mitochondrial fractions, respectively. ChChd3-shRNA1 and shRNA2 represent two different clones derived from a single shRNA sequence.
Depletion of Sam50 has been shown to cause defects in the assembly of VDAC and TOM40 (36, 38). To determine whether the loss of Sam50 in ChChd3 knockdown cells resulted in defects in the assembly and stability of these proteins, we initially assessed the total cell lysates of control-shRNA- and ChChd3-shRNA-expressing cells for changes in these protein levels. No changes in TOM40 protein levels were observed. However, VDAC protein levels were reduced in ChChd3-shRNA cells (data not shown). To look for specific changes in mitochondrial protein levels, we separated soluble (cytoplasmic) and insoluble fractions (containing mitochondria) from the control-shRNA and ChChd3-shRNA cells and analyzed for VDAC and Tom40 levels (Fig. 8B). ChChd3 knockdown cells showed an ∼40% reduction in VDAC levels in the membrane fraction, whereas Tom40 levels were not affected.
In addition to the reduction in VDAC levels, we found that IM proteins, AIF, and prohibitin levels are also reduced in the mitochondrial fraction of ChChd3 knockdown cells (Fig. 8B). When the oxidative phosphorylation complexes on the IM were analyzed in these samples, cytochrome c oxidase subunits, COX-II and COX-IV, showed a clear reduction in the mitochondrial fraction of ChChd3 cells (Fig. 8B).
Loss of ChChd3 Leads to Reduced Cell Proliferation and Increased Autophagy
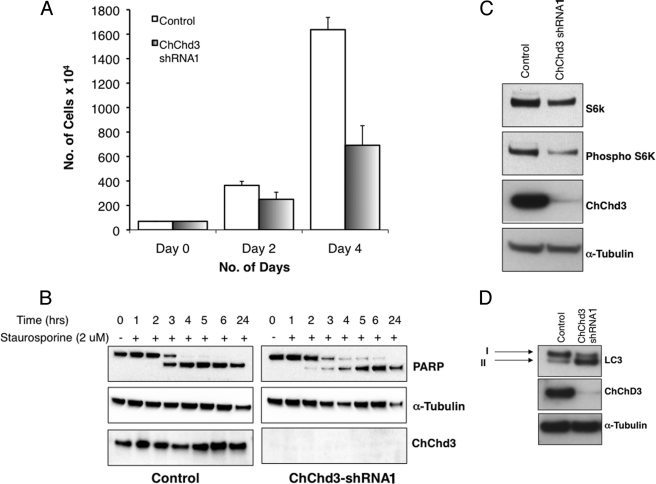
Defects in mitochondrial structure, reduced crista content, and reduced cellular bioenergetics in ChChd3 knockdown cells are further reflected in a reduced growth rate. When an equal number of control and ChChd3 knockdown cells were plated and counted manually, a 60% reduction in cell number was observed after 4 days (Fig. 9A). To determine the cause for reduced cell number, we analyzed the control and ChChd3 knockdown cells for changes in apoptosis. No significant difference was observed when the cells were stained with annexin V to visualize apoptotic cells (data not shown). Also, no detectable changes in cleavage of poly(ADP-ribose) polymerase were found in both the untreated as well as staurosporine-treated conditions (Fig. 9B). As there was no indication of increased apoptosis, the reduction in ChChd3 knockdown cell number is most likely attributable to decreased cellular proliferation. Consistent with this, immunoblotting of cell lysates for phosphorylated and total p70 S6 kinase revealed a decrease in both the protein levels in the ChChd3 knockdown cells (Fig. 9C), suggesting that these cells spend a longer time in the G1 phase and may be accelerating the processes leading to cellular senescence.
FIGURE 9.
Loss of ChChd3 results in reduced cellular proliferation. A, rate of cellular proliferation in control and ChChd3 knockdown cells measured by plating an equal number of cells and counting them periodically for 4 days. Error bars represent standard deviation from triplicate samples. B, reduced growth rate is not due to the increased apoptosis or sensitivity toward apoptosis in ChChd3 knockdown cells. Control and ChChd3 knockdown cells were treated with staurosporine to induce apoptosis, and the cells were lysed after the indicated time periods and analyzed for the changes in poly(ADP-ribose) polymerase (PARP) cleavage. C, ChChd3 knockdown cells show reduced p70 S6 kinase (S6k) and phospho-p70 S6 kinase protein levels. Equal amounts of protein from the control and ChChd3 knockdown cells were analyzed for p70 S6 kinase and phospho-p70 S6 kinase. D, ChChd3 knockdown cells show elevated autophagy. Equal amounts of protein from the total cell lysates of control and ChChd3 knockdown cells were separated on a SDS-PAGE and immunoblotted against LC3 antibody. Tubulin is used as the loading control.
Because EM images of ChChd3 knockdown cells showed an increase in number of the vesicles, multivesicular bodies, lysosomes, and autophagosomes (Fig. 5A, panels f and g), we investigated a possible increase in autophagy in these cells by probing for the endogenous LC3 protein, a well known marker for autophagy (39). Upon initiation of autophagy, LC3 is cleaved to form LC3-II, and ChChd3 knockdown cells showed a clear increase in the LC3-II band compared with the control cells (Fig. 9D). Although growing evidence points toward the role of autophagy in maintaining homeostasis by degrading the dysfunctional organelles and proteins, autophagy has also been specifically shown to associate with the type II (nonapoptotic) programmed cell death (40). Because ChChd3 knockdown cells display severe defects in mitochondrial IM structure, it is likely that these abnormal mitochondria are being targeted for destruction by autophagy, and the elevated autophagy is also a possible reason for the decreased cell number observed in ChChd3 knockdown cells.
DISCUSSION
ChChd3 belongs to the subset of chch domain family proteins that are known to be primarily in the IMS of mitochondria and are characterized for their roles in protein import and as metallochaperones (22, 41). Although ChChd3 was identified in our laboratory a few years ago as a substrate for protein kinase A (15), the function of this protein is so far completely unknown. In this study, we describe the characterization of ChChd3 as a novel protein essential for maintaining crista integrity and mitochondrial function. We provide evidence suggesting that ChChd3 is one of the key proteins that stabilizes protein complexes involved in maintaining crista architecture and protein import, the two primary processes that regulate mitochondrial structure and function.
Down-regulation of ChChd3 in HeLa cells resulted in fragmentation and clustering of the mitochondrial network (Fig. 2A). Studies with the dominant negative mutant of Drp1, DRP1K38A, which is resistant to fission, showed that the mitochondria in ChChd3 knockdown cells have impaired fusion activity. Additionally, ChChd3 interacts preferentially with the soluble IMS form of OPA1. Recent studies have suggested the essential role of OPA1 not only in mitochondrial fusion but also in remodeling of cristae (33). Even though both the long and short isoforms have been shown to be essential in regulating mitochondrial fusion (42) and crista morphology (10, 14), the individual role of these isoforms is not clearly understood. A recent immuno-EM study identified a differential distribution of the two yeast isoforms of OPA1 (Mgm1) across the IM (43). Long Mgm1 was found strongly enriched in the crista membrane, and the short Mgm1 was preferentially localized on the OM/inner boundary membrane. Furthermore, by using various mutations in the GTPase domain, these authors showed that the functional GTPase domain is only required in short Mgm1 but not in long Mgm1 suggesting the idea of distinct function of each of these isoforms in mitochondria. It is likely that the fusion activity of OPA1 along with mitofusins on the OM is controlled by the s-OPA1 on the inner boundary membrane, whereas the crista morphology is regulated by long OPA1 on the crista membrane. Interaction of ChChd3 preferentially with soluble OPA1 and impaired fusion in ChChd3 knockdown cells indicate toward the potential involvement of ChChd3 in regulating the function of this isoform.
Depletion of ChChd3 resulted in major remodeling of the cristae and the CJ architecture (Figs. 5 and 6). Previous studies have suggested that mitofilin and OPA1 are located at the CJs and involved in regulating crista morphology (12, 34). We demonstrate that ChChd3 interacts with mitofilin and OPA1, and loss of ChChd3 results in near loss of mitofilin and 50% reduction in OPA1 protein levels (Fig. 2C). This suggests the possibility that all three proteins are in a complex at the CJs, where ChChd3 is a scaffolding protein that stabilizes the protein complexes and thus is involved in maintaining the integrity of CJs and in the formation/stabilization of cristae.
We also found that ChChd3 and mitofilin interact with Sam50 (Fig. 7), the component of the SAM complex on the OM, involved in β-barrel protein assembly. Very few proteins have so far been known to regulate both the protein import and organelle morphology (44). The yeast protein, Mdm10, originally characterized for its role in mitochondrial dynamics and morphology has been shown to interact with SAM complex and is required for the β-barrel protein assembly (45). To date, no mammalian proteins have been identified with dual functions in biogenesis of mitochondrial proteins and organelle morphology. Our results suggest the possible involvement of ChChd3 in protein import and/or assembly via SAM complex in addition to its role in regulating crista architecture.
The N-terminal POTRA domain of Sam50 is located in the IMS and has been shown to bind β-barrel precursor proteins in vitro (46). In the IMS, ChChd3 may have a role in transporting the precursor proteins to the SAM complex. A similar function was suggested for IMS-translocase of inner membrane proteins (47). However, so far there is no evidence showing the association of translocase of inner membrane proteins with the SAM complex.
As ChChd3 is localized to both the OM and IM fractions in the fractionated mitochondria and associates with both OM and IM proteins, we predict that ChChd3, together with mitofilin and Sam50, is located at contact sites where both the inner and outer membranes come together. These membrane contacts are known to be essential for various mitochondrial functions, including metabolite channeling, coordinated fusion, fission, protein transport, and also for maintaining structural integrity of outer and inner membranes (48, 49). Because CJs and contact sites are suggested to be two distinct structures on the IM and are not in preferential proximity to each other (49, 50), we propose that ChChd3 may be localized in discrete complexes at CJs and contact sites (Fig. 7I, model). At the CJs, ChChd3 is likely to be associated with mitofilin and OPA1, and at the contact sites it is expected to be in a complex with mitofilin and Sam50. It is likely that ChChd3 is dynamically distributed between these two distinct complexes under different conditions in the mitochondria.
The distinct morphological changes observed due to the knockdown of ChChd3 are more severe than the knockdown phenotypes of mitofilin, OPA1, or MICS1, the mitochondrial proteins previously known to be involved in regulating crista morphology (11–13). Loss of mitofilin resulted in an increase in the surface area of IM, with the cristae having degraded to the form of concentric sheets (12). Down-regulation of OPA1 caused fragmentation of mitochondria and remodeling of cristae to reticular, curved, or ring-shaped structures (11). The cristae in MICS1-depleted mitochondria were observed to be curved and ring-like invaginations of the IM (13). In contrast, mitochondria in ChChd3 knockdown cells often did not contain any cristae. Those with cristae transitioned partly from lamellar to tubular cristae and overall showed a 50% reduction in the crista membrane surface area, indicating that either cristae were not formed properly or were disassembled more rapidly. Furthermore, loss of ChChd3 leads to a remarkable change in the CJ opening diameter without any significant changes in CJ number. This suggests that ChChd3 is not necessary for the formation of CJs; however, it is important for maintaining CJ architecture. On the contrary, loss of mitofilin was shown to affect the formation of CJs. Recent studies have established that CJ opening size depends on the OPA1 oligomers (10, 14), which are important to our finding that ChChd3 associates with OPA1 and are thus likely to have a role in OPA1 oligomerization and in maintaining CJ architecture. Consistent with the reduction in crista surface area, cells lacking ChChd3 also showed reduction in several IM protein levels. At this point, it is not clear if reduced crista content has triggered the reduction in IM proteins or the decreased crista content is the result of reduced levels of the proteins necessary for the proper formation and stabilization of the cristae.
The severe defect in mitochondrial morphology of ChChd3 knockdown cells is also reflected in strikingly lower rates of cellular respiration indicating a clear defect in mitochondrial function. Furthermore, the decrease in mitochondrial respiration is not compensated by an increase in glycolysis. It is likely that the deficiency of ChChd3 resulted in an overall reduced rate of substrate oxidation, energy production, and energy utilization that is consistent with the observed decreased rate of cell proliferation.
Our studies clearly suggest that ChChd3 is indispensable for maintaining crista structure and cellular metabolism. In a recent review, Perkins et al. (51) compiled findings showing that the central nervous system has significantly greater crista to mitochondrial surface area compared with the peripheral nervous system. Accordingly, ChChd3 (referred to as FLJ20420), which we showed to be essential for crista integrity, is highly abundant at synaptic membranes and in the neurons of rat brain throughout the gray matter, spinal cord, and at the dorsal root ganglion (16). Furthermore, recent studies have shown altered levels of ChChd3 in disease models of familial amyotrophic lateral sclerosis and ischemia (17, 52). Increasing lines of evidence have now indicated that the impairment of mitochondrial structure and function is one of the primary mechanisms behind motor neuron death in familial amyotrophic lateral sclerosis and cell death and injury in ischemia. Hence, it will be interesting to further understand whether ChChd3 plays a role in disease pathogenesis.
Our characterization of ChChd3 as a scaffolding protein for the SAM complex on the OM and mitofilin and OPA1 complex on the IM suggests that ChChd3 could be one of the key proteins that coordinate protein import and mitochondrial morphology, the two essential processes involved in proper functioning of mitochondria. Further studies are warranted to understand the dynamic localization of ChChd3 in these protein complexes and how ChChd3 regulates each of these protein complexes mechanistically. This will provide a better insight into the possible link between the essential processes that regulate organelle structure and function.
Supplementary Material
Acknowledgments
We appreciate the gift of DRP1 GFP cDNA from Dr. Stefan Strack, University of Iowa, and mito-RFP cDNA from Dr. Luca Scorano, Venetian Institute of Molecular Medicine. We thank Susanna Petrosyan for assistance with the mitochondrial preparations and Seahorse extracellular flux measurements; Michelle Yoon for assistance with the statistical measurements of mitochondrial morphometry; Ohkyung Kwon for assistance with the confocal microscopy experiments; and Juniper Pennypacker for assistance with the site-directed mutagenesis. We also thank Drs. K. S. Madhusoodanan, Mira Sastri, and Christopher Eggers for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 DK54441 (to S. S. T.) and P21 RR04050 (to M. H. E.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S3, Methods, and additional references.
M. Darshi and S. S. Taylor, manuscript in preparation.
- OM
- outer membrane
- IM
- inner membrane
- IMS
- intermembrane space
- CJ
- crista junction
- SAM
- sorting and assembly machinery
- VDAC
- voltage-dependent anion channel
- chch
- coiled-coil helix-coiled-coil helix
- OCR
- oxygen consumption rate
- ECAR
- extracellular acidification rate
- RFP
- red fluorescent protein.
REFERENCES
- 1. Chan D. C. (2006) Annu. Rev. Cell Dev. Biol. 22, 79–99 [DOI] [PubMed] [Google Scholar]
- 2. Hoppins S., Lackner L., Nunnari J. (2007) Annu. Rev. Biochem. 76, 751–780 [DOI] [PubMed] [Google Scholar]
- 3. Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E., Chan D. C. (2003) J. Cell Biol. 160, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olichon A., Emorine L. J., Descoins E., Pelloquin L., Brichese L., Gas N., Guillou E., Delettre C., Valette A., Hamel C. P., Ducommun B., Lenaers G., Belenguer P. (2002) FEBS Lett. 523, 171–176 [DOI] [PubMed] [Google Scholar]
- 5. Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. (2001) Mol. Biol. Cell 12, 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoon Y., Krueger E. W., Oswald B. J., McNiven M. A. (2003) Mol. Cell. Biol. 23, 5409–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knott A. B., Bossy-Wetzel E. (2008) Ann. N.Y. Acad. Sci. 1147, 283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen H., Chan D. C. (2009) Hum. Mol. Genet. 18, R169–R176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D. M., Brèthes D., di Rago J. P., Velours J. (2002) EMBO J. 21, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G. V., Rudka T., Bartoli D., Polishuck R. S., Danial N. N., De Strooper B., Scorrano L. (2006) Cell 126, 177–189 [DOI] [PubMed] [Google Scholar]
- 11. Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. (2003) J. Biol. Chem. 278, 7743–7746 [DOI] [PubMed] [Google Scholar]
- 12. John G. B., Shang Y., Li L., Renken C., Mannella C. A., Selker J. M., Rangell L., Bennett M. J., Zha J. (2005) Mol. Biol. Cell 16, 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oka T., Sayano T., Tamai S., Yokota S., Kato H., Fujii G., Mihara K. (2008) Mol. Biol. Cell 19, 2597–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamaguchi R., Lartigue L., Perkins G., Scott R. T., Dixit A., Kushnareva Y., Kuwana T., Ellisman M. H., Newmeyer D. D. (2008) Mol. Cell 31, 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schauble S., King C. C., Darshi M., Koller A., Shah K., Taylor S. S. (2007) J. Biol. Chem. 282, 14952–14959 [DOI] [PubMed] [Google Scholar]
- 16. Dreger M., Mika J., Bieller A., Jahnel R., Gillen C., Schaefer M. K., Weihe E., Hucho F. (2005) J. Proteome Res. 4, 238–249 [DOI] [PubMed] [Google Scholar]
- 17. Fukada K., Zhang F., Vien A., Cashman N. R., Zhu H. (2004) Mol. Cell. Proteomics 3, 1211–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Banci L., Bertini I., Ciofi-Baffoni S., Janicka A., Martinelli M., Kozlowski H., Palumaa P. (2008) J. Biol. Chem. 283, 7912–7920 [DOI] [PubMed] [Google Scholar]
- 19. Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D. P., Katrakili N., Tokatlidis K. (2009) Nat. Struct. Mol. Biol. 16, 198–206 [DOI] [PubMed] [Google Scholar]
- 20. Banci L., Bertini I., Ciofi-Baffoni S., Tokatlidis K. (2009) FEBS Lett. 583, 1699–1702 [DOI] [PubMed] [Google Scholar]
- 21. Rigby K., Zhang L., Cobine P. A., George G. N., Winge D. R. (2007) J. Biol. Chem. 282, 10233–10242 [DOI] [PubMed] [Google Scholar]
- 22. Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. (2005) Cell 121, 1059–1069 [DOI] [PubMed] [Google Scholar]
- 23. Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. (2003) Cell 115, 629–640 [DOI] [PubMed] [Google Scholar]
- 24. Pagliarini D. J., Wiley S. E., Kimple M. E., Dixon J. R., Kelly P., Worby C. A., Casey P. J., Dixon J. E. (2005) Mol. Cell 19, 197–207 [DOI] [PubMed] [Google Scholar]
- 25. Wu M., Neilson A., Swift A. L., Moran R., Tamagnine J., Parslow D., Armistead S., Lemire K., Orrell J., Teich J., Chomicz S., Ferrick D. A. (2007) Am. J. Physiol. Cell Physiol. 292, C125–C136 [DOI] [PubMed] [Google Scholar]
- 26. Gerencser A. A., Mark K. A., Hubbard A. E., Divakaruni A. S., Mehrabian Z., Nicholls D. G., Polster B. M. (2009) J. Neurochem. 110, 990–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kremer J. R., Mastronarde D. N., McIntosh J. R. (1996) J. Struct. Biol. 116, 71–76 [DOI] [PubMed] [Google Scholar]
- 28. Lawrence A., Bouwer J. C., Perkins G., Ellisman M. H. (2006) J. Struct. Biol. 154, 144–167 [DOI] [PubMed] [Google Scholar]
- 29. Perkins G. A., Renken C. W., Song J. Y., Frey T. G., Young S. J., Lamont S., Martone M. E., Lindsey S., Ellisman M. H. (1997) J. Struct. Biol. 120, 219–227 [DOI] [PubMed] [Google Scholar]
- 30. Sun M. G., Williams J., Munoz-Pinedo C., Perkins G. A., Brown J. M., Ellisman M. H., Green D. R., Frey T. G. (2007) Nat. Cell Biol. 9, 1057–1065 [DOI] [PubMed] [Google Scholar]
- 31. Resh M. D. (1999) Biochim. Biophys. Acta 1451, 1–16 [DOI] [PubMed] [Google Scholar]
- 32. Smirnova E., Shurland D. L., Ryazantsev S. N., van der Bliek A. M. (1998) J. Cell Biol. 143, 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamaguchi R., Perkins G. (2009) Biochim. Biophys. Acta 1787, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Odgren P. R., Toukatly G., Bangs P. L., Gilmore R., Fey E. G. (1996) J. Cell Sci. 109, 2253–2264 [DOI] [PubMed] [Google Scholar]
- 35. Xie J., Marusich M. F., Souda P., Whitelegge J., Capaldi R. A. (2007) FEBS Lett. 581, 3545–3549 [DOI] [PubMed] [Google Scholar]
- 36. Kozjak-Pavlovic V., Ross K., Benlasfer N., Kimmig S., Karlas A., Rudel T. (2007) EMBO Rep. 8, 576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Young J. C., Hoogenraad N. J., Hartl F. U. (2003) Cell 112, 41–50 [DOI] [PubMed] [Google Scholar]
- 38. Humphries A. D., Streimann I. C., Stojanovski D., Johnston A. J., Yano M., Hoogenraad N. J., Ryan M. T. (2005) J. Biol. Chem. 280, 11535–11543 [DOI] [PubMed] [Google Scholar]
- 39. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klionsky D. J., Emr S. D. (2000) Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palumaa P., Kangur L., Voronova A., Sillard R. (2004) Biochem. J. 382, 307–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song Z., Chen H., Fiket M., Alexander C., Chan D. C. (2007) J. Cell Biol. 178, 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zick M., Duvezin-Caubet S., Schäfer A., Vogel F., Neupert W., Reichert A. S. (2009) FEBS Lett. 583, 2237–2243 [DOI] [PubMed] [Google Scholar]
- 44. Stojanovski D., Rissler M., Pfanner N., Meisinger C. (2006) Biochim. Biophys. Acta 1763, 414–421 [DOI] [PubMed] [Google Scholar]
- 45. Meisinger C., Rissler M., Chacinska A., Szklarz L. K., Milenkovic D., Kozjak V., Schönfisch B., Lohaus C., Meyer H. E., Yaffe M. P., Guiard B., Wiedemann N., Pfanner N. (2004) Dev. Cell 7, 61–71 [DOI] [PubMed] [Google Scholar]
- 46. Habib S. J., Waizenegger T., Niewienda A., Paschen S. A., Neupert W., Rapaport D. (2007) J. Cell Biol. 176, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wiedemann N., Truscott K. N., Pfannschmidt S., Guiard B., Meisinger C., Pfanner N. (2004) J. Biol. Chem. 279, 18188–18194 [DOI] [PubMed] [Google Scholar]
- 48. Reichert A. S., Neupert W. (2002) Biochim. Biophys. Acta 1592, 41–49 [DOI] [PubMed] [Google Scholar]
- 49. Perkins G., Renken C., Martone M. E., Young S. J., Ellisman M., Frey T. (1997) J. Struct. Biol. 119, 260–272 [DOI] [PubMed] [Google Scholar]
- 50. Perkins G. A., Renken C. W., Frey T. G., Ellisman M. H. (2001) J. Neurosci. Res. 66, 857–865 [DOI] [PubMed] [Google Scholar]
- 51. Perkins G., Bossy-Wetzel E., Ellisman M. H. (2009) Exp. Neurol. 218, 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kavazis A. N., Alvarez S., Talbert E., Lee Y., Powers S. K. (2009) Am. J. Physiol. Heart Circ. Physiol. 297, H144–H152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.