Abstract
Apolipoprotein A-I (apoA-I) is the major protein component of high density lipoproteins (HDL) and a critical element of cholesterol metabolism. To better elucidate the role of the apoA-I structure-function in cholesterol metabolism, the conformation of the apoA-I N terminus (residues 6–98) on nascent HDL was examined by electron paramagnetic resonance (EPR) spectroscopic analysis. A series of 93 apoA-I variants bearing single nitroxide spin label at positions 6–98 was reconstituted onto 9.6-nm HDL particles (rHDL). These particles were subjected to EPR spectral analysis, measuring regional flexibility and side chain solvent accessibility. Secondary structure was elucidated from side-chain mobility and molecular accessibility, wherein two major α-helical domains were localized to residues 6–34 and 50–98. We identified an unstructured segment (residues 35–39) and a β-strand (residues 40–49) between the two helices. Residues 14, 19, 34, 37, 41, and 58 were examined by EPR on 7.8, 8.4, and 9.6 nm rHDL to assess the effect of particle size on the N-terminal structure. Residues 14, 19, and 58 showed no significant rHDL size-dependent spectral or accessibility differences, whereas residues 34, 37, and 41 displayed moderate spectral changes along with substantial rHDL size-dependent differences in molecular accessibility. We have elucidated the secondary structure of the N-terminal domain of apoA-I on 9.6 nm rHDL (residues 6–98) and identified residues in this region that are affected by particle size. We conclude that the inter-helical segment (residues 35–49) plays a role in the adaptation of apoA-I to the particle size of HDL.
Keywords: Apolipoproteins, Cholesterol Metabolism, High Density Lipoprotein (HDL), Lipoprotein Structure, Protein Structure, Apolipoprotein A-I
Introduction
A key element of mammalian cholesterol homeostasis is reverse cholesterol transport wherein excess cholesterol in peripheral tissues is conveyed to the liver by lipoproteins for excretion into the intestine as bile (1). High density lipoproteins (HDL) are the primary mediator of reverse cholesterol transport (2). Although a significant portion of the anti-atherosclerotic character of HDL has been ascribed to its anti-inflammatory, anti-oxidant, and anti-thrombotic capacity (for reviews see Refs. 3, 4), a significant portion of the anti-atherosclerotic nature of HDL is due to its ability to mediate mobilization of cholesterol from macrophages in the arterial wall (5). Apolipoprotein A-I (apoA-I)4 is the primary protein component of HDL (70% of total protein content) and imparts HDL with a majority of its biological activities.
During reverse cholesterol transport, apoA-I transitions from lipid-free protein to spherical HDL through a two-step process. First, lipid free/lipid-poor apoA-I acquires phospholipid and cholesterol from the ATP-binding cassette transporter A1 (ABCA1) (6, 7) and generates nascent HDL. Second, cholesterol on nascent HDL is esterified by lecithin:cholesterol acyltransferase to yield cholesteryl ester (8, 9) and mature spherical HDL. ApoA-I must be structurally adaptive to accommodate changes in HDL size and geometry as it transitions from one subclass to another. An understanding of apoA-I structure will provide insight into how the conformational dynamics of apoA-I enable the formation of distinct HDL subclasses and impart these subclasses with specific lipid and receptor/enzyme binding traits.
Of the forms of apoA-I studied, the structure of apoA-I on 9.6 nm reconstituted nascent HDL (rHDL) is perhaps the best understood. Several models describing the conformation of apoA-I on 9.6 nm rHDL have been suggested. Of these, the “double-belt” model is the most widely supported by the literature (10–18) wherein two apoA-I molecules circumscribe a circular lipid bilayer in an antiparallel alignment. Recently, Wu et al. (19, 20) have proposed the double superhelix model, based on small angle neutron scattering data, wherein apoA-I assumes a left-handed double helix, spiraling around an elliptical/rod-like lipid particle. Although compelling, this model remains to be validated by other methods.
Through our understanding of apoA-I on nascent HDL, the molecular details of the belt model have provided insight into how apoA-I mediates biological activity of HDL. For instance, a portion of helix 5 has been postulated to form a “hinge” domain and play a role in HDL particle remodeling upon lipid loading (21, 22). By electron paramagnetic resonance (EPR) spectroscopy analysis, we have determined the location of this “hinge domain” as a 12-amino acid-long segment centered on residue 139. Because this portion of apoA-I aligns with its counterpart in a paired apoA-I on HDL, we hypothesized this stretch of residues forms a pore-like structure, described in the “looped belt” model (14), wherein this pore provides lecithin:cholesterol acyltransferase access to cholesterol and the acyl chain of POPC. Recently, this has been supported by molecular dynamic simulation computational analysis, wherein Jones et al. (23) determined that this region could form an amphipathic presentation tunnel for the acyl chains of POPC. Interestingly, Jones et al. (23) observed that the sn-2 acyl chain of POPC, the preferred acyl chain that is transacylated to cholesterol by lecithin:cholesterol acyltransferase (24), can insert into the tunnel at a significantly higher frequency than the sn-1 acyl chain, further substantiating a functional role for this apoA-I conformational feature on HDL.
In contrast to the central domain of apoA-I, the structure of the N-terminal 43 amino acids of lipid-bound apoA-I remains incomplete. Models of apoA-I on nascent HDL commonly do not depict these residues. This is likely due to the fact that a majority of belt models were derived from the initial observation of Borhani et al. (11) who proposed the “belt” model based on x-ray crystallographic results from lipid-free Δ1–43 apoA-I, which lacks the first 43 amino acids of apoA-I but remains capable of producing rHDL up to 9.6 nm in diameter (25). Despite being overlooked, we believe the apoA-I N terminus plays a substantial role in stabilizing HDL and guiding the conformational transition of apoA-I between particle sizes (26). Indeed, the N-terminal amino acids are necessary for formation of larger rHDL complexes, and their absence leads to less stable HDL particles (25).
We have previously used EPR spectroscopy to describe the structural organization of the N-terminal domain of apoA-I in the lipid-free state. Here, we extend those studies, reporting on the structure of residues 6–98 on 9.6 nm rHDL and, at select sites, on 7.8 and 8.4 nm rHDL. Our analyses of 9.6 nm rHDL reveals a secondary structure composed of random coil and β-strand positioned between two α-helices. These secondary structure elements have been placed into a tertiary and quaternary context using chemical cross-link/mass spectroscopy (CCL/MS) inter-/intramolecular distance data from Davidson and co-workers (17, 28) and Thomas and co-workers (10, 27). Furthermore, we have identified N-terminal residues important in apoA-I structural rearrangement that occur in response to changes in HDL lipid cargo and particle size. These insights into the structure and dynamics of apoA-I advance our understanding of the role of apoA-I structural elements in HDL function.
EXPERIMENTAL PROCEDURES
Materials
Thio-specific nitroxide spin label (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate) was received as a kind gift from Dr. K. Hideg (University of Pecs, Hungary). POPC and cholesterol were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL).
Production of Recombinant and Spin-labeled ApoA-I Protein
Ninety three single Cys substitutions within apoA-I cDNA (S6C-Q98C) were created using either primer-directed PCR mutagenesis or by the megaprimer PCR method (29). The mutations were verified by dideoxy automated fluorescent sequencing. The proteins were expressed and purified as described previously (30). Proteins were (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate spin-labeled as described previously (31). In brief, 8 mg of protein was sequentially incubated with 100 μm Tris-(2-carboxyethyl)phosphine and 300 μm (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate spin label on a Ni2+-chelated HiTrap column (GE Healthcare) under denaturing conditions (3 m guanidine HCl), then extensively washed with PBS (20 mm phosphate, 500 mm NaCl), and eluted by imidazole. Protein purity (>95%) was confirmed by SDS-PAGE analysis.
HDL Reconstitution
rHDL were prepared by the deoxycholate method (32, 33). Dried POPC and free cholesterol were resuspended in TBS (8.2 mm Tris-HCl, 150 mm NaCl, 0.1 mm EDTA), pH 8.0, with 19 mm sodium deoxycholate. The mixture was vortexed and incubated at 37 °C until clarified. Spin-labeled apoA-I was added to the mixture and incubated for 1 h at 37 °C. Deoxycholate was removed by extensive dialysis against TBS, pH 8. Protein to lipid molar ratios (apoA-I/free cholesterol/POPC) were 1:4:80 for 9.6 nm rHDL and 1:2:30 for 8.4 and 7.8 nm rHDL. Residual lipid-free protein was removed from the preparation by KBr density gradient ultracentrifugation at 50,000 × g for 3 h in a Beckman Optima TLA 100.4 rotor. Homogeneous rHDL subclasses were isolated by size exclusion chromatography as described previously (34). The size and purity of lipidated discoidal complexes were confirmed by nondenaturing gradient gel electrophoresis (NDGGE) (see supplemental Fig. S1 for NDGGE of representative samples).
Electron Paramagnetic Resonance (EPR) Analysis
EPR measurements were carried out in a JEOL X-band spectrometer fitted with a loop-gap resonator (35, 36). Aliquots (5 μl) of purified rHDL (60 μm spin-labeled protein) were placed in sealed quartz capillaries and loaded in the resonator. Spectra were acquired at room temperature (20–22 °C) from a single 2-min scan over a field of 100 G at a microwave power of 2 milliwatts and a modulation amplitude optimized to the natural line width of the individual spectrum (0.5–1.5 G). Spectra obtained in the presence of a final concentration of 2% (w/v) SDS were double-integrated and normalized. Molecular accessibility of spin-labeled side chains to NiEDDA or CrOx (depending on experiment) and O2 was determined using successive power saturation scans as described previously (37). Π½ values (which also were used to calculate the contrast function (Φ)) were calculated using software provided by C. Altenbach.
Molecular Modeling
Accelrys discovery studio (Discovery Studio Client version 2.5.0.9164, Accelrys Software Inc., San Diego) was used to build a tertiary/quaternary context for the N-terminal 1–98 residues on 9.6 nm rHDL. An initial model was generated by aligning apoA-I monomers constructed from the continuous α-helical ring-shaped coordinates of residues 44–243 from Segrest et al. (38), wherein amino acids 44–243 were assigned a curved α-helical secondary structure with a 3.6-residue periodicity. This assignment is consistent with existing double-belt models for 9.6 nm rHDL apoA-I. The primary sequence of the N-terminal 43 amino acids was added to residues 44–243, and the secondary structure determined by this study applied to residues 6–98. Similar to the model of Bhat et al. (27), the 6–34 α-helical domain had the same curvature and aligned antiparallel to residues 99–243. Amino acids 1–5 were oriented randomly.
To account for the pattern of residue immobilization and solvent accessibility of helix 6-34 (both sides of the apolar face, see “Results”), residues 1–43 were folded back upon the remainder of the protein. The two apoA-I monomers were positioned according to the model of Bhat et al. (10, 27) and Martin et al. (14), wherein the two monomers align at position 132 (at a distance of 12.2 Å). According to the model of Bhat et al. (10, 27), the C termini were arranged at greater intermolecular distance (22.6 Å between positions 232 and 237 at the C termini), which is sufficient space to accommodate the interleaving of residues 1–43. The intermolecular distances for Lys-Lys residues within residues 99–243 of this initial model are consistent with the CCL/MS results of Bhat et al. (10, 27) and Silva et al. (17, 28) (Table 2).
TABLE 2.
Molecular modeling constrains and final distances (Å) of significant residues in the three folding scenarios explored and comparison with double-super helix model predicted distances
a Cross-links were observed by Thomas and co-workers (10, 27) and Davidson and co-workers (17, 28). Cα-lysine-(cross-linker)-Cα-lysine distances depend on the cross-linker used as follows: disuccinimidyl glutarate, 22.3 Å; bis(sulfosuccinimidyl) suberate, 26.0 Å; and dithiobis(succinimidyl propionate), 26.6 Å.
b Cα-lysine-(cross-linker)-Cα-lysine distances from our initial model were derived as described under “Experimental Procedures.” The positions of the corresponding residues (more than residue 50) were fixed during minimization and molecular dynamics of the N-terminal residues (1–49).
c Cα-lysine-(cross-linker)-Cα-lysine distances from our initial model (see under “Experimental Procedures“) were before minimization and molecular dynamics of the N-terminal residues (1–49).
d Imposed distance constraints were during minimization and molecular dynamics of the N-terminal residues (1–49). The distance values were selected based on the experimental cross-links observed by Thomas and co-workers (10, 27) and Davidson and co-workers (17, 28).
e Cα-lysine-(cross-linker)-Cα-lysine final distances after minimization and molecular dynamics of the N-terminal residues (1–49) are shown.
f Averages ± S.D. of two intermolecular distance values for the two monomers of apoA-I are reported.
g The two numbers are distances between the indicated positions within monomer A (intramolecular) or between A–B (intermolecular) and within monomer B (intramolecular) or between B–A (intermolecular), respectively.
In the context of three different constraints scenarios for the N-terminal Lys-Lys distances (see under “Results”), we applied a variation of CHARMm forcefield (39, 40). We executed a total of three energy minimization protocols. In two protocols, the steepest descent method was applied, with maximum number of steps 10,000 and 200,000, respectively; root mean square gradient was 0.001 and energy change was 0.0 kT/e. In the third protocol, the conjugate gradient method was used, and maximum number of steps was 200,000, and root mean square gradient 0.001 and energy change were 0.0 kT/e. Heating and equilibration energy minimization protocols were further applied to the minimized initial model (on average 15% energy decrease of the initial conformation in kcal/mol), until the average temperature and structure remained stable. Both of these stages were executed with 200,000 steps of 0.001 ps each and a target temperature of 310 K. Within the imposed time scales of 200 ps, we can predict bond stretching and interdomain bending but not globular protein tumbling. To determine whether significant globular tumbling occurs at the N terminus, molecular dynamics were also simulated for 2 ns.
RESULTS
Site-directed spin labeling of apoA-I followed by EPR spectroscopy was used to analyze the structure of the N-terminal domain of apoA-I on rHDL. A series of apoA-I substitution variants were created, wherein residues 6–98 (inclusive) were individually cysteine-substituted and labeled with a thiol-specific paramagnetic nitroxide spin label (Fig. 1A). The 93 spin-labeled apoA-I variants were converted to 9.6 nm rHDL, prepared by combining spin-labeled proteins with POPC lipids and cholesterol at a protein to lipid molar ratio of 1:4:80 (apoA-I/free cholesterol/POPC), as described under “Experimental Procedures.” rHDL particles with a diameter of 9.6 nm were isolated by size exclusion chromatography purification, as described previously (34). The EPR spectra obtained for all positions (residues 6–98) are shown in supplemental Fig. S2.
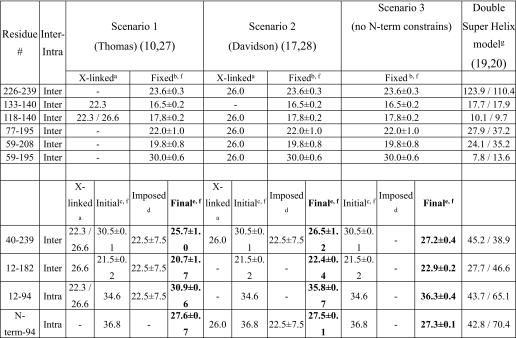
FIGURE 1.
EPR analysis of apoA-I N-terminal 6–98 residues on 9.6 nm rHDL. A, nitroxide spin-probe covalently bound to the thiol group of apoA-I variants containing a single cysteine residue (left panel). The EPR spectra obtained from the apoA-I variants (see supplemental Fig. S2 for the entire data set) describe the local environment of the individually spin-labeled residues. Side chains that exhibit a high degree of immobilization display broadening of the spectra (exemplified by K23C, middle panel), whereas sharper spectral shapes (e.g. D24C, right panel) indicate high motional freedom of the spin-labeled side chains. The central line width (indicated by arrows in middle and right panels) is inversely related to the degree of side-chain mobility. B, inverse central line width (δ−1; G−1) values are plotted as a function of residue number. Green sinusoids indicate regions that display a periodicity which correspond to an α-helical secondary structure. Dashed lines indicate regions where the helical structure pattern is less obvious in the mobility score analysis. For comparison, gray boxes indicate previously proposed helical regions (57).
Steric Environment of ApoA-I N-terminal Residue Side Chains on 9.6 nm rHDL
The extent of inhomogeneous broadening of the X-band EPR spectrum is highly dependent upon the motional freedom of the spin label, and as a result, the spectral characteristics of each spin-labeled side chain provide direct information on the level of structural order at each targeted site (see Fig. 1A for representative spectra of labeled side chains with low and high mobility, respectively). When a series of sites is considered, the periodicity of change in side chain mobility over consecutive spin-labeled residues can be used to identify the position of secondary structure elements in a protein.
We employed the mobility parameter, δ−1 (41), to provide a model-independent evaluation of the side chain mobility at individual sites. δ−1 is derived from the inverse of the central line width (Fig. 1A, arrows) of the spectra. By examining δ−1 magnitude as a function of residue number, we identified periodicities that reflect the presence of secondary structural elements. The spectral characteristics of residues 6–98 vary from highly mobile to immobilized side chains, exhibited by narrow or broad spectral line shapes, respectively. Of the 93 residues, 43 have δ−1 >0.27, with few very highly mobile residues (δ−1 ≥0.38) as follows: 13, 24, 35, 62 and 95; 49 residues have δ−1 ≤0.27, with few very highly immobilized residues (δ−1 ≤0.22) as follows: 57, 71, 74, 75, 79, and 86. Analysis of δ−1 magnitude periodicity patterns defines several helical regions of the apoA-I primary sequence (Fig. 1B). However, there are regions (residues 6–9, 15–18 and 37–49, see Fig. 1B, dashed lines) that display no apparent pattern, indicative of a loop or random coil. Similarly, at positions 69 and 81, there is a clear deviation from the predicted periodic pattern. These positions may therefore represent bending or disruption of the backbone fold, within the otherwise helical region at residues 50–98.
In addition to the secondary structure assignment, the mobility data reveal clusters of highly sterically hindered side chains (δ−1 ≤0.27) at positions 16–23, 56–65, and 86–93. In particular, all intervening residues between positions 16 and 23 have δ−1 ≤0.27. This region is also characterized by a condensed range of mobility (Δδ−1 ≤0.05). This continuous restriction in mobility may arise from protein-lipid contact, or from intra- or intermolecular protein-protein contact (or a combination thereof).
Molecular Accessibility of ApoA-I N-terminal Residue Side Chains on 9.6 nm rHDL
Molecular accessibility analyses were performed employing the diffusible relaxation agents O2 and CrOx, to correlate patterns of side chain mobility with differences in molecular accessibility. In addition to mapping the secondary structure distribution in a protein, these measurements provide insight into the chemical environment surrounding each spin label (14, 31, 42). The relative polarity of the environment surrounding the spin-labeled side chain can be assessed by the different solubility preferences of the two relaxers (hydrophobic for O2 and hydrophilic for CrOx). This determination is especially useful for the analysis of proteins that interact with lipids, as the logarithmic ratio of the accessibility parameters (Π) for the two relaxers provides a polarity index or contrast function (Φ), which is dependent upon the membrane penetration depth of the spin label (43, 44). Results from ΠCrOx and Φ molecular accessibility analyses are presented in Fig. 2 and supplemental Table S1.
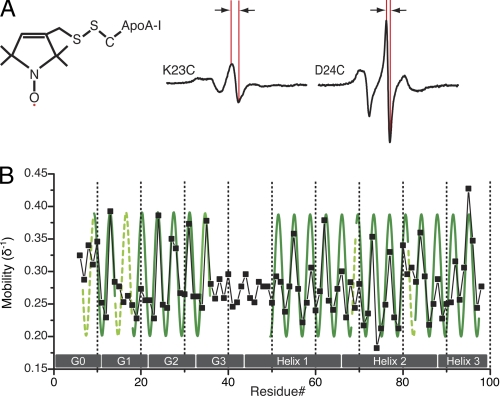
FIGURE 2.
Molecular accessibility analysis of apoA-I N-terminal 6–98 residues on 9.6 nm rHDL. The accessibility parameter data for the hydrophilic relaxer (ΠCrOx) and the contrast value (Φ) were jointly used to evaluate the secondary structure (A and B, respectively). A periodicity of 3.67 residues per turn (36) was used to identify the regions displaying the α-helical character, which are indicated by blue and orange sinusoids in A and B, respectively. A periodicity of 2 was used to identify β-strand structure. C, linear representation of the resulting secondary structure model, α-helical structure (α), is shown in green, random coil (rc) in yellow, and β-strand structure (β) in red. As a reference, gray boxes (G0–3 and helix 1–3) indicate the location of previously suggested helical structures (57).
The pattern of polar accessibility (ΠCrOx, Fig. 2A) and contrast function amplitudes (Φ, Fig. 2B) identifies two major regions of contiguous α-helical structure, which encompasses residues 6–34 and 50–98, respectively (Fig. 2C). Helical wheel projections of the two α-helices illustrate the high degree of correlation between the distribution of hydrophobic/hydrophilic residues and the molecular accessibility patterns (Fig. 3).
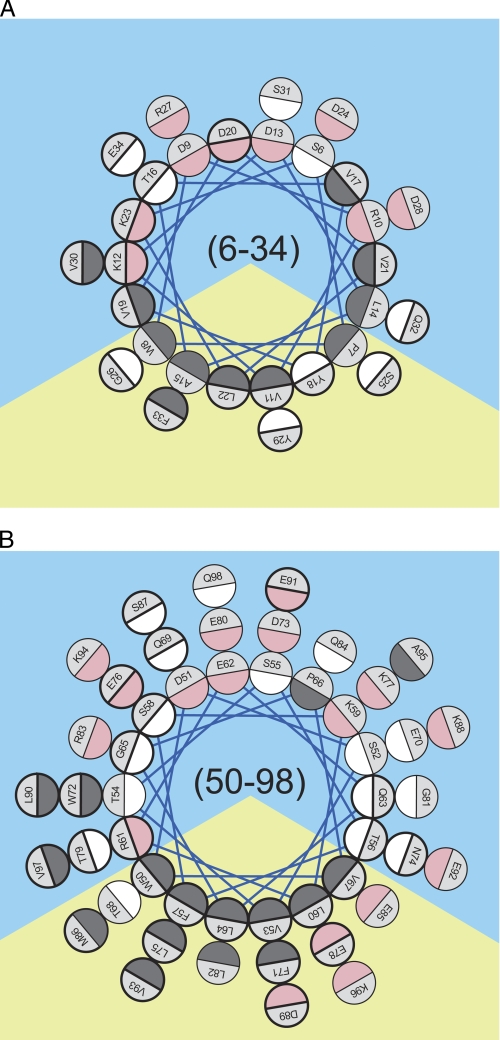
FIGURE 3.
Edmundson helical wheel analysis of residues 6–34 (A) and 50–98 (B). Helical projections were generated assuming a perfect α-helical periodicity of 3.6 residues per helical turn (58). The color code is dark gray for hydrophobic, white for polar and uncharged, and pink for charged residues. Prediction of the orientation of the amphipathic wheels was solely based on clustering of the hydrophobic residue on a sector of the wheel. The apolar and polar solvation space is represented by yellow and blue backgrounds, respectively. Residues that display a spectral characteristic corresponding to low motional freedom (δ−1≤ 0.27) (supplemental Table S1) are circled with bold lines.
Notably, comparison of side-chain molecular accessibility and mobility reveal that the vast majority of the sterically restricted side chains in helix 50–98 are localized on the hydrophobic face of the amphipathic α-helix. Therefore, in this region of apoA-I, reductions in side chain mobility are likely due to protein-lipid contact. In contrast, highly immobilized positions in helix 6-34 are evenly distributed along the helical axis between residues 16 and 23 (Fig. 3), independent of hydrophobicity of the side chain environment. This difference in correlation between mobility and hydrophobicity suggests that the immobilization of residues 16–23 is likely due to protein-lipid and protein-protein contacts (see “Discussion”).
Both molecular accessibility and side chain mobility analyses reveal that a nonhelical segment (residues 35–49) lies between the two helices. In this inter-helical region, a stretch of 10 amino acids (residues 40–49) exhibits a pattern of molecular accessibility consistent with a β-strand (alternating periodicity), whereas the contrast function value of the preceding residues (35–39) bears a more random pattern. As a result, we assign the latter a random coil structure although interactions with other residues and/or a twisting or bending of a β-strand could also produce a random pattern of solvent accessibility. This complex pattern therefore limits us from excluding the possibility that residues 35–39 form a β-strand. Moreover, molecular accessibility to the hydrophilic relaxer, CrOx, is low for the labeled side chains in this region, with an accessibility (ΠCrOx) value of 0.4 or below. This finding indicates that residues 35–49 are either in direct protein-lipid contact or buried within a protein-protein contact surface. The low accessibility of residues 35–49 to aqueous relaxer is consistent except for the side chain at residues 41 and 43, which display fairly high ΠCrOx values of 0.70 and 0.77, respectively. The positioning of these residues in hydrophilic environments is supported by the low contrast function values observed at these positions (Φ approximately −1.7), which also takes O2 accessibility into account.
Molecular Modeling of the N-terminal 1–98-Residue Domain on 9.6 nm rHDL
The tertiary/quaternary folding of the N-terminal 1–98 residues was tested in three scenarios of energy minimization followed by molecular dynamics simulation. The primary objective of this study was to determine whether residues 1–43 were amenable to the interleaving between apoA-I molecules. All scenarios shared the same initial model, as described under “Experimental Procedures,” wherein amino acids 50–243 matched intermolecular constraints reported by both Davidson and co-workers (17, 28) and Thomas and co-workers (10, 27) (Table 2), along with EPR-based intermolecular alignment data (14). The tertiary and quaternary conformation of residues 1–40 was configured to account for the distribution of side chain immobility and the CCL/MS-based molecular distances observed by Davidson and co-workers (17, 28) and Thomas and co-workers (10, 27), wherein the lengths of the chemical cross-linkers employed were taken into account.
By combining cross-linker length (disuccinimidyl glutarate 7.7 Å, dithiobis(succinimidyl propionate) 12.0 Å (27), and bis(sulfosuccinimidyl) suberate 11.4 Å (10)) and lysine side chain length, we determined the maximum distance of Cα-lysine-(cross-linker)-Cα-lysine as follows: disuccinimidyl glutarate 22.3 Å, dithiobis(succinimidyl propionate) 26.6 Å, and bis(sulfosuccinimidyl) suberate 26.0 Å. Because disuccinimidyl glutarate, dithiobis(succinimidyl propionate), and bis(sulfosuccinimidyl) suberate all yielded cross-links, we chose to impose distance constraints of 22.5 ± 7.5 Å for the Cα-lysine-(cross-linker)-Cα-lysine cross-linked distances.
In the first scenario, the N-terminal intermolecular Lys40–Lys239 and Lys12–Lys182 and the intramolecular Lys12–Lys94 distance constraints were imposed in accordance with the model of Thomas and co-workers (27). In the second scenario, only intermolecular Lys40–Lys239 and intramolecular N-terminal amino group-Lys94 distance constraints were imposed, corresponding with observations made by the Davidson and co-workers (17). In the third scenario, all tertiary constraints N-terminal to residue 40 were omitted. Molecular dynamics was simulated for 200 ps. Scenario 1 and scenario 2 were also simulated up to 2 ns (2 × 106 time steps of 0.001 ps), wherein protein domain tumbling can be calculated. No significant differences were observed between the resulting structures from the two simulation times.
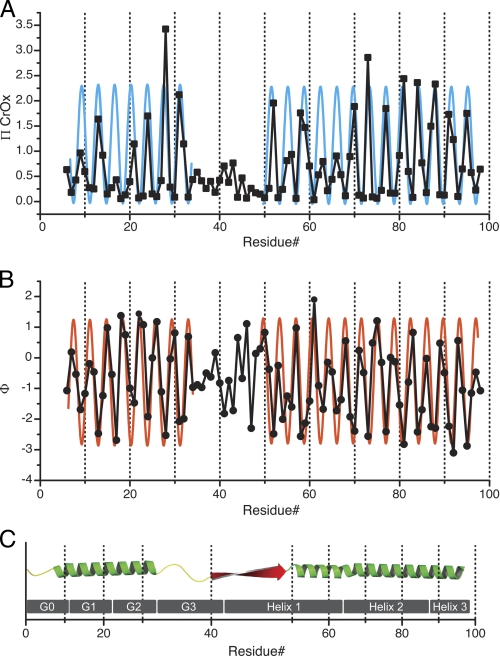
Energy minimization and molecular dynamics simulation of the three scenarios converged to yield similar structures (Table 2). In all cases, residues 1–43 maintained their position and overall secondary structure assignment. The tumble rate for this region did not exceed that of the whole protein suggesting that this region maintained its relative position. These computational results support the assertion that interleaving residues 1–43 between apoA-I molecules (Fig. 4) is a stable conformation.
FIGURE 4.
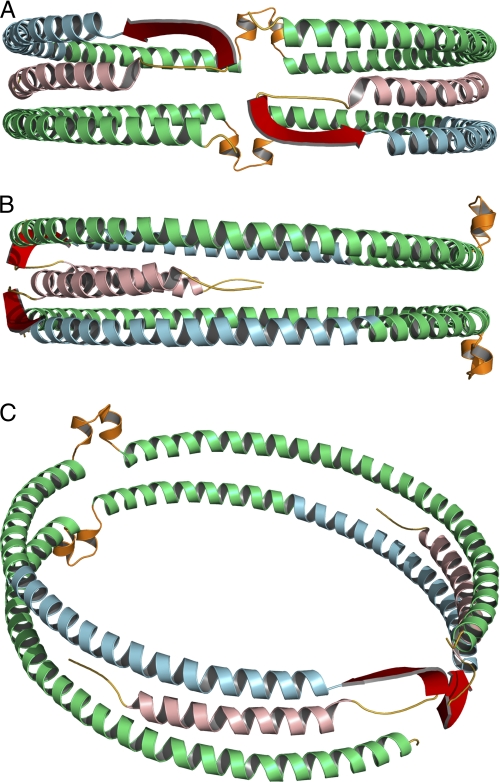
Full-length model of apoA-I on 9.6 nm rHDL. Green, residues derived from Segrest et al. (38) model (99–243). Secondary structure domains derived from the current EPR analysis are color-coded as follows: yellow, unstructured residues 1–5 and 35–39; light red, helical residues 6–34; blue, helical residues 50–98; red, β-strand residues 40–49. The loop (orange) of the looped-belt model is at position 133–146 (14). A, front view, front of the protein complex is arbitrarily designated as the N-terminal region of the protein. B, side view, side of the protein complex is arbitrarily designated as a position parallel to the lipid bilayer of the rHDL disc and 90° relative to the N terminus. C, oblique view.
Adaptation of Conformation of ApoA-I N Terminus to Changes in rHDL Size
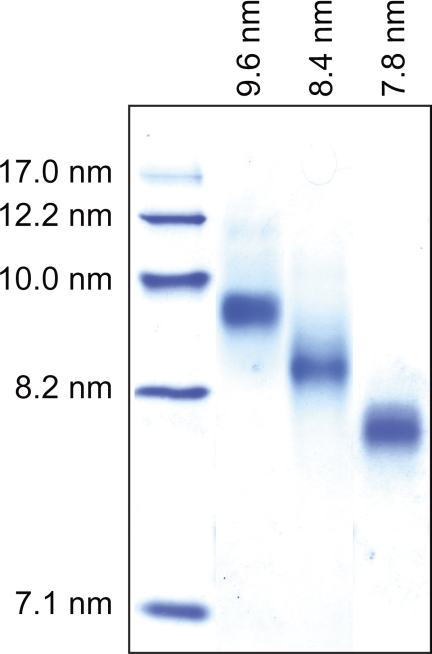
In addition to our analysis of the apoA-I N-terminal domain structure on 9.6 nm rHDL, we investigated whether sites within the N-terminal domain were sensitive to variations in particle size. Spin labels were positioned at residues 14, 19, 34, 37, 41, and 58 and incorporated into 7.8, 8.4, and 9.6 nm apoA-I rHDL to examine the effect of particle size on the conformation of apoA-I at these residues. These positions are representative of each of the identified secondary structural domain. Residue 34 lies at the interface between helix 6-34 and random coil (residues 35–39). Residue 37 is positioned in the random coil segment. Residue 41 is in the β-strand at residues 40–49. Residues 14 and 19 are in helix 6-34. The high degree of immobilization of these two residues (δ−1 of 0.28 and 0.23, respectively) suggests they are sensitive to structural changes. Similarly, the highly immobilized residue 58 (δ−1 = 0.25) was selected to probe changes occurring at the N-terminal portion of helix 50–98. The purity of isolated rHDL particles (7.8, 8.4, and 9.6 nm) for all apoA-I variants was confirmed by NDGGE (see Fig. 5, for a representative gel).
FIGURE 5.
rHDL particles of defined sizes. Representative NDGGE gel illustrating homogeneous populations of 7.8, 8.4, and 9.6 nm rHDL containing the spin-labeled V19C apoA-I variant. Similar levels of purity were obtained for all rHDL samples used throughout this study.
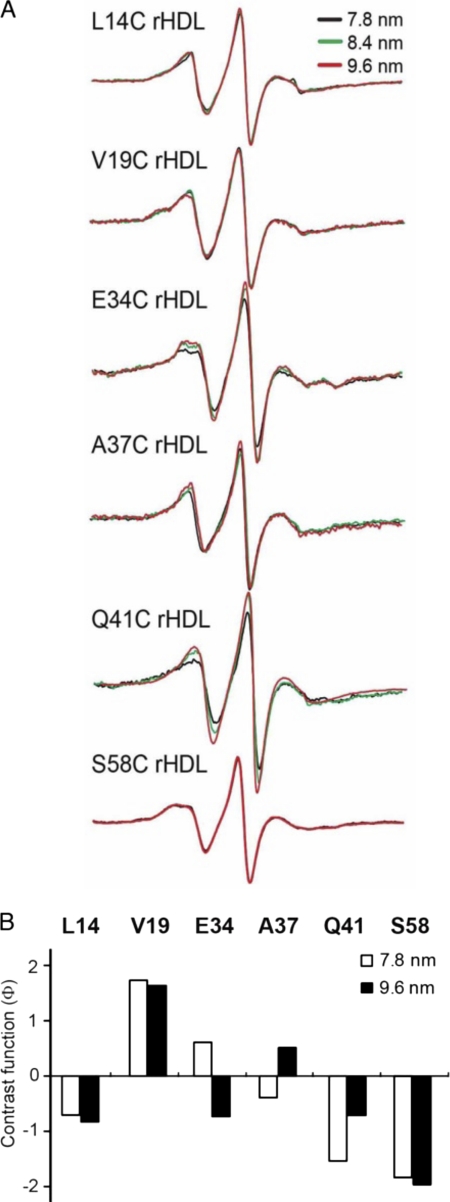
A comparison of EPR spectra from rHDL subclasses is shown in Fig. 6A. Only positions 34 and 41 display significant differences in their EPR line shapes relative to particle size (Fig. 6A). Changes in line shape for position 41 were the most prominent, narrowing as the rHDL expands from 7.8 to 9.6 nm (Fig. 6A), indicative of a less-restricted side chain in the larger particle. Molecular accessibility for these residues on different sized particles was also determined (Fig. 6B). Because the CrOx relaxer carries a net negative charge, it has negligible partitioning around aliphatic chains and provides excellent contrast for scanning positions along a lipid-associated protein. However, alterations in local electrostatics may also bias the CrOx collision frequency. Therefore, to ascertain conformational changes in the surroundings of the reporting residues, we evaluated the polarity at positions 14, 19, 34, 37, 41, and 58 on 7.8 and 9.6 nm rHDL by using a combination of the hydrophilic relaxer, CrOx, and the uncharged polar relaxer, NiEDDA, that is unaffected by local electrostatic variability bias. The molecular accessibility of position 41 demonstrated that this residue was one of the sites most affected by rHDL particle size. The primary difference was an increase in O2 accessibility with increasing particle size, whereas the collision frequency with the polar NiEDDA remained unchanged. Thus, the contrast function (Φ) indicates that position 41 enters a less polar environment as rHDL particle diameter increases from 7.8 to 9.6 nm (Fig. 6B; Table 1). Similarly, the polarity around residue 37 side chain decreased with increasing particle size. The changes in hydrophobicity of the environment at positions 37 and 41 relative to particle size is consistent with a progressive exposure of these two residues to phospholipid acyl chains as particle diameter increases.
FIGURE 6.
Adaptation of the N-terminal domain of apoA-I to different rHLD sizes. Single spin-labeled apoA-I in rHDL particles of defined sizes (7.8, 8.4, and 9.6 nm diameter) were analyzed by EPR spectroscopy. Spin-labeled positions were chosen to represent regions of random coil (residues 34 and 37) and β-strand (residue 41) structure, as well as two highly immobilized residues (14 and 19) in the hydrophobic/hydrophilic interface of helix 6-34 and a highly immobilized residue (58) in helix 50-98. A, EPR spectra of spin-labeled proteins on different rHDL sizes. B, bars represent the contrast value (Φ) (Table 1) for the different rHDL sizes. A low, negative value indicates a polar environment, and a positive value represents a hydrophobic milieu.
TABLE 1.
Molecular accessibility analysis of spin-labeled apoA-I variants in rHDL of different sizes
| Residue | rHDL size | Фa | ΠO2 | ΠNiEDDA | δ−1 |
|---|---|---|---|---|---|
| nm | |||||
| 14 | 7.8 | −0.71 | 0.14 | 0.29 | 0.30 |
| 14 | 9.6 | −0.82 | 0.19 | 0.43 | 0.28 |
| 19 | 7.8 | 1.73 | 0.19 | 0.03 | 0.25 |
| 19 | 9.6 | 1.63 | 0.23 | 0.05 | 0.25 |
| 34 | 7.8 | 0.62 | 0.17 | 0.09 | 0.24 |
| 34 | 9.6 | −0.72 | 0.07 | 0.14 | 0.24 |
| 37 | 7.8 | −0.39 | 0.15 | 0.22 | 0.33 |
| 37 | 9.6 | 0.52 | 0.21 | 0.13 | 0.34 |
| 41 | 7.8 | −1.53 | 0.11 | 0.50 | 0.27 |
| 41 | 9.6 | −0.68 | 0.25 | 0.50 | 0.25 |
| 58 | 7.8 | −1.81 | 0.13 | 0.81 | 0.26 |
| 58 | 9.6 | −1.97 | 0.14 | 0.99 | 0.26 |
a The contrast function (Φ), a measure of the hydrophobicity of the immediate environment to the spin-labeled side chain, was calculated from the polar (ΠNiEDDA) and nonpolar (ΠO2) accessibility parameters. The empirically determined Φ value ranges from −1.5 (hydrophilic) to 4 (hydrophobic) (41).
In contrast to residues 37 and 41, the environment of residue 34 changed from hydrophobic to hydrophilic, as indicated by both a decreased contrast function (Φ) value and a decreased accessibility by O2 (ΠO2 in Table 1), with increasing particle diameter. The changes observed at position 34 are consistent with a reduction in protein-protein contacts and a partitioning of the Glu side chain to solvent on 9.6 nm rHDL. We hypothesize that residue 34 is located within a hinge that responds to increases in rHDL size by rotating away from a protein-protein contact, facilitating the phospholipid bilayer association of upstream positions. Molecular dynamics simulations of the apoA-I N terminus on 9.6 nm rHDL indicate that the random coil region (35–39) is extremely flexible upon heating and equilibration. Thus, we hypothesize that upon expansion of the phospholipid bilayer, conformational changes in the random coil facilitate the remodeling of upstream α-helical residues and changes in their protein-protein and protein-phospholipid interactions.
Whereas the non-α-helical residues 34, 3, and 41 exhibited marked changes in molecular accessibility, residues 14 and 19 in the 6-34 α-helical domain and residue 58 in the 50-98 α-helical domain showed no significant changes relative to rHDL particle size. For the rHDL size range analyzed, the α-helical residues (14, 19, and 58) are not apparently affected by particle size.
DISCUSSION
Structural studies of apoA-I are hampered by the conformational plasticity of this protein, which is especially manifested in the heterogeneity of HDL particle sizes. To overcome this obstacle and enable studies of apoA-I in solution, various biophysical approaches have been employed. EPR spectroscopy combined with site-directed spin labeling is one such approach that has proven particularly useful. We employed EPR spectroscopy to analyze the structural features of the N-terminal domain of apoA-I (residue 6–98) on nascent HDLs of different size.
The N-terminal domain of apoA-I is important for stabilizing its lipid-free structure. Stability was originally thought to be due to the contribution of the N-terminal conformation to the overall protein fold (45, 46). We previously proposed that the stabilizing function of the N terminus is mediated by direct contact between β-strands found in the N-terminal (42) and C-terminal (31) domains. This direct contact between the N- and C-terminal domains (31, 47–49) is also thought to modulate the lipid-binding properties of the N terminus (50, 51). Moreover, although naturally occurring mutations in the central domain of human apoA-I are typically associated with low plasma HDL concentrations and abolished lecithin:cholesterol acyltransferase activation, mutations in the N-terminal domain have a preponderance of amyloidogenic activity (for a review see Ref. 52). The apoA-I found in amyloid plaques adopt a β-strand secondary structure that promotes stacking and fibril formation. Little is known of the molecular details behind what initiates and promotes amyloidogenic apoA-I fibril formation, but a β-strand formation-mediated mechanism has been previously described by our laboratories for the amyloidogenic “Iowa” (G26R) variant of apoA-I (53). Overall, findings from wild-type apoA-I and variants demonstrate the critical role the N-terminal domain plays in apoA-I stability and response to lipid environment. However, our understanding of the conformation of the N-terminal domain of apoA-I on 9.6 nm nascent HDL is limited (10, 17, 27, 28).
Two distinct but corroborative pieces of information can be derived from EPR spectroscopy; the first is molecular accessibility, and the second is side chain mobility. The former provides a measure of the polarity of the environment surrounding the labeled site, and the latter provides a measure of the degree of contact at the site of labeling. Combined, these data give an indication of how much molecular contact exists at a particular site and insight into whether the contact is protein-protein or protein-lipid. Through EPR spectroscopic analysis, we observed two α-helical domains at positions 6–34 and 50–98 (Fig. 2).
Molecular accessibility analysis of helices 6-34 and 50-98 reveals that solvent accessibility of residues for both helices is reduced along their apolar helical face. This change in solvent accessibility is commonly found when amphipathic α-helices are associated with lipid bilayers. As a result, we concluded that both helical segments make protein-lipid contact along their apolar face.
Mobility analysis of residues in both helices indicated that interactions for helix 50-98 are different from that observed for helix 6-34. In helix 50-98, immobilized side chains cluster along the apolar face of helix 50-98. In contrast, the immobilized residues in helix 6-34 distribute to all angular directions (specifically residues 16–23) (Fig. 3), indicative of a more complex structural organization. This complex pattern of molecular accessibility and residue immobilization is likely the result of lipid association (along the apolar face) in addition to hindrances arising from protein-protein contacts on either side of the apolar face. This observation is consistent with the position of residues 1–43 proposed by Bhat et al. (10, 27), which suggests that both protein-lipid and protein-protein interactions maintain the position of residues 6–34 on the edge of the bilayer.
We turned to CCL/MS data to arrange the N-terminal secondary structures into a tertiary/quaternary context. Two possible tertiary contexts presented themselves initially, the traditional double-belt model and the more recently proposed double superhelix model. Remarkably, despite the clear differences in the overall shapes of the double superhelix (19) and the double-belt model, the intermolecular Lys-Lys distances for the central domain of apoA-I (133–140, 118–140, 77–195, 59–208, and 59–195) are similar (Table 2). However, inter- and intramolecular Lys-Lys distances at the C and N termini (226–239, 40–239, 12,182, 12,94, N-terminal 94) of the double superhelix model do not correspond to CCL/MS results of the double-belt models (Table 2). Thus, our initial model for residues 1–98 was based upon the double-belt model, inspired by the “belt buckle” model (see Thomas and co-workers (10, 27)), wherein residues 1–43 are folded back upon the rest of the protein. This conformation positions residues 1–43 proximal to residues 50–90 of the same apoA-I and to the C terminus (residues 170–243) of the paired apoA-I. This orientation is based upon CCL/MS Lys40–Lys239 and Lys12–Lys182 intermolecular and the Lys12–Lys94 intramolecular cross-links. But unlike the belt buckle model, which positions residues 1–43 off from the rHDL disc edge, we interleaved helix 6-34 between the two antiparallel helices (50–98 and 243–170). This arrangement is consistent with protein-protein and protein-lipid contact observed by EPR (Fig. 4) in addition to CCL/MS data.
Energy minimization and molecular dynamics simulation analysis of three folding scenarios for the 1–98 N-terminal domains on 9.6 nm rHDL (Table 2 and Fig. 4) confirmed that interleaving the N-terminal short helix (6-34) between domain 50–98 of the same molecule and C-terminal domain 170–243 of the paired apoA-I is not catastrophically unfavorable. The absence of lipids in our simulation eliminates the contribution of protein-lipid interactions, one of the main driving forces for apolipoprotein stabilization on lipoprotein particles. Despite this limitation, our results confirm that this arrangement of residues 6–34 is not energetically prohibited, and three helices can align to approximately the thickness of a phospholipid bilayer ∼35 Å (54). Interestingly, the helix 6-34 maintains its position even in the absence of N-terminal constraints (scenario 3), further suggesting that this may be the proper position for this region of apoA-I. We acknowledge that although supportive of our conclusion, molecular dynamics simulation, as employed here, is not definitive proof.
Both random coil and β-strand are present at residues 35–49 on 9.6 nm rHDL. The presence of these elements suggests a functional role within the HDL particle. We hypothesized that these non-α-helical elements may participate in the ability of apoA-I to adapt to changes in HDL particle diameter and shape. EPR analysis reveals that the non-α-helical segment (residues 35–49) undergoes significant conformational adaptation to accommodate changes in rHDL lipid content. Noteworthy is that in previous EPR analysis we determined this region is α-helical in lipid-free apoA-I (42). Although a majority of apoA-I increases in helical content upon nascent HDL particle formation, residues 35–49 transition from α-helical to non-α-helical. Moreover, residues 34, 37, and 41 on 7.8 and 9.6 nm diameter rHDL showed polarity changes suggesting a region of variable lipid association. In contrast, residues 14, 19 in helix 6-34, and 58 in helix 50-98 were not affected by particle size changes. These results suggest that the α-helical domains identified in the 9.6 nm rHDL are present on smaller rHDL particles, although more thorough analysis is required to confirm this conclusion. Recently, mutational analysis of Leu residues (33, 42, 46, and 47) indicated that this flexible and partially unstructured region is important in lipid binding (55). However, lipid-free apoA-I variants maintained wild type levels of ABCA1-mediated cholesterol efflux activity, suggesting that this region is specifically involved in nascent HDL particle expansion by phospholipid accrual, rather than in the initial lipidation of the lipid-free molecule.
Furthermore, the percentage of α-helicity for residues 1–98 is 80% (assuming residues 1–5 are non-α-helical). These values fall within the expected range of apoA-I α-helicity on 9.6 nm rHDL as measured by circular dichroism spectroscopy (78–82%) (28, 56). This suggests that ∼30 residues of the remainder of apoA-I (20% of residues 99–243) are non-α-helical. We hypothesize that these residues are highly mobile non-lipid-associated residues and, as we had observed at the N terminus, likely to be adaptive to increases in rHDL particle diameter and HDL cargo composition. These portions of apoA-I may play additional roles in HDL function and thus are important to identify.
Through EPR analyses of the N-terminal domain of apoA-I on nascent HDL, we were able to define the secondary structure of the rHDL-associated apoA-I N-terminal residues (6–98) and position these structural elements in a tertiary/quaternary context. Analysis of apoA-I in different rHDL subclasses revealed that the random coil and β-strand structures identified are involved in the structural response to HDL size changes. Because of the positioning of this portion of apoA-I near a partially exposed region of phospholipid acyl chain, it is tempting to hypothesize that the N terminus in conjunction with the C terminus serves to modulate the degree of phospholipid acyl chain exposure and thus the rate of HDL remodeling and particle stability. Further analysis of the apoA-I structure and protein-protein interaction in this region will be required to elucidate the role the apoA-I N terminus plays in HDL stabilization and particle size transition.
Supplementary Material
Acknowledgments
We thank Nicole C. DeValle for technical assistance. We thank Michael J. Thomas, Mary G. Sorci-Thomas, and Jere P. Segrest for providing their models of apoA-I on discoidal rHDL.
This work was supported, in whole or in part, by National Institutes of Health Grants HL77268 and HL78615. This work was also supported by grants from the Swedish Research Council, the Petrus and Augusta Hedlund Foundation, the Crafoord Foundation, the Carl Trygger Foundation for Scientific Research, and American Heart Association Scientist Development Grant 0235222N.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- apoA-I
- apolipoprotein A-I
- CCL/MS
- cross-linking coupled with mass-spectrometry
- CrOx
- chromium oxalate
- NiEDDA
- nickel ethylenediaminediacetic acid
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-phosphocholine
- NDGGE
- nondenaturing gradient gel electrophoresis
- rHDL
- reconstituted HDL.
REFERENCES
- 1. Glomset J. A. (1968) J. Lipid Res. 9, 155–167 [PubMed] [Google Scholar]
- 2. Miller G. J., Miller N. E. (1975) Lancet 1, 16–19 [DOI] [PubMed] [Google Scholar]
- 3. Sviridov D., Mukhamedova N., Remaley A. T., Chin-Dusting J., Nestel P. (2008) J. Atheroscler. Thromb. 15, 52–62 [DOI] [PubMed] [Google Scholar]
- 4. Tabet F., Rye K. A. (2009) Clin. Sci. 116, 87–98 [DOI] [PubMed] [Google Scholar]
- 5. Cuchel M., Rader D. J. (2006) Circulation 113, 2548–2555 [DOI] [PubMed] [Google Scholar]
- 6. Adorni M. P., Zimetti F., Billheimer J. T., Wang N., Rader D. J., Phillips M. C., Rothblat G. H. (2007) J. Lipid Res. 48, 2453–2462 [DOI] [PubMed] [Google Scholar]
- 7. Mulya A., Lee J. Y., Gebre A. K., Thomas M. J., Colvin P. L., Parks J. S. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 1828–1836 [DOI] [PubMed] [Google Scholar]
- 8. Fielding C. J., Shore V. G., Fielding P. E. (1972) Biochem. Biophys. Res. Commun. 46, 1493–1498 [DOI] [PubMed] [Google Scholar]
- 9. Glomset J. A. (1962) Biochim. Biophys. Acta 65, 128–135 [DOI] [PubMed] [Google Scholar]
- 10. Bhat S., Sorci-Thomas M. G., Alexander E. T., Samuel M. P., Thomas M. J. (2005) J. Biol. Chem. 280, 33015–33025 [DOI] [PubMed] [Google Scholar]
- 11. Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davidson W. S., Thompson T. B. (2007) J. Biol. Chem. 282, 22249–22253 [DOI] [PubMed] [Google Scholar]
- 13. Li H., Lyles D. S., Thomas M. J., Pan W., Sorci-Thomas M. G. (2000) J. Biol. Chem. 275, 37048–37054 [DOI] [PubMed] [Google Scholar]
- 14. Martin D. D., Budamagunta M. S., Ryan R. O., Voss J. C., Oda M. N. (2006) J. Biol. Chem. 281, 20418–20426 [DOI] [PubMed] [Google Scholar]
- 15. Panagotopulos S. E., Horace E. M., Maiorano J. N., Davidson W. S. (2001) J. Biol. Chem. 276, 42965–42970 [DOI] [PubMed] [Google Scholar]
- 16. Segrest J. P. (1977) Chem. Phys. Lipids 18, 7–22 [DOI] [PubMed] [Google Scholar]
- 17. Silva R. A., Hilliard G. M., Li L., Segrest J. P., Davidson W. S. (2005) Biochemistry 44, 8600–8607 [DOI] [PubMed] [Google Scholar]
- 18. Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., 3rd, Smith J. D., Gogonea V., Hazen S. L. (2007) Nat. Struct. Mol. Biol. 14, 861–868 [DOI] [PubMed] [Google Scholar]
- 19. Wu Z., Gogonea V., Lee X., Wagner M. A., Li X. M., Huang Y., Undurti A., May R. P., Haertlein M., Moulin M., Gutsche I., Zaccai G., Didonato J. A., Hazen S. L. (2009) J. Biol. Chem. 284, 36605–36619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gogonea V., Wu Z., Lee X., Pipich V., Li X. M., Ioffe A. I., Didonato J. A., Hazen S. L. Biochemistry 49, 7323–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Córsico B., Toledo J. D., Garda H. A. (2001) J. Biol. Chem. 276, 16978–16985 [DOI] [PubMed] [Google Scholar]
- 22. Davidson W. S., Hilliard G. M. (2003) J. Biol. Chem. 278, 27199–27207 [DOI] [PubMed] [Google Scholar]
- 23. Jones M. K., Catte A., Li L., Segrest J. P. (2009) Biochemistry 48, 11196–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Subbaiah P. V., Liu M., Paltauf F. (1994) Biochemistry 33, 13259–13266 [DOI] [PubMed] [Google Scholar]
- 25. Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., Segrest J. P. (2010) J. Biol. Chem. 285, 4652–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li L., Chen J., Mishra V. K., Kurtz J. A., Cao D., Klon A. E., Harvey S. C., Anantharamaiah G. M., Segrest J. P. (2004) J. Mol. Biol. 343, 1293–1311 [DOI] [PubMed] [Google Scholar]
- 27. Bhat S., Sorci-Thomas M. G., Tuladhar R., Samuel M. P., Thomas M. J. (2007) Biochemistry 46, 7811–7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kammann M., Laufs J., Schell J., Gronenborn B. (1989) Nucleic Acids Res. 17, 5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ryan R. O., Forte T. M., Oda M. N. (2003) Protein Expr. Purif. 27, 98–103 [DOI] [PubMed] [Google Scholar]
- 31. Oda M. N., Forte T. M., Ryan R. O., Voss J. C. (2003) Nat. Struct. Biol. 10, 455–460 [DOI] [PubMed] [Google Scholar]
- 32. Nichols A. V., Gong E. L., Blanche P. J., Forte T. M. (1983) Biochim. Biophys. Acta 750, 353–364 [DOI] [PubMed] [Google Scholar]
- 33. Nichols A. V., Gong E. L., Blanche P. J., Forte T. M., Shore V. G. (1987) J. Lipid Res. 28, 719–732 [PubMed] [Google Scholar]
- 34. Cavigiolio G., Shao B., Geier E. G., Ren G., Heinecke J. W., Oda M. N. (2008) Biochemistry 47, 4770–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Froncisz W., Hyde J. S. (1982) J. Magn. Reson. 47, 515–521 [Google Scholar]
- 36. Hubbell W. L., Froncisz W., Hyde J. S. (1987) Rev. Sci. Instrum. 58, 1879–1886 [Google Scholar]
- 37. Oh K. J., Altenbach C., Collier R. J., Hubbell W. L. (2000) Methods Mol. Biol. 145, 147–169 [DOI] [PubMed] [Google Scholar]
- 38. Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., Harvey S. C. (1999) J. Biol. Chem. 274, 31755–31758 [DOI] [PubMed] [Google Scholar]
- 39. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shao B., Pennathur S., Pagani I., Oda M. N., Witztum J. L., Oram J. F., Heinecke J. W. (2010) J. Biol. Chem. 285, 18473–18484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Columbus L., Kálai T., Jekö J., Hideg K., Hubbell W. L. (2001) Biochemistry 40, 3828–3846 [DOI] [PubMed] [Google Scholar]
- 42. Lagerstedt J. O., Budamagunta M. S., Oda M. N., Voss J. C. (2007) J. Biol. Chem. 282, 9143–9149 [DOI] [PubMed] [Google Scholar]
- 43. Altenbach C., Greenhalgh D. A., Khorana H. G., Hubbell W. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1667–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klug C. S., Su W., Feix J. B. (1997) Biochemistry 36, 13027–13033 [DOI] [PubMed] [Google Scholar]
- 45. Davidson W. S., Hazlett T., Mantulin W. W., Jonas A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13605–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rogers D. P., Roberts L. M., Lebowitz J., Datta G., Anantharamaiah G. M., Engler J. A., Brouillette C. G. (1998) Biochemistry 37, 11714–11725 [DOI] [PubMed] [Google Scholar]
- 47. Behling Agree A. K., Tricerri M. A., Arnvig McGuire K., Tian S. M., Jonas A. (2002) Biochim. Biophys. Acta 1594, 286–296 [DOI] [PubMed] [Google Scholar]
- 48. Fang Y., Gursky O., Atkinson D. (2003) Biochemistry 42, 6881–6890 [DOI] [PubMed] [Google Scholar]
- 49. Tricerri M. A., Behling Agree A. K., Sanchez S. A., Jonas A. (2000) Biochemistry 39, 14682–14691 [DOI] [PubMed] [Google Scholar]
- 50. Kono M., Tanaka T., Tanaka M., Vedhachalam C., Chetty P. S., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. (2010) J. Lipid Res. 51, 804–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koyama M., Tanaka M., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. (2009) Biochemistry 48, 2529–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sorci-Thomas M. G., Thomas M. J. (2002) Trends Cardiovasc. Med. 12, 121–128 [DOI] [PubMed] [Google Scholar]
- 53. Lagerstedt J. O., Cavigiolio G., Roberts L. M., Hong H. S., Jin L. W., Fitzgerald P. G., Oda M. N., Voss J. C. (2007) Biochemistry 46, 9693–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Catte A., Patterson J. C., Jones M. K., Jerome W. G., Bashtovyy D., Su Z., Gu F., Chen J., Aliste M. P., Harvey S. C., Li L., Weinstein G., Segrest J. P. (2006) Biophys. J. 90, 4345–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith L. E., Davidson W. S. (2010) Biochim. Biophys. Acta 1801, 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rogers D. P., Brouillette C. G., Engler J. A., Tendian S. W., Roberts L., Mishra V. K., Anantharamaiah G. M., Lund-Katz S., Phillips M. C., Ray M. J. (1997) Biochemistry 36, 288–300 [DOI] [PubMed] [Google Scholar]
- 57. Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. (1992) J. Lipid Res. 33, 141–166 [PubMed] [Google Scholar]
- 58. Schiffer M., Edmundson A. B. (1967) Biophys. J. 7, 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.