Abstract
Taspase1 is a threonine protease responsible for cleaving intracellular substrates. As such, (de)regulated Taspase1 function is expected not only to be vital for ordered development but may also be relevant for disease. However, the full repertoires of Taspase1 targets as well as the exact biochemical requirements for its efficient and substrate-specific cleavage are not yet resolved. Also, no cellular assays for this protease are currently available, hampering the exploitation of the (patho)biological relevance of Taspase1. Here, we developed highly efficient cell-based translocation biosensor assays to probe Taspase1 trans-cleavage in vivo. These modular sensors harbor variations of Taspase1 cleavage sites and localize to the cytoplasm. Expression of Taspase1 but not of inactive Taspase1 mutants or of unrelated proteases triggers proteolytic cleavage and nuclear accumulation of the biosensors. Employing our assay combined with scanning mutagenesis, we identified the sequence and spatial requirements for efficient Taspase1 processing in liquid and solid tumor cell lines. Collectively, our results defined an improved Taspase1 consensus recognition sequence, Q3(F/I/L/V)2D1↓G1′X2′D3′D4′, allowing the first genome-wide bioinformatic identification of the human Taspase1 degradome. Among the 27 most likely Taspase1 targets are cytoplasmic but also nuclear proteins, such as the upstream stimulatory factor 2 (USF2) or the nuclear RNA export factors 2/5 (NXF2/5). Cleavage site recognition and proteolytic processing of selected targets were verified in the context of the biosensor and for the full-length proteins. We provide novel mechanistic insights into the function and bona fide targets of Taspase1 allowing for a focused investigation of the (patho)biological relevance of this type 2 asparaginase.
Keywords: Anticancer Drug, Cancer Therapy, Carcinogenesis, Leukemia, Protease, MLL, Cellular Assay, Chemical Biology, Small Molecule Inhibitor, Virtual Screening
Introduction
By cleaving proteins, proteases are involved in the control of a large number of key physiological processes such as development, metabolism, tissue remodeling, cell proliferation, and apoptosis (1–3). Protease signaling therefore differs from the majority of other signaling pathways by being mostly irreversible (3). Protease signaling is strictly regulated, and the deregulation of protease activity can contribute to various pathologies, including cancer (3).
The human Taspase1 gene encodes a protein of 420 amino acids (aa),4 which is the proenzyme of Taspase1. It belongs to a family of enzymes possessing an asparaginase-2 homology domain. In the MEROPS database, Taspase1 is found as T02.004, classifying this protein as a class PB, subclass PB(T), and T2 family protease. In contrast to the other cis-active type 2 asparaginases, such as amidohydrolases, l-asparaginase, and glycosylasparaginase, only Taspase1 is able to cleave other substrates in trans (4). Therefore, Taspase1 represents a distinct class of proteolytic enzymes. Taspase1-mediated cleavage of proteins follows distinct aspartate residues, suggesting that Taspase1 evolved from hydrolyzing asparagines and glycosylasparagines to recognize a conserved peptide motif with an aspartate at the P1 position (4). Thus, the discovery of Taspase1 founded a new class of endopeptidases that utilize the N-terminal threonine of its mature β-subunit as the active site (4). The N-terminal threonine (Thr234) is generated by autoproteolysis of the Taspase1 proenzyme (referred to as cis-cleavage) into the two subunits α and β, which assemble into an asymmetric α2/β2-heterotetramer, representing the active protease (5). Mutation of the catalytic nucleophile, Thr234, significantly attenuates the catalytic activity of Taspase1 (4, 5). Interestingly, Khan et al. (5) reported that during Taspase1 crystallization, a chloride ion was found bound to amino acids Gly49, Gln100, and Thr234. This interaction was suggested to function as a reversible competitive inhibitor already at physiological relevant concentrations (IC50 = 26 nm). However, the detailed biochemical and structural requirements for Taspase1 cis- and/or trans-cleavage still remain to be resolved.
Taspase1 was first identified as the protease responsible for the cleavage of the mixed lineage leukemia (MLL) protein at conserved (Q3X2D1↓G1′) sites (4). Besides its relevance for proper Hox gene expression, MLL is found as a translocation partner in a variety of acute leukemias and hence is considered as a proto-oncogene (4, 6–9). Proteolytic cleavage of mature MLL is considered a crucial constituent of MLL-mediated tumorigenesis (6, 10–12). Recent data indicate also that other essential proteins, such as the precursor of the transcription factor IIA (TF2A) (13), are considered as Taspase1 targets. However, the molecular mechanisms of how Taspase1 affects TF2A or MLL protein function through site-specific proteolysis and what other (patho)physiological pathways are regulated in humans by Taspase1 remain to be determined.
Besides genetic instruments, chemical decoys allowing the targeted inhibition/activation of proteins are powerful tools to dissect complex biological pathways in general. Unfortunately, catalytic activity of Taspase1 is not affected by common protease inhibitors, and no small molecule inhibitors of this enzyme are currently available (4, 14). Consequently, we developed and employed here cell-based assays combined with bioinformatic approaches to provide novel insights into Taspase1 (patho)biological functions.
EXPERIMENTAL PROCEDURES
Antibodies (Ab) and Reagents
The Ab used are as follows: α-GFP (sc-8334; Santa Cruz Biotechnology, Heidelberg, Germany); α-Myc tag and α-ClevCasp3 (New England Biolabs); α-Taspase1 (AP1330b, directed against the C terminus; BioCat GmbH, Heidelberg, Germany); α-GAPDH (sc-47724), α-TF2A-β (sc-5315), and α-Casp3 (sc-7272) (Santa Cruz Biotechnology). Appropriate HRP-, Cy3-, or FITC-conjugated secondary antibodies (Sigma and Santa Cruz Biotechnology) and α-USF2 (sc-81645) (Santa Cruz Biotechnology) and α-NXF5 (sc-133860) (Santa Cruz Biotechnology) were used. Reagents were from Sigma unless stated otherwise.
Cell Culture, Microscopy, and Imaging
Cell lines used in the study (supplemental Table S2) were maintained and transfected as described previously (12, 15–17). Twelve-bit black and white images were captured using a digital Axiocam CCD camera (Carl Zeiss, Jena, Germany). Quantitation, image analysis, and presentation were performed as described previously (2, 18). The nuclear signal was similarly obtained by measuring the pixel intensity in the nucleus. Nuclei were marked by Hoechst 33258 staining as described previously (2, 19). To determine the average intracellular protein localization, at least 200 fluorescent cells from three separate images were examined in three independent experiments. The number of cells exhibiting cytoplasmic (cytoplasmic signal >75% of the total cellular signal), cytoplasmic and nuclear, or nuclear (nuclear signal >75% of the total cellular signal) fluorescence was counted.
Plasmids
To generate plasmid p_NLS-GFP/GST-CS2-NESRev (p_BioTasp), encoding a fusion composed of the SV40 large T-antigen NLS, GST, and GFP, the Taspase1 cleavage site from MLL (CS2; aa 2713KISQLDGVDD2722), and a Myc epitope-tagged NES from the HIV-1 Rev protein (NESRev) (20), the CS2 coding sequence was inserted into vector pNLS-GFP/GST-CS3-RevNES (p_BioCasp3), replacing CS3. p_BioCasp3 encodes a biosensor harboring the cleavage site for caspase3 (CS3: aa KRKGDEVDGVDE) (2). pCasp3 allows ectopic expression of caspase3 (2). p_BioTaspR encodes a red fluorescent biosensor (NLS-mCherry/GST-CS2-NESRev), in which GFP was replaced by mCherry (2). Expression plasmids for BioTasp variants, in which CS2 was mutated (p_BioTaspmut; see supplemental Table S2 and supplemental Table S1 for oligonucleotides used), were generated by oligonucleotide-annealing and cloning into the NotI/XhoI restriction sites of p_BioTasp as described previously (2).
Likewise, the coding sequences for full-length TF2A (p_BioTaspTF), its cleavage site (p_BioTaspTFCS), full-length USF2 (p_BioTaspUSF2), or the cleavage site from myosin1F (aa 152–268) (p_BioTaspMyo) or NXF5 (aa 626–692) (p_BioTaspNXF5) were inserted into p_BioTasp, thereby replacing the CS2. pTRE-NLS-GFP/GST-CS2-NESRev (p_BioTaspi) allows the inducible expression of the biosensor (“Tet-Off”) and was constructed by inserting the NLS-GFP/GST-CS2-NESRev coding sequence into pTRE-NLS-GFP/GST-NESRev (2).
The Taspase1, TF2A, or Myo1F coding sequence was amplified from cDNA obtained from a human head and neck tumor. mRNA preparation and cDNA synthesis from tumor tissue was performed as described previously (21). Cloning of the Taspase1 coding sequence into expression vectors pc3, pc3-GFP, pc3-BFP, and pc3-mCherry using BamHI/EcoRI or BamHI/NheI restriction sites, respectively, allowed the expression of Taspase1, alone or as a fusion with fluorescent proteins as described previously (17, 22). Plasmid p_TaspTV-BFP, p_TaspTA-BFP, and p_TaspTV-mCherry encoding the catalytically inactive Taspase1 mutant, TaspTV, were generated by splice overlap extension PCR as reported previously (2, 23). p_TF2A-GFP, encoding a TF2A-GFP fusion, was generated by PCR amplification and cloning into pc3-GFP as reported previously (18). pc3_RevM10BL-BFP, encoding a mutant HIV-1 Rev protein, was described previously (24). The NXF5 or USF2 coding sequence was derived by PCR from plasmids described (25, 26).
Plasmids were verified by sequence analysis as described previously (21). Oligonucleotides used for PCR amplification and cloning are listed in supplemental Table S1.
Generation of Inducible Tet-Off BioTaspi Cell Lines
MEF3T3 stably expressing the Tet-controlled transactivator (27) were transfected with the p_ BioTaspi, in which the expression of the biosensor is under the control of the tetracycline-responsive promoter element. Transfectants were selected by G418 (800 μg/ml) and puromycin (2 μg/ml) selection, as well as by fluorescence-activated cell sorting as reported previously (15). Cells were cultured in medium containing 1 μg/ml doxycycline (Dox) (15) to repress expression of the biosensor.
Protein Extraction, Immunoblot Analysis, and Immunofluorescence
Preparation of whole cell lysates and immunoblotting was carried out as described previously (19, 28). Equal loading of lysates was controlled by reprobing blots for GAPDH as described previously (29). Immunofluorescence was performed as reported in detail (2, 16, 30).
Database Searches and Pathway Analysis
For the in silico identification of potential human Taspase1 targets, ScanProsite searches were performed using the patterns Q(FILV)DGXDD, Q(FILV)DGXXD, and Q(FILV)DGXDX as queries. Searches were performed in the human taxon of the UniProtKB/SwissProt database, and all other parameters were used at default settings. Pathway analysis was performed using GeneSpringGX11.0.2 (Waldbronn, Germany) and MAPPFinder, a component of the software package GenMAPP version 2.1 (31).
Statistical Analysis
For experiments stating p values, a paired Student's t test was performed. Unless stated otherwise, p values represent data obtained from three independent experiments done in triplicate. p values <0.05 were considered as significant.
RESULTS
Development of Cell-based Assays to Probe Taspase1 Activity
Previous studies employed mainly in vitro assays to analyze Taspase1 function. As biochemical data must be verified at the cellular level, we here focused on the most relevant test tube, the living cell. To develop such a cell-based assay, we first cloned Taspase1 from a human head and neck tumor. Sequence analysis revealed an open reading frame (ORF) identical to the one described by Hsieh et al. (4). The same ORF could be amplified from the BCR-Abl+ chronic myeloic leukemia cell line K562. Upon ectopic expression as a Taspase1-GFP, -BFP, or -mCherry fusion, the protease localized predominantly to the nucleus, accumulating at the nucleoli in living cells (Figs. 1 and 2). Localization was confirmed by staining with α-Taspase1 Ab (Fig. 1a). A similar localization, although with less prominent nucleolar accumulation, was observed for a Taspase1 mutant, in which the catalytic nucleophile, Thr234, was changed into Val or Ala (TaspTV, TaspTA) (Figs. 1d, 2, and 4 and supplemental Fig. S1a, and data not shown). These mutations abolished the enzymatic activity of Taspase1 (4, 5), which was confirmed by immunoblot analysis (Fig. 1e).
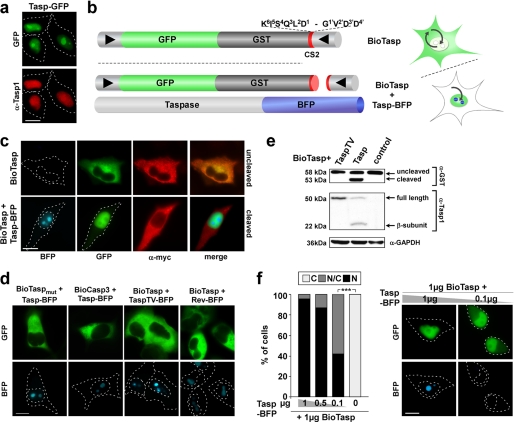
FIGURE 1.
In vivo assays for Taspase1 activity. Green/blue fluorescent proteins were visualized by fluorescence microscopy. Scale bars, 10 μm. Dashed lines mark the nuclear/cytoplasmic cell boundaries obtained from the corresponding phase contrast images. a, nuclear/nucleolar localization of Taspase1-GFP (Tasp-GFP) in HeLa cell transfectants visualized by GFP fluorescence (upper panel) or immunofluorescence using α-Taspase1 Ab (lower panel). b, principal and domain organization of the translocation sensor to probe Taspase1 activity. BioTasp is composed of GST, GFP, combinations of a nuclear import (NLS), and a Myc epitope-tagged export (NES) signal, combined with the Taspase1 cleavage site from MLL (CS2; aa 2713KISQLDGVDD2722). BioTasp localizes predominantly to the cytoplasm but is continuously shuttling between the nucleus and the cytoplasm. Coexpression of Taspase1-BFP (Tasp-BFP) results in the removal of the NES by proteolytic cleavage of the biosensor, allowing nuclear accumulation of the green fluorescent fusion.▶, NLS; ◀, NES. c, expression of Taspase1 triggers nuclear accumulation of BioTasp in living HeLa transfectants. The autofluorescent biosensor is predominantly cytoplasmic (upper panel), and coexpression of Taspase1-BFP results in the proteolytic cleavage of the Myc-NESRev and nuclear accumulation of BioTasp (lower panel). Myc-NESRev was detected using α-Myc tag Ab. d, biosensors containing a nonfunctional Taspase1 cleavage site (BioTaspmut) or a cleavage site for caspase3 (BioCasp3) remained cytoplasmic upon coexpression of Tasp-BFP. No nuclear accumulation of BioTasp was observed upon coexpression of inactive TaspTV-BFP or the nucleolar Rev-BFP protein. e, trans-cleavage of BioTasp shown by immunoblot analysis of whole cell lysates. 293T cells were transfected with BioTasp together with the indicated Taspase1 expression plasmids or the empty vector (control). Expression of proteins and cleavage products was visualized using α-GST and α-Taspase1 Ab. GAPDH served as loading control. f, efficacy of BioTasp processing in vivo. HeLa cells were transfected with the indicated amounts of BioTasp and Tasp-BFP expression plasmid. 24 h later, the numbers of cells showing cytoplasmic (C), cytoplasmic and nuclear (N/C), or nuclear (N) fluorescence were counted in at least 200 sensor-expressing cells. Results and images from a representative experiment are shown. The numbers of cell showing cytoplasmic fluorescence significantly decreased upon cotransfection of as little as 0.1 μg of Tasp-BFP expression plasmid (***, p < 0.0001).
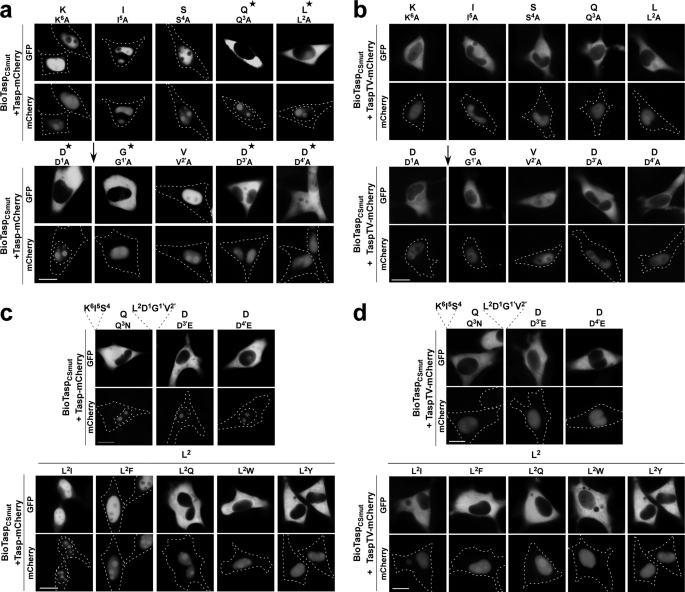
FIGURE 2.
In vivo mapping of residues critical for trans-cleavage activity of Taspase1. HeLa cells were transfected with the indicated BioTasp cleavage site mutants (BioTaspCSmut) together with the Tasp- or TaspTV-mCherry expression plasmid. 24 h later, nuclear translocation of the BioTaspCSmut variants was analyzed in at least 200 fluorescent living cells. Representative examples are shown. a and b, alanine-scanning mutagenesis using active (a) or inactive Taspase (b). c and d, similar exchange mutagenesis using active (c) or inactive Taspase (d). Scale bars, 10 μm. *, residues required for Taspase1 cleavage. ↓ indicates position of Taspase1 cleavage.
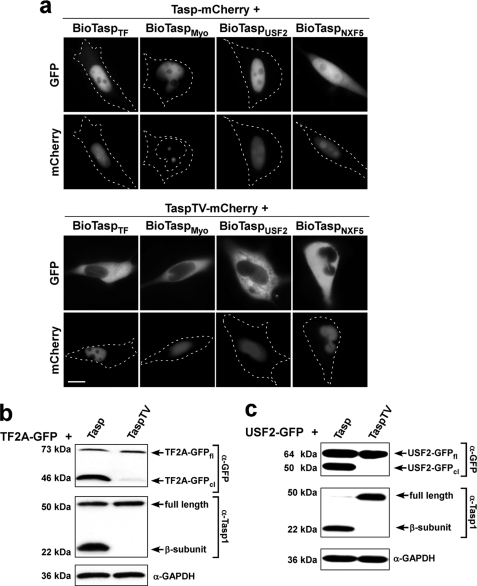
FIGURE 4.
Verification of in silico predicted Taspase1 targets. a, HeLa cells were transfected with biosensors harboring full-length TF2A (BioTaspTF) or USF2 (BioTaspUSF2) or the predicted cleavage site from myosin1F (BioTaspMyo) or NXF5 (BioTaspNXF5) together with the Tasp-mCherry expression plasmid. 24 h later, trans-cleavage resulting in nuclear translocation of the biosensors was analyzed in at least 200 fluorescent living cells. Representative examples are shown (upper panel). Coexpression of inactive TaspTV-mCherry did not affect the cytoplasmic localization of the biosensors (lower panel). Scale bars, 10 μm. b and c, proteolytic processing of full-length TF2A-GFP (b) or USF2-GFP (c) upon coexpression of Taspase1 but not of the inactive TaspTV mutant demonstrated by immunoblot analysis of transfected 293T cell lysates. Protein expression and cleavage was visualized by α-GFP and α-Taspase1 Ab. GAPDH served as loading control.
Translocation-based autofluorescent biosensors are powerful tools to assess protein-protein interaction and nucleo-cytoplasmic transport in vivo (2, 15). Hence, we generated a Taspase1 protease biosensor by integrating the Taspase1 cleavage site from MLL (CS2; aa 2713KISQLD↓GVDD2722) into a biosensor backbone composed of GST, GFP, the SV40 large T-antigen nuclear import signal (NLS), and a Myc epitope-tagged nuclear export signal from the HIV-1 Rev protein (NESRev) (Fig. 1b). The resulting NLS-GFP/GST-CS2-NESRev fusion protein (BioTasp) localizes predominantly to the cytoplasm (Fig. 1c), but it is continuously shuttling between the nucleus and the cytoplasm, as confirmed by treatment with the export inhibitor leptomycin B (supplemental Fig. S3).
Importantly, cotransfection of either Taspase1-BFP (Tasp-BFP) or red fluorescent Taspase1-mCherry (Tasp-mCherry) fusions resulted in the proteolytic cleavage of NESRev and the subsequent nuclear accumulation of BioTasp in various epithelial and liquid cancer cell lines (Fig. 1, c and f; supplemental Fig. S1b; supplemental Table S2). As a control, a construct containing a nonfunctional Taspase1 cleavage site (BioTaspmut, aa 2713KISQLAAVDD2722) or a cleavage site for caspase3 (BioCasp3) remained cytoplasmic under identical experimental conditions (Fig. 1d). Also, no nuclear accumulation was observed upon coexpression of the inactive TaspTV-BFP fusion, the nucleolar HIV-1 Rev-BFP protein, or the unrelated proteases caspase3 or -9, further confirming the specificity of the assay (supplemental Fig. S1c) (data not shown). Proteolytic processing of the biosensor upon expression of untagged or tagged Taspase1 was confirmed by immunoblot analysis (Fig. 1e) (data not shown). In contrast to the high amounts of Taspase1 required for cleavage in vitro (14), cotransfection of 1 μg of BioTasp with as little as 0.1 μg of the Tasp-BFP expression plasmid was sufficient to significantly catalyze cleavage and nuclear accumulation of the biosensor (Fig. 1f). These results clearly underline the practical advantages and biological relevance of the BioTasp cell-based assays to investigate Taspase1 function in living cells.
Cell lines inducibly expressing biosensors would circumvent the need for cotransfection of Taspase1 and probe the activity of the endogenous protein. Hence, we generated stable cell lines, in which the expression of the biosensor can be regulated by Dox (27). Dox interacts with Tet-controlled transactivator preventing its binding and activation of the tetracycline-responsive promoter element-containing CMV promoter. Removal of Dox allows Tet-controlled transactivator binding triggering transcriptional activation and expression of the biosensor, BioTaspi (supplemental Fig. S2a). In the presence of Dox, no expression of the biosensor is visible (supplemental Fig. S2b, left panel). Upon Dox removal, expression of the cytoplasmic BioTaspi can be detected (supplemental Fig. S2b, middle panel), and cleavage by endogenous Taspase1 results in its nuclear accumulation (supplemental Fig. S2b, right panel). Notably, the levels of BioTaspi are lower compared with transient expression, allowing its almost quantitative cleavage by endogenous Taspase1.
In Vivo Mapping of Residues Critical for trans-Cleavage Activity of Taspase1
Next, to characterize the sequence and spatial requirements for Taspase1 processing in vivo, we first performed Ala-scan mutagenesis of the MLL cleavage site (CS2; aa 2713KISQLD↓GVDD2722). As depicted in Fig. 2, coexpression of the BioTasp mutants (BioTaspCSmut) with Tasp-mCherry resulted in proteolytic cleavage and nuclear accumulation of only those biosensors in which nonessential residues were mutated. In contrast, changing critical aa into Ala almost completely prevented cleavage and nuclear accumulation of the autofluorescent proteins, leading to the following consensus sequence: K6I5S4Q3L2D1↓G1′V2′D3′D4′ (essential aa are shown in boldface). Notably, even replacing critical residues by chemically similar aa could not rescue cleavage, except for residue P2. Leu at this position can also be exchanged for the hydrophobic aa Ile, Val, or Phe (Fig. 2c; data not shown). Notably, an intermediate size in combination with hydrophobicity was required for productive cleavage, as replacing Leu with polar amino acids such as Trp, Gln, or Tyr completely prevented cleavage (Fig. 2c). These results could be confirmed by immunoblot analysis (data not shown). Again, specificity of the assay was verified by cotransfection of inactive TaspTV-mCherry, which did not result in nuclear accumulation of the biosensors (Fig. 2, b and d). Nuclear accumulation of all the BioTaspCSmut variants upon leptomycin B treatment further excluded the formal possibility that mutagenesis had affected the shuttling capability of the biosensors (supplemental Fig. S3). Similar results were obtained upon expression in other adherent or liquid cancer cell lines (data not shown).
Genome-wide in Silico Identification of the Taspase1 Degradome
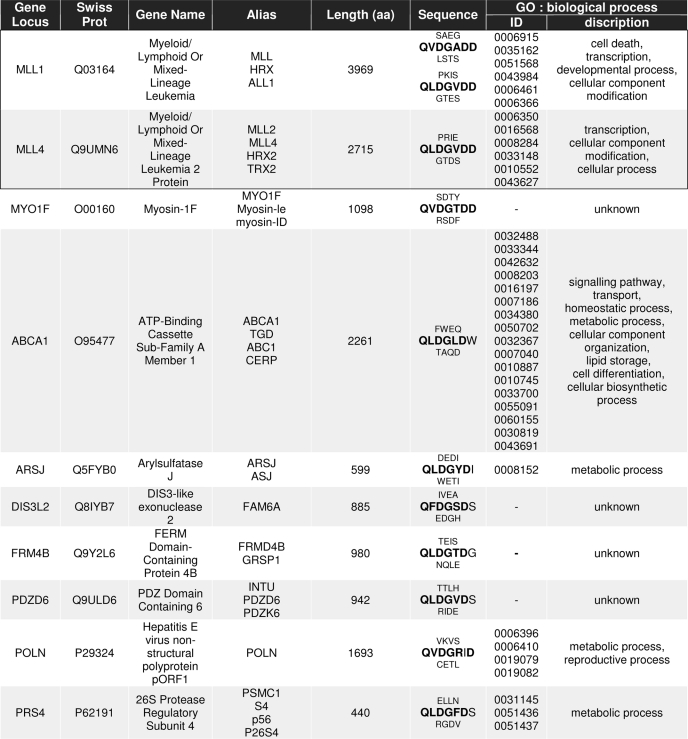
Previous studies suggested the motif Q3X2D1↓G1′ (X = any residue) already as an improved Taspase1 consensus cleavage site (4, 5). However, employing this motif to identify potential Taspase1 targets by database searches resulted in a list of more than 1400 potential targets (data not shown). As the experimental validation of such an extensive list of putative Taspase1 substrates is almost impossible, we thus exploited the knowledge obtained from our mutational analysis. Consequently, we used the motif Q3(F/I/L/V)2D1↓G1′X2′D3′D4′ to scan the Swiss-Prot database. Our finding that a cleavage site containing Ala4′ instead of Asp4′ was still partially processed by Taspase1 in trans (Fig. 2a) and that the TF2A cleavage site contains Gly3′ instead of Asp3′ (Table 1) suggested that some variability at these positions is tolerated. Hence, the motifs Q3(F/I/L/V)2D1↓G1′X2′X3′D4′ and Q3(F/I/L/V)2D1↓G1′X2′D3′X4′ were included in our search, resulting in 27 putative Taspase1 targets, for which diverse molecular functions and intracellular localizations were predicted bioinformatically using MAPPFinder analysis (Table 1 and Fig. 3a) (14).
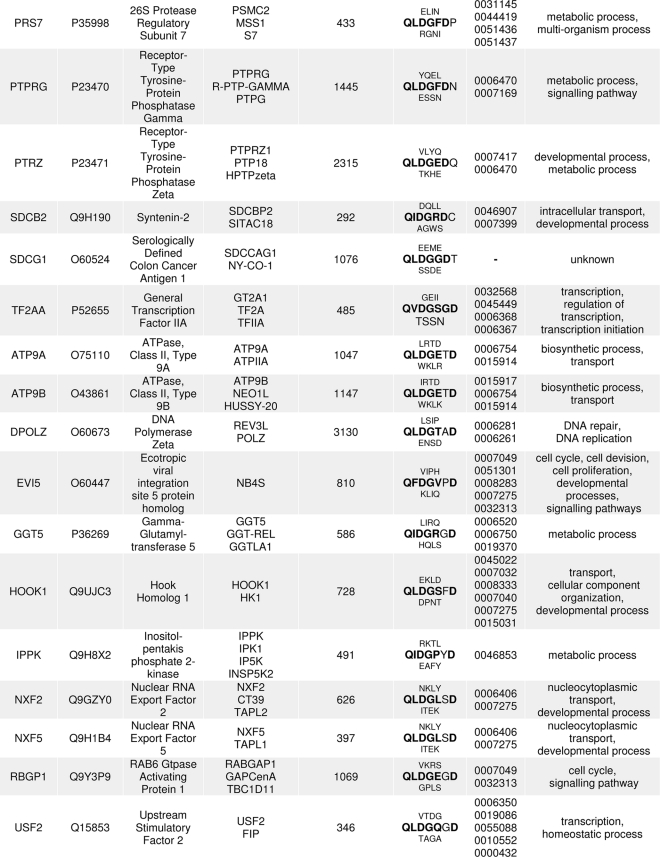
TABLE 1.
Characteristics of potential human Taspase1 targets identified by ScanProsite
Gene loci, SwissProt accession numbers, gene names, and aliases in accordance with the HUGO Gene Nomenclature Committee are given together with the aa sequence containing the predicted Taspase1 cleavage site (consensus marked in boldface). Gene ontology (GO) identifiers and descriptions are listed. Previously known Taspase1 targets are framed, and target probability decreases with line order.
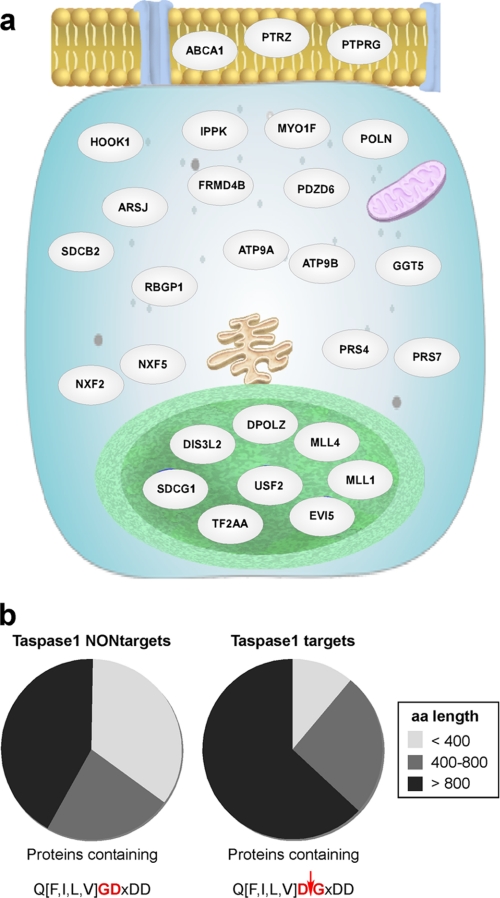
FIGURE 3.
Bioinformatical analysis of potential Taspase1 targets. a, predicted cellular localization. Target proteins (gray, see Table 1 for details) are depicted in their respective cellular compartment according to their gene ontology term classification using the GeneSpring GX “Pathway and Network Analysis” tool. Cell membrane (yellow) with pores (blue), cytoplasm (light blue) containing endoplasmic reticulum (brown), mitochondria (violet), and the nucleoplasm (green) surrounded by the nuclear membrane (light green) are represented schematically. b, length distribution of potential Taspase1 targets versus proteins randomly found in a partially scrambled motif search. Results are obtained from database searches employing the sequence Q3(F/I/L/V)2D1↓G1′X2′D3′D4′/X3′D4′/D3′X4′ (Taspase1 targets) versus the partially scrambled motif Q3(F/I/L/V)2G1D1′X2′D3′D4′/X3′D4′/D3′X4′ (Taspase1 NONtargets).
Collectively, the cleavage sites were scattered throughout the respective proteins, and no specific pattern was evident (supplemental Fig. S4b). However, as the average length of protein entries in the UniProt database is 321 aa (supplemental Fig. S4a), we noticed that the majority of identified Taspase1 targets are large proteins composed of more than 800 aa (Fig. 3b). Next, we tested whether this result is merely based on the fact that the chance of finding any random sequence is higher in larger compared with smaller proteins. Thus, we analyzed the results obtained from database searches employing the sequences Q3(F/I/L/V)2D1↓G1′X2′D3′D4′ versus Q3(F/I/L/V)2G1D1′X2′D3′D4′. The latter represents an identical but nonfunctional cleavage site, as the critical aspartate at the P1 and glycine at the P1′ position were interchanged. Remarkably, the search results differ in showing a significant enrichment of larger proteins as putative Taspase1 targets (Fig. 3b).
Validation of Predicted Taspase1 Targets
To experimentally verify that the predicted proteins represent most likely biologically relevant Taspase1 substrates, we first tested the cleavage sites of several targets in the biosensor system. Indeed, integration of the cleavage sites from MLL1, TF2A, MLL4, MYO1F, USF2, as well as NXF2 or -5 (Table 1) resulted in cytoplasmic biosensor proteins, which efficiently accumulated in the nucleus upon trans-cleavage by Taspase1 (Fig. 4a) (data not shown). However, we observed that cleavage of NXF2 or -5 was less efficient compared with the other sites tested (Fig. 4a) (data not shown). Again, coexpression of the inactive TaspTV mutant and leptomycin B treatment confirmed the functionality of the assay (Fig. 4a) (data not shown).
Second, to additionally underline the significance of our in silico analysis, we set out to demonstrate that the predicted full-length proteins are also recognized and proteolytically processed by Taspase1, as has been shown for MLL1 (4). Actually, immunoblot analysis demonstrated that full-length TF2A-GFP and the nuclear protein USF2-GFP were also efficiently cleaved upon coexpression of Taspase1 but not by inactive TaspTV (Fig. 4, b and c). Because of the large size of the ORF, we failed to clone and thus test full-length MYO1F. Notably, upon ectopic overexpression of full-length NXF5-GFP or untagged NXF5, we observed massive cell death in transfectants (supplemental Fig. S5a), which precluded investigating the effect of Taspase1 on NXF5. As NXF5 has been reported to interact with FG-repeat-containing nucleoporins, NXF5 overexpression may interfere with the transport capability of the nuclear pore thereby affecting cell viability (32). Similar effects have been reported for other proteins involved in nucleo-cytoplasmic transport (33).
These findings demonstrate again the utility and flexibility of our biosensor system allowing it to swiftly test putative Taspase1 target sites originating from toxic, large, or yet uncharacterized proteins. However, to finally uncover the biological relevance of their processing by Taspase1, comprehensive experimental investigations tailored to the particular needs of the respective target are required.
DISCUSSION
Besides their critical role in intra- and intercellular “waste management,” proteases are accepted as important signaling molecules involved in numerous biological and pathological functions (3). To dissect the biological processes a given protease participates in, one has to understand the mechanisms of protease activity, as well as the biochemistry that relates their structure to function (34, 35). Having obtained this information about a protease in its normal physiological setting, it is further important to understand how these protease properties are modified in a disease state to develop targeted intervention strategies. In contrast to other disease-relevant proteases, such as matrix metalloproteinases, which were the first protease targets considered for combating cancer because of their role in extracellular matrix degradation (3), relatively little is known about the (patho)biological relevance and function of Taspase1. A key part of understanding protease signaling in both health and disease is to identify the physiological substrates of a protease, i.e. the protease degradome. Various strategies, including chemical and in silico biology, genetics, and proteomics, are employed to achieve this goal (3). For Taspase1, the so-called “bottom-up” approach has been successfully used to identify Taspase1 as the protease responsible for the cleavage of the MLL protein (4, 6, 7).
So far, the genome-wide in silico identification of additional Taspase1 targets has clearly been hampered by lacking a detailed understanding of the substrate specificity of Taspase1. Although the sequence Q3X2D1↓G1′ has been proposed as an improved Taspase1 consensus cleavage site (5), employing this motif resulted in a list of more than 1400 putative human targets by searching the Swiss-Prot database. As the experimental validation of such an extensive number of putative Taspase1 substrates is practically impossible, we first identified residues critical for cleavage in living cells. In contrast to in vitro assays using recombinant proteins or combinatorial fluorescent substrate libraries (3), our biosensor assay combined with positional scanning mutagenesis allowed us to pinpoint residues essential for trans-cleavage activity in vivo. As Taspase1 is an aspartase, Asp at the P1 position is mandatory for cleavage. The unique chemical characteristics of Gly at P1′ does not even tolerate its replacement by Ala. Also, Gln at position P3 is required for substrate recognition, as an exchange of this uncharged polar amino acid by the smaller hydrophobic residue Ala or even the highly similar but slightly smaller amino acid Asn completely blocks trans-cleavage. Biologically, this finding is supported by the fact that all previously described Taspase1 substrates contain a Gln at this position (13). Previous reports characterize position P2 as irrelevant for cleavage, proposing that P2 can host any amino acid (4, 5). In contrast, our data now convincingly demonstrate that although Leu can be substituted by hydrophobic residues of similar size (Phe, Ile, and Val), other aa such as the smaller hydrophobic amino acid Ala are not tolerated. Even though the alike hydrophobic amino acid Met at P2 cannot be excluded biochemically, no human Taspase1 substrates containing Met2 were identified bioinformatically. Hence, hydrophobicity in combination with size is required for productive cleavage, as replacing Leu with charged aa such as Trp, Gln, or Tyr completely prevented cleavage.
On the other hand, position P2′ is highly flexible, whereas the residues at positions P3′ and P4′ seem to be interdependent. We found that at least one of these residues must be Asp but that a small amino acid at the other position, like Gly present in TF2A or Ala at either position, does not completely abrogate substrate cleavage. Glu at either position, however, prevents cleavage, indicating that not only charge but also size is important to maintain a secondary structure allowing docking and productive substrate recognition by the protease. Collectively, our in vivo analysis now defines the motif Q3(F/I/L/V)2D1↓G1′V2′D3′D4′ as an improved Taspase1 cleavage consensus sequence, allowing for the first time a rational genome-wide bioinformatic identification of potential human Taspase1 targets. As some variability at positions P3′ and P4′ is tolerated, the motifs Q3(F/I/L/V)2D1↓G1′X2′X3′D4′ and Q3(F/I/L/V)2D1↓G1′X2′D3′X4′ were additionally included in our search.
The obtained list of the 27 most likely human Taspase1 targets not only contains known Taspase1 substrates such as MLL, but several candidates, which have not been considered as potential targets for this protease. Noteworthy, we also identified a putative Taspase1 target site in a human hepatitis E virus. As proteolytic processing is essential for replication and pathogenesis of many viruses, it is tempting to speculate that also Taspase1 might play a role in these processes. Interestingly, according to bioinformatic predictions, the Taspase1 targets localize to various intracellular compartments, such as the cell membrane, the cytoplasm, and/or the nucleus (Fig. 3). Because also shuttling proteins, such as NXF2/5 (36), have been suggested, it will be critical to investigate whether Taspase1 itself might be able to actively shuttle between the nucleus and the cytoplasm allowing the processing of substrates in both compartments (15).
Our results clearly indicate a nonrandom evolutionary enrichment of large proteins (>800 aa) among the list of putative Taspase1 targets. It might be assumed that those targets are preferentially involved in the formation of multiprotein complexes, for which processing by Taspase1 represents a critical (fine) tuning mechanism. Such examples are not only the MLL protein (4) but also disease-relevant MLL fusions containing a functional Taspase1 cleavage site. Proteolytic cleavage stabilizes these MLL fusion proteins and appears to trigger their subnuclear localization (37, 38), ultimately promoting cell cycle progression (10). Hence, cleavage-induced formation of stable MLL-containing complexes is considered a crucial constituent of MLL-mediated tumorigenesis (11). As such, we found that only AF4·MLL but not the reciprocal translocation product, MLL·AF4, lacking the Taspase1 cleavage site, can cause pro-B acute lymphocytic leukemia in a murine model (39). It is also striking that in the MLL·MYO1F translocation product, detectable in infant acute monocytic leukemia (40, 41), the unconventional myosin type 1F (MYO1F) partner provides the functional Taspase1 cleavage site. Hence, proteolytic processing of Taspase1 of the MLL fusion proteins in general appears to be critical for disease development, underlining the pathobiological and therapeutic relevance of this protease.
The suggested impact of Taspase1 for neoplastic diseases is further supported by the identification of additional targets, which have been reported to play a role in cancer development and progression, such as the oncogene Evi5 (42, 43). As an additional example, we show that the oncological relevant transcription factor USF2 (44–46) is a novel bona fide Taspase1 target. USF2 not only contains a functional cleavage site but also the full-length protein is cleaved in trans by Taspase1. As USF2 dimerizes with USF1 to regulate transcription through E-box motifs in target genes (44), cleavage of USF2 might be sufficient to attenuate the activity of this transcriptional regulator complex, as no cleavage site could be detected in USF1 (supplemental Fig. S5).
Likewise, cleavage of TF2A appears not to be required for the activation of this transcription factor but rather seems to affect its turnover as part of an intricate degradation mechanism that allows fine-tuning of TF2A functions (13, 47).
Also our data indicate that nuclear shuttle proteins involved in mRNA transport and processing such as NXF2 and -5 are Taspase1 targets. Again, enhanced or reduced levels of NXF2 have been linked to disease such as chronic lymphocytic leukemia (48) or mental retardation (32). Of note, we could only identify a functional Taspase1 cleavage site in NXF2 and -5 but not in the other family members NXF1 or NXF3 (supplemental Fig. S5). Several studies showed that NXF members differ significantly in their ability to export and guide the cytoplasmic trafficking of mRNAs (32, 48). Thus, Taspase1 cleavage might represent a potential mechanism to selectively regulate the biological activity of NXF family members.
Collectively, our study underlines the complexity of the (patho)biological processes in which Taspase1 might be involved. However, we are well aware that the above-mentioned suggestions are currently lacking experimental evidence. Besides genetic and biochemical approaches, small molecules that allow a (transient) chemical knock-out of Taspase1 in a specific biological system would be highly valuable. Unfortunately, no effective small molecule inhibitors of this enzyme are currently available. Hence, we also suggest employing our translocation biosensor, BioTasp, for high throughput screening assays to identify potent Taspase1 inhibitors, which could finally help to dissect the (patho)biological impact of Taspase1.
Supplementary Material
Acknowledgments
We thank S. Friedel for excellent technical assistance and G. Froyen and A. J. M. Verhoeven for providing the cDNAs.
This work was supported by German Cancer Aid Grant FKZ102362 (to R. H. S. and R. M.), Head and Neck Cancer Foundation (to C. B.), Wilhelm-Sander Foundation, Funds of the Chemical Industry, Stiftung Rheinland-Pfalz für Innovationen Grants DFG KN973/1-1 and DFG INST371/5-1FUGG, Mainz Screening Center, donation from Dr. Krieg, and the Inneruniversity and MAIFOR Mainz Support Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
- aa
- amino acid
- α
- anti
- Dox
- doxycycline
- MLL
- mixed lineage leukemia
- NES
- nuclear export signal
- NLS
- nuclear import signal.
REFERENCES
- 1. Overall C. M., Dean R. A. (2006) Cancer Metastasis Rev. 25, 69–75 [DOI] [PubMed] [Google Scholar]
- 2. Knauer S. K., Moodt S., Berg T., Liebel U., Pepperkok R., Stauber R. H. (2005) Traffic 6, 594–606 [DOI] [PubMed] [Google Scholar]
- 3. Turk B. (2006) Nat. Rev. Drug Discov. 5, 785–799 [DOI] [PubMed] [Google Scholar]
- 4. Hsieh J. J., Cheng E. H., Korsmeyer S. J. (2003) Cell 115, 293–303 [DOI] [PubMed] [Google Scholar]
- 5. Khan J. A., Dunn B. M., Tong L. (2005) Structure 13, 1443–1452 [DOI] [PubMed] [Google Scholar]
- 6. Takeda S., Chen D. Y., Westergard T. D., Fisher J. K., Rubens J. A., Sasagawa S., Kan J. T., Korsmeyer S. J., Cheng E. H., Hsieh J. J. (2006) Genes Dev. 20, 2397–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu H., Cheng E. H., Hsieh J. J. (2007) Genes Dev. 21, 2385–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marschalek R. (2010) FEBS J. 277, 1822–1831 [DOI] [PubMed] [Google Scholar]
- 9. Meyer C., Schneider B., Jakob S., Strehl S., Attarbaschi A., Schnittger S., Schoch C., Jansen M. W., van Dongen J. J., den Boer M. L., Pieters R., Ennas M. G., Angelucci E., Koehl U., Greil J., Griesinger F., Zur Stadt U., Eckert C., Szczepañski T., Niggli F. K., Schäfer B. W., Kempski H., Brady H. J., Zuna J., Trka J., Nigro L. L., Biondi A., Delabesse E., Macintyre E., Stanulla M., Schrappe M., Haas O. A., Burmeister T., Dingermann T., Klingebiel T., Marschalek R. (2006) Leukemia 20, 777–784 [DOI] [PubMed] [Google Scholar]
- 10. Capotosti F., Hsieh J. J., Herr W. (2007) Mol. Cell. Biol. 27, 7063–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hsieh J. J., Ernst P., Erdjument-Bromage H., Tempst P., Korsmeyer S. J. (2003) Mol. Cell. Biol. 23, 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaussmann A., Wenger T., Eberle I., Bursen A., Bracharz S., Herr I., Dingermann T., Marschalek R. (2007) Oncogene 26, 3352–3363 [DOI] [PubMed] [Google Scholar]
- 13. Zhou H., Spicuglia S., Hsieh J. J., Mitsiou D. J., Høiby T., Veenstra G. J., Korsmeyer S. J., Stunnenberg H. G. (2006) Mol. Cell. Biol. 26, 2728–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J. T., Chen D. Y., Yang Z., Ramos A. D., Hsieh J. J., Bogyo M. (2009) Bioorg. Med. Chem. Lett. 19, 5086–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fetz V., Knauer S. K., Bier C., Kriess J. P., Stauber R. H. (2009) Sensors 7, 5423–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knauer S. K., Bier C., Habtemichael N., Stauber R. H. (2006) EMBO Rep. 7, 1259–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knauer S. K., Bier C., Schlag P., Fritzmann J., Dietmaier W., Rödel F., Klein-Hitpass L., Kovács A. F., Döring C., Hansmann M. L., Hofmann W. K., Kunkel M., Brochhausen C., Engels K., Lippert B. M., Mann W., Stauber R. H. (2007) Cell Cycle 6, 1502–1509 [PubMed] [Google Scholar]
- 18. Engels K., Knauer S. K., Metzler D., Simf C., Struschka O., Bier C., Mann W., Kovács A. F., Stauber R. H. (2007) J. Pathol. 211, 532–540 [DOI] [PubMed] [Google Scholar]
- 19. Engels K., Knauer S. K., Loibl S., Fetz V., Harter P., Schweitzer A., Fisseler-Eckhoff A., Kommoss F., Hanker L., Nekljudova V., Hermanns I., Kleinert H., Mann W., du Bois A., Stauber R. H. (2008) Cancer Res. 68, 5159–5166 [DOI] [PubMed] [Google Scholar]
- 20. Heger P., Lohmaier J., Schneider G., Schweimer K., Stauber R. H. (2001) Traffic 2, 544–555 [DOI] [PubMed] [Google Scholar]
- 21. Schlingemann J., Habtemichael N., Ittrich C., Toedt G., Kramer H., Hambek M., Knecht R., Lichter P., Stauber R., Hahn M. (2005) Lab. Invest. 85, 1024–1039 [DOI] [PubMed] [Google Scholar]
- 22. Knauer S. K., Stauber R. H. (2005) Anal. Chem. 77, 4815–4820 [DOI] [PubMed] [Google Scholar]
- 23. Knauer S. K., Krämer O. H., Knösel T., Engels K., Rödel F., Kovács A. F., Dietmaier W., Klein-Hitpass L., Habtemichael N., Schweitzer A., Brieger J., Rödel C., Mann W., Petersen I., Heinzel T., Stauber R. H. (2007) FASEB J. 21, 207–216 [DOI] [PubMed] [Google Scholar]
- 24. Knauer S. K., Carra G., Stauber R. H. (2005) Mol. Cell. Biol. 25, 2573–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jun L., Frints S., Duhamel H., Herold A., Abad-Rodrigues J., Dotti C., Izaurralde E., Marynen P., Froyen G. (2001) Curr. Biol. 11, 1381–1391 [DOI] [PubMed] [Google Scholar]
- 26. van Deursen D., van Leeuwen M., Vaulont S., Jansen H., Verhoeven A. J. (2009) Biochim. Biophys. Acta 1791, 229–237 [DOI] [PubMed] [Google Scholar]
- 27. Gossen M., Freundlieb S., Bender G., Müller G., Hillen W., Bujard H. (1995) Science 268, 1766–1769 [DOI] [PubMed] [Google Scholar]
- 28. Fetz V., Bier C., Habtemichael N., Schuon R., Schweitzer A., Kunkel M., Engels K., Kovács A. F., Schneider S., Mann W., Stauber R. H., Knauer S. K. (2009) Int. J. Cancer 124, 2033–2041 [DOI] [PubMed] [Google Scholar]
- 29. Krämer O. H., Knauer S. K., Greiner G., Jandt E., Reichardt S., Gührs K. H., Stauber R. H., Böhmer F. D., Heinzel T. (2009) Genes Dev. 23, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krämer O. H., Knauer S. K., Zimmermann D., Stauber R. H., Heinzel T. (2008) Oncogene 27, 732–740 [DOI] [PubMed] [Google Scholar]
- 31. Dahlquist K. D. (2004) Curr. Protoc. Bioinformatics 7.5, 1–26 [DOI] [PubMed] [Google Scholar]
- 32. Lai D., Sakkas D., Huang Y. (2006) RNA 12, 1446–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marg A., Shan Y., Meyer T., Meissner T., Brandenburg M., Vinkemeier U. (2004) J. Cell Biol. 165, 823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schilling O., Overall C. M. (2008) Nat. Biotechnol. 26, 685–694 [DOI] [PubMed] [Google Scholar]
- 35. López-Otín C., Overall C. M. (2002) Nat. Rev. Mol. Cell Biol. 3, 509–519 [DOI] [PubMed] [Google Scholar]
- 36. Tretyakova I., Zolotukhin A. S., Tan W., Bear J., Propst F., Ruthel G., Felber B. K. (2005) J. Biol. Chem. 280, 31981–31990 [DOI] [PubMed] [Google Scholar]
- 37. Yokoyama A., Kitabayashi I., Ayton P. M., Cleary M. L., Ohki M. (2002) Blood 100, 3710–3718 [DOI] [PubMed] [Google Scholar]
- 38. Bursen A., Moritz S., Gaussmann A., Moritz S., Dingermann T., Marschalek R. (2004) Oncogene 23, 6237–6249 [DOI] [PubMed] [Google Scholar]
- 39. Bursen A., Schwabe K., Ruster B., Henschler R., Ruthardt M., Dingermann T., Marschalek R. (2010) Blood 115, 3570–3579 [DOI] [PubMed] [Google Scholar]
- 40. Taki T., Akiyama M., Saito S., Ono R., Taniwaki M., Kato Y., Yuza Y., Eto Y., Hayashi Y. (2005) Oncogene 24, 5191–5197 [DOI] [PubMed] [Google Scholar]
- 41. Zadro C., Alemanno M. S., Bellacchio E., Ficarella R., Donaudy F., Melchionda S., Zelante L., Rabionet R., Hilgert N., Estivill X., Van Camp G., Gasparini P., Carella M. (2009) Biochim. Biophys. Acta 1792, 27–32 [DOI] [PubMed] [Google Scholar]
- 42. Jacob B., Osato M., Yamashita N., Wang C. Q., Taniuchi I., Littman D. R., Asou N., Ito Y. (2010) Blood 115, 1610–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eldridge A. G., Loktev A. V., Hansen D. V., Verschuren E. W., Reimann J. D., Jackson P. K. (2006) Cell 124, 367–380 [DOI] [PubMed] [Google Scholar]
- 44. Ocejo-Garcia M., Baokbah T. A., Ashurst H. L., Cowlishaw D., Soomro I., Coulson J. M., Woll P. J. (2005) J. Pathol. 206, 151–159 [DOI] [PubMed] [Google Scholar]
- 45. Landa I., Ruiz-Llorente S., Montero-Conde C., Inglada-Pérez L., Schiavi F., Leskelä S., Pita G., Milne R., Maravall J., Ramos I., Andía V., Rodríguez-Poyo P., Jara-Albarrán A., Meoro A., del Peso C., Arribas L., Iglesias P., Caballero J., Serrano J., Picó A., Pomares F., Giménez G., López-Mondéjar P., Castello R., Merante-Boschin I., Pelizzo M. R., Mauricio D., Opocher G., Rodríguez-Antona C., González-Neira A., Matías-Guiu X., Santisteban P., Robledo M. (2009) PLoS Genet. 5, e1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen N., Szentirmay M. N., Pawar S. A., Sirito M., Wang J., Wang Z., Zhai Q., Yang H. X., Peehl D. M., Ware J. L., Sawadogo M. (2006) Oncogene 25, 579–587 [DOI] [PubMed] [Google Scholar]
- 47. Høiby T., Zhou H., Mitsiou D. J., Stunnenberg H. G. (2007) Biochim. Biophys. Acta 1769, 429–436 [DOI] [PubMed] [Google Scholar]
- 48. Dubovsky J. A., McNeel D. G., Powers J. J., Gordon J., Sotomayor E. M., Pinilla-Ibarz J. A. (2009) Clin. Cancer Res. 15, 3406–3415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.