Abstract
Eukaryotic initiation factor 6 (eIF6), a highly conserved protein from yeast to mammals, is essential for 60 S ribosome biogenesis and assembly. Both yeast and mammalian eIF6 are phosphorylated at Ser-174 and Ser-175 by the nuclear isoform of casein kinase 1 (CK1). The molecular basis of eIF6 phosphorylation, however, remains elusive. In the present work, we show that subcellular distribution of eIF6 in the nuclei and the cytoplasm of mammalian cells is mediated by dephosphorylation and phosphorylation, respectively. This nucleo-cytoplasmic shuttling is dependent on the phosphorylation status at Ser-174 and Ser-175 of eIF6. We demonstrate that Ca2+-activated calcineurin phosphatase binds to and promotes nuclear localization of eIF6. Increase in intracellular concentration of Ca2+ leads to rapid translocation of eIF6 from the cytoplasm to the nucleus, an event that is blocked by specific calcineurin inhibitors cyclosporin A or FK520. Nuclear export of eIF6 is regulated by phosphorylation at Ser-174 and Ser-175 by the nuclear isoform of CK1. Mutation of eIF6 at the phos-phorylatable Ser-174 and Ser-175 to alanine or treatment of cells with the CK1 inhibitor, D4476 inhibits nuclear export of eIF6 and results in nuclear accumulation of eIF6. Together, these results establish eIF6 as a substrate for calcineurin and suggest a novel paradigm for calcineurin function in 60 S ribosome biogenesis via regulating the nuclear accumulation of eIF6.
Keywords: Calcineurin, Nuclear Translocation, Phosphatase, Phosphorylation Enzymes, Ribosomes, Nuclear Import and Export, Ribosome Biogenesis Factor
Introduction
Eukaryotic translation initiation factor 6 (eIF6)3 is a highly conserved protein between yeast and mammals. Both the mammalian and yeast proteins are each 245 amino acid long and 72% identical in amino acid sequence (1, 2). Although eIF6 was originally isolated and characterized from the postribosomal supernatant of both wheat germ (3, 4) and mammalian cell extracts including anucleated rabbit reticulocyte lysates (1, 5, 6), most of our knowledge of the functional properties of eIF6 have been derived from molecular genetic studies examining the yeast eIF6 ortholog Tif6p in the yeast Saccharomyces cerevisiae (2, 7). These studies have provided compelling evidence that, at least in yeast cells, Tif6p, encoded by a single copy essential gene, does not function as a canonical translation initiation factor (2). Rather, Tif6p is essential for the biogenesis of 60 S ribosomal subunits in S. cerevisiae (2, 7, 8). Specifically, the lack of Tif6p prevents the processing of the 27SB pre-rRNA to form the mature 25 S and 5.8 S rRNAs, the constituents of the 60 S ribosomal particle (8). In agreement with the essential requirement of Tif6p in pre-ribosomal RNA processing, Tif6p is found to be a constituent of a multiprotein assembly complex associated with the pre-60 S ribosomal particles in the nucleolus, where biogenesis and maturation of 60 S ribosomal subunits take place (2, 9, 10). Indeed, in exponentially growing yeast cells, Tif6p is localized primarily in the nucleolus where most of the steps of 60 S ribosome biogenesis occur (11, 12).
In previous studies, we have observed that in both mammalian and yeast cells, eIF6 (Tif6p) is phosphorylated at Ser-174 (major site) and Ser-175 (minor site) (10, 13). Purification and characterization of the protein kinase from rabbit reticulocyte lysates identified casein kinase 1α (CK1α) as the enzyme responsible for phosphorylation of mammalian eIF6 (13). We also demonstrated that the yeast CK1 ortholog Hrr25p binds to and phosphorylates Tif6p at Ser-174 and Ser-175 both in vivo and in vitro (10). The sites of in vitro phosphorylation in both mammalian (13) and yeast eIF6 (10) were identified as the serine residues at positions 174 (major site) and 175 (minor site), that are present in a highly conserved CK1 consensus sequence (14). More importantly, Hrr25p-mediated phosphorylation of Tif6p is required for efficient processing of pre-ribosomal RNAs to form the mature 25 S and 5 S rRNAs (10). Conversely, depletion of Hrr25p from yeast cells or Ala replacement of Ser-174 and Ser-175 of Tif6p abolished cell growth and viability (10, 13). Taken together, these results suggested that phosphorylation of Tif6p at Ser-174 and Ser-175 plays an important regulatory role in the function of Tif6p. However, the molecular basis of phosphorylation of eIF6 (Tif6p) was not apparent from these studies.
Under steady state growth conditions of S. cerevisiae, Tif6p, tagged with GFP at the genomic locus, is localized predominantly in the nucleolus and bound to the pre-60 S ribosomal particles (2, 9–12). However, indirect immunofluorescence studies, as well as Western blot analysis, showed that significant level of eIF6 was also present in the cytosol (8, 13). It has also been reported that Tif6p bound to the pre-60 S ribosomal particles can escort pre-60 S complex through the nuclear pore to the cytoplasm (15, 16). Once in the cytosol, Efl1p and Sdo1p, which have been shown to genetically interact with Tif6p (11, 12), facilitate the release of Tif6p from the pre-60 S particles in the cytoplasm via a structural rearrangement. It has been postulated that the released cytosolic eIF6 is rapidly imported to the nucle(ol)us for continued 60 S ribosome biogenesis, which is essential to sustain rapid exponential growth of yeast cells (11, 12). However, neither the nuclear import nor the recycling of eIF6 has so far been directly demonstrated either in yeast or mammalian cells.
In the present work, we have used mammalian cells to investigate the nuclear import of eIF6. This decision has been governed by our consideration that unlike Tif6p in yeast cells, mammalian eIF6 is distributed both in the cytosol and nuclei (1, 5, 6, 17). Furthermore, the nuclei and the cytosol of mammalian cells can easily be separated from each other and analyzed for the relative distribution of eIF6 between the two cellular compartments.
The goal of this study was to directly test whether mammalian eIF6 shuttles between the cytoplasm and nucleus and whether phosphorylation status of eIF6 at Ser-174 and Ser-175 contributes to the shuttling process. Here, we show that eIF6 indeed shuttles between the cytosol and nucleus of mammalian cells. We find that phosphorylation at Ser-174 and Ser-175 by the nuclear isoform of protein kinase CK1 promotes cytoplasmic localization of eIF6. Dephosphorylation mediated by the Ca2+/calmodulin-dependent protein phosphatase calcineurin facilitates nuclear import and accumulation of eIF6. The implications of this work in relation to the regulation and function of eIF6 in 60 S ribosome biogenesis are discussed.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
A variety of antibodies used in this study were purchased as follows: Mouse monoclonal anti-Myc antibody and anti-HA antibody from Santa Cruz Biotechnology; Rabbit monoclonal anti-His antibody, goat anti-mouse, and anti-rabbit antibodies from Roche Diagnostics Corporation; Rabbit polyclonal anti-eIF6 antibody from Cell Signaling Technology; Alexa Fluor goat anti-mouse antibody from Molecular Probes Inc. The other reagents used in this work: cyclosporin A (CsA), FK-520 (analog of FK-506), ionomycin, D4476, and leptomycin B were purchased from Calbiochem; Hoechst 33258 was purchased from Molecular Probes Inc.; Protease inhibitors were purchased from Roche Diagnostics Corporation.
Recombinant eIF6 Plasmid Constructs and Purified Recombinant eIF6
The wild-type mammalian eIF6 expression construct was generated by inserting PCR-amplified human eIF6-encoding sequences (1) at the XhoI/BamHI sites of the mammalian expression vector pcDNA3.1-Myc-His such that the Myc-His epitopes were fused in-frame with the eIF6 coding sequence at the C-terminal end to yield the pcDNA-eIF6-Myc-His expression construct. The eIF6 phosphorylation site mutant constructs were generated by one-step PCR using QuikChange site-directed mutagenesis kit (Stratagene). Appropriate mutagenic oligonucleotide primers were designed to create the desired serine-to-alanine mutations at positions Ser-174 and/or Ser-175 to yield the pcDNA-eIF6-S174A-Myc-His single mutant, pcDNA-eIF6-S175A-Myc-His single mutant and pcDNA-eIF6-S174,175A-Myc-His double mutant of mammalian eIF6. All mutations and constructs were sequenced before use to ensure the proper reading frames.
Recombinant human His6-eIF6 was purified from Escherichia coli BL21 (DE3) cells carrying the open reading frame of human eIF6 in the plasmid pRSET-A by following a procedure as previously described (13). The procedure involved affinity purification from a Ni-NTA column followed by gradient elution from a fast-protein liquid chromatography-Mono Q column. The final preparation was >95% pure as judged by SDS-polyacrylamide gel electrophoresis followed by Coomassie Blue Staining.
Recombinant Calcineurin Plasmid Constructs
HA-tagged calcineurin A subunit and untagged calcineurin B subunit constructs have been described before (18).
Cell Culture and Expression of eIF6
COS-7 African Green Monkey, human 293T, and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mm glutamine, 50 units/ml of penicillin G-sodium and 50 μg/ml of streptomycin sulfate at 37 °C in humidified incubators containing 5% CO2. For expression of eIF6 from various recombinant pcDNA3.1-Myc-His expression plasmids, cells were seeded 18–24 h before transfection and grown to 75–80% confluence. Transfections were carried out using Effectene transfection reagent (Qiagen) following the manufacturer's protocol. The transfection efficiency varied from 50–70% of the total cell population. Transfected cells were harvested 24–48-h post-transfection as needed and further analyzed as indicated.
Preparation of Nuclear and Cytoplasmic Fractions from Mammalian Cell Extracts and Western Blotting
Cells were harvested at 4 °C and resuspended in buffer NE (10 mm HEPES pH 7.9, 100 mm KCl, 0.5 mm EDTA, 1 mm DTT, 1 mm PMSF, 1.5 mm MgCl2), containing a mixture of protease inhibitors. The suspension was incubated on ice for 15 min, lysed by adding 1% Nonidet P-40 followed by vortexing for 10 s. The lysate was immediately centrifuged at 1000 × g for 5 min and the postnuclear supernatant (“Cytosolic Fraction”) was kept at 0–4 °C. The nuclear pellet was resuspended in the same volume of NE buffer as that of the cytosolic fraction and the resuspended nuclei were vortexed, briefly sonicated, incubated on ice for 30 min and centrifuged at 1000 × g for 5 min. The clarified supernatant was designated as the “Nuclear Fraction.”
For Western blot analysis of eIF6, an equal amount of each sample was subjected to SDS-polyacrylamide gel (10% gel) electrophoresis (SDS-PAGE) followed by transfer to Immobilon-P membranes (Millipore Corp.). Membranes were blocked in 5% nonfat milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 1 h and then treated with appropriate antibodies. After extensive washing, the immunoreactive bands were visualized by Enhanced Chemiluminescence (Amersham Biosciences). The Western bands were scanned, analyzed by densitometry using Image J software (National Institutes of Health, Bethesda, MD), and the results plotted as bar graphs in Microsoft Excel. In each case, data are the average of three individual experiments. The error bars show the standard deviation from the average value, thus reflecting the range of the three independent values obtained in each case.
Immunofluorescence Microscopy
All cells were grown on poly-l-lysine-coated glass cover slips and transfected with the appropriate mammalian eIF6 expression construct (wild-type or the phosphorylation-defective eIF6, as indicated) using Effectene transfection reagent (Qiagen). Following the indicated treatments, cells were fixed in 3% paraformaldehyde at room temperature and processed for immunofluorescence microscopy as described (19). Briefly, cells were permeabilized with solution I (0.2% saponin in 1× phosphate-buffered saline containing 1% fetal calf serum and 0.5% BSA as blocking agents). To detect Myc-tagged eIF6, samples were incubated at room temperature for 1 h with mouse monoclonal anti-Myc antibody at a dilution of 1:50 followed by incubation with Alexa Fluor goat anti-mouse IgG (secondary antibody) at a dilution of 1:200 for 1h. Nuclear staining with Hoechst 33258 was carried out after incubation with the secondary antibody for 10 min. Confocal images were taken using the Leica TCS SP2 AOBS Confocal microscope (Leica, Dearfield, IL) using a 63 oil immersion objective (1.4 NA). Laser lines at 405 and 546 nm were provided by 20 milliwatt Diode and 1.5 milliwatt HeNe lasers, respectively. The images (1024 × 1024 pixel dimension) were taken with a step size of 0.4 micron. Images were Z-stacked and saved as TIFF files. They were processed using the Image J software and Adobe Photoshop. The number of transfected cells showing nuclear, cytoplasmic or both nuclear and cytoplasmic staining for Myc-tagged eIF6 was counted in each case, and the percentage was calculated. At least 100–120 transfected cells were counted in a blindly fashion from a large number of random fields independently by three individuals for each set of experiments so that the percentage calculated was unbiased. Data were plotted in Microsoft Excel and, in each case, data are the average of three individual experiments. The error bars show the standard deviation from the average value, thus reflecting the range of the three independent values obtained in each case. It should be noted that in each immunofluorescence analysis, ∼15–20% of the transfected cell populations contained eIF6 both in the nucleus and in the cytoplasm. Except for Fig. 2, this population is not shown.
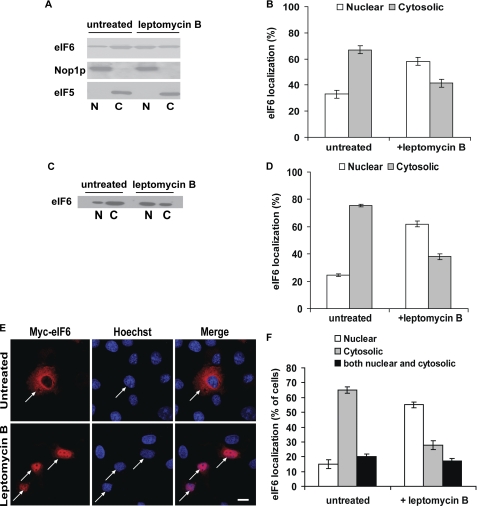
FIGURE 2.
Leptomycin B blocks nuclear export of eIF6 in mammalian cells. A, COS-7 cells, either treated with leptomycin B (10 ng/ml) for 1–2 h or untreated (control) as indicated, were fractionated into nuclear (N) and cytoplasmic (C) fractions. Each fraction was subjected to Western blot analysis to detect endogenous eIF6 using anti-eIF6 antibody as a probe. Nuclear and cytoplasmic fractions were also subjected to Western blot analysis using anti-Nop1p (a nuclear marker) and anti-eIF5 (a cytosolic marker), antibodies, respectively. B, relative distribution of endogenous eIF6 in the cytoplasmic and nuclear fractions of COS-7 cells, as presented in panel A, was quantified. C, COS-7 cells were transiently transfected with the Myc-His-tagged wild-type eIF6 construct. Cells were treated or not (untreated) with leptomycin B (10 ng/ml) as indicated. Nuclear (N) and cytoplasmic (C) fractions derived from extracts of above-mentioned COS-7 cells, were subjected to Western blot analysis using anti-eIF6 antibody as a probe. D, results presented in panel C were quantified. E, indirect immunofluorescence was performed with transiently transfected COS-7 cells, untreated or treated with leptomycin B, as indicated. Bar, 10 μm. F, number of cells showing nuclear, cytoplasmic or both nuclear and cytoplasmic staining under each condition was counted, and the percentage calculated. At least 100–120 transfected cells were examined in each case. Each experiment depicted in this figure was performed three separate times and data plotted are the average of the three independent experiments. For details of each experiment and the quantification of data, see sections on “Western blotting” (panels A–D) and “Immunofluorescence Microscopy” (panels E and F) under “Experimental Procedures.”
Immunoprecipitation Studies
COS-7 cells were transiently transfected with combinations of wild-type Myc-His-tagged eIF6 construct and HA-tagged calcineurin A and untagged calcineurin B constructs using Effectene transfection reagent (Qiagen) following the manufacturer's protocol. After 48 h, one set of cells was treated with 2 μm ionomycin for 1 h. Cells (ionomycin treated or untreated) were washed in cold PBS (phosphate buffered-saline) and harvested in TLB buffer (20 mm Tris pH 7.4, 137 mm NaCl, 25 mm β-glycerophosphate, 2 mm sodium pyrophosphate, 2 μm EDTA, 10% glycerol and 1% Triton X-100). Cells were incubated on ice for 15 min, scraped and centrifuged at 13000 rpm at 4° C. After preclearing, the lysate was used to immunoprecipitate eIF6 using rabbit polyclonal anti-eIF6 antibody, overnight at 4° C. Normal rabbit IgG was used as negative control in the immunoprecipitation reaction. HA- tagged calcineurin A and Myc-His-tagged eIF6 were detected by Western blot analysis using anti-HA and anti-His antibodies, respectively.
RESULTS
Intracellular Localization of eIF6 in Mammalian Cells
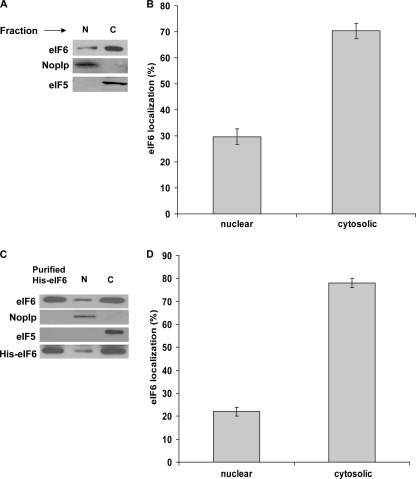
To investigate the relative intracellular distribution of endogenous eIF6, COS-7 cells were fractionated into nuclear and cytosolic fractions (“Experimental Procedures”). The validity of the fractionation was verified by using antibodies for Nop1p and eIF5 as markers for nuclei and cytoplasm, respectively (13) (Fig. 1, panel A). Western blot analysis using rabbit polyclonal anti-eIF6 antibodies showed that endogenous eIF6 was distributed between nucleus and cytoplasm. In agreement with the results previously published (3, 17), a major fraction of endogenous eIF6 was found in cytosol (∼70%), while a smaller but significant fraction of endogenous eIF6 (∼30%) was located in nucleus (Fig. 1, panels A and B). Likewise, when COS-7 cells were transiently transfected with an eIF6 cDNA construct that overexpressed Myc-His-tagged eIF6, the relative distribution of eIF6 (endogenous as well as expressed eIF6 taken together) between the cytoplasm and nuclei was found to be similar to that of the endogenous eIF6 (∼75–80% cytosolic) (Fig. 1, panels C and D). Nuclear and cytoplasmic fractions were also subjected to Western blot analysis using anti-His antibody to detect only the Myc-His-tagged eIF6 (Fig. 1, panel C). It is to be noted that the endogenous and the Myc-His-tagged eIF6 co-migrated together in 10% SDS-PAGE. Similar results were observed using 293T or HeLa cells (data not shown), excluding the possibility that eIF6 localization is a function of a particular cell type.
FIGURE 1.
Subcellular distribution of eIF6 in mammalian cells. A, COS-7 cells were fractionated into nuclear (N) and cytoplasmic (C) fractions. An aliquot of each fraction was subjected to Western blot analysis to detect endogenous eIF6 using anti-eIF6 antibody as a probe. Nuclear and cytoplasmic fractions were also subjected to Western blot analysis using anti-Nop1p antibodies, a nuclear marker, and anti-eIF5 antibodies, a bona fide cytosolic protein marker, respectively. B, relative distribution of endogenous eIF6 in the cytoplasmic and nuclear fractions of COS-7 cells, as presented in panel A, was quantified. C, nuclear (N) and cytoplasmic (C) fractions derived from extracts of COS-7 cells, transfected with the Myc-His-tagged wild type eIF6 construct, were subjected to Western blot analysis using anti-eIF6 antibody as a probe. Purified recombinant His-tagged-eIF6 was run in a parallel lane as a positive control. Nop1p and eIF5 served as nuclear and cytosolic markers, respectively. The nuclear and cytoplasmic fractions were also subjected to Western blot analysis using anti-His antibody as a probe. It should be noted that the endogenous and the Myc-His-tagged eIF6 migrated together in 10% SDS-PAGE. D, results presented in panel C were quantified. Each experiment depicted in this figure was performed three separate times, and data plotted are the average of the three independent experiments. The error bars show standard deviation from the average value, thus reflecting the range of the three independent values obtained in each case.
To test if eIF6 shuttles between the cytoplasm and the nuclei of mammalian cells, we first investigated whether endogenous eIF6 is exported from the nucleus to the cytosol. For this purpose, COS-7 cells were treated for 1–2 h with leptomycin B, a specific inhibitor of the nuclear export receptor protein CRM1 (20), and the intracellular localization of endogenous eIF6 in these cells was then compared with the untreated control cells by Western blot analysis using rabbit polyclonal anti-eIF6 antibodies (Fig. 2A). In agreement with the cell fractionation data described above (Fig. 1), Western blot analysis showed that in the control untreated cells, the majority of endogenous eIF6 was cytosolic (∼70%). In the leptomycin B-treated cells, however, endogenous eIF6 was found to be predominantly nuclear (∼60%) (Fig. 2, panels A and B). The subcellular distribution of the nuclear marker Nop1p and cytosolic marker eIF5 were not affected by leptomycin treatment (Fig. 2A). Likewise, when COS-7 cells were transiently transfected with Myc-His-tagged eIF6 construct, eIF6 (endogenous as well as expressed eIF6 taken together) was found to be predominantly nuclear (∼60%) in the presence of leptomycin B while the protein was mainly cytosolic (∼75%) in untreated transfected cells (Fig. 2, panels C and D). The subcellular distribution of the loading controls Nop1p and eIF5 (not shown) remained unaffected by leptomycin treatment, as expected.
We also examined the subcellular distribution of Myc-His-tagged eIF6 by indirect immunofluorescence microscopy using anti-Myc antibody (Fig. 2E). The indirect immunoflourescence studies revealed that Myc-His-tagged eIF6 was localized mainly in the cytosol in majority (∼65%) of the untreated transfected cell populations. However, there was also a significant transfected cell population where Myc-His-tagged eIF6 was either present both in the cytoplasm and the nuclei (∼20% of the transfected cell population) or primarily in the nuclei (∼15% of transfected cells). In contrast, in leptomycin-treated transfected cells, nearly 55% of the cell population examined had Myc-His-tagged eIF6 localized in the nuclei within 1 h of treatment (Fig. 2, panels E and F), while in the remaining transfected cell population Myc-His-tagged eIF6 was either distributed both in the nuclei and the cytoplasm or present primarily in the cytosol (Fig. 2, panels E and F). These results suggested that eIF6 is exported from the nucleus to the cytoplasm of mammalian cells.
Phosphorylation of eIF6 at Ser-174 and Ser-175 Regulates Its Subcellular Localization
In both mammalian and yeast cells, eIF6 is phosphorylated at Ser-174 (major site) and Ser-175 (minor site) by the nuclear isoforms of CK1 (CK1α or CK1δ in mammalian cells (13), and Hrr25p in yeast (10)). Ablation of eIF6 phosphorylation, either by replacement of Ser-174 and Ser-175 with alanine or by depletion of Hrr25p, the nuclear isoform of CK1 kinase in yeast cells, causes loss of yeast cell growth and viability (10, 13). These results demonstrate the physiological importance of eIF6 phosphorylation. However, the precise role of phosphorylation of eIF6 at these two serine sites was not clear.
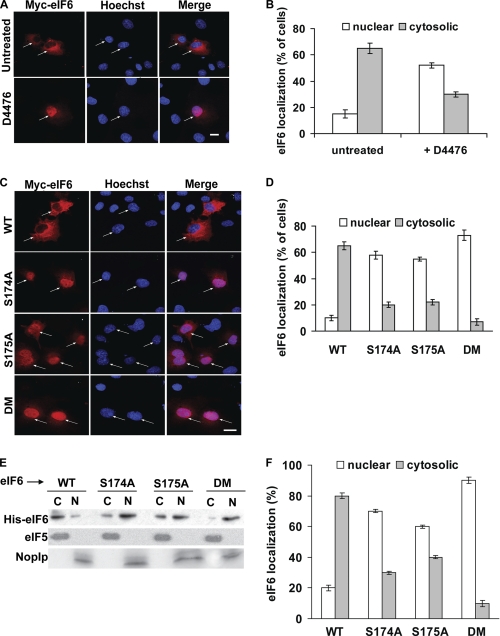
Previous studies indicated that in both mammalian and yeast cells, alteration in nucleo-cytoplasmic shuttling is a common consequence of CK1-mediated phosphorylation. For example, subcellular distribution of NFAT family of transcription factors in vertebrates (21–25) and a stress-responsive transcription factor Crz1p in yeast (26–28) is regulated by CK1 kinase. To understand the molecular basis of eIF6 phosphorylation by CK1 kinase, we first examined the subcellular distribution of Myc-His-tagged eIF6 in the presence of CK1 inhibitor D4476, by indirect immunofluorescence microscopy using anti-Myc antibody. Administration of D4476 resulted in the nuclear accumulation of the expressed Myc-His-tagged eIF6 in majority (∼55%) of the transiently transfected COS-7 cells (Fig. 3, panels A and B). These data indicate that CK1 regulates subcellular distribution of eIF6.
FIGURE 3.
Phosphorylation at Ser-174 and Ser-175 regulates nuclear export of eIF6. A, COS-7 cells, transiently transfected with the wild-type Myc-His-eIF6 construct were treated or not (untreated) for 2 h with 1 μm of CK1 inhibitor D4476 as indicated. Fixed cells were subjected to indirect immunofluorescence analysis to detect Myc-His-tagged eIF6 localization. Bar, 10 μm. B, quantification of the data of panel A. C, COS-7 cells were transiently transfected with Myc-His-tagged wild-type (WT) and mutant S174A, S175A, or S174,175A double mutant (DM) eIF6 constructs. Fixed cells were subjected to indirect immunofluorescence analysis to detect Myc-His-tagged eIF6 localization. Bar, 10 μm. D, number of cells showing nuclear, cytosolic, and both nuclear-cytosolic (not shown) staining was counted and expressed as the percentage of total cells viewed. E, aliquots of nuclear (N) and cytoplasmic (C) fractions derived from COS-7 cells transiently transfected with Myc-His-tagged wild-type and mutant S174A, S175A, or S174,175A double mutant (DM) eIF6 were subjected to Western blot analysis using anti-His antibody as a probe. Each fraction was also separately subjected to Western blot analysis using anti-eIF5 and anti-Nop1p antibodies to represent cytosolic and nuclear compartments, respectively. F, quantification of the data of panel E. Each experiment depicted in this figure was performed three separate times and data plotted are the average of the three independent experiments. For details of each experiment and the quantification of data, see sections on “Immunofluorescence Microscopy” (panels A–D) and “Western blotting” (panels E and F) under “Experimental Procedures.”
To explore the hypothesis that CK1-catalyzed phosphorylation at Ser-174 and Ser-175 regulates the nuclear export of eIF6, we prepared several pcDNA3.1-Myc-His-eIF6 expression constructs in which either Ser-174 or Ser-175 or both amino acids were mutated to alanine (“Experimental Procedures”). COS-7 cells were transiently transfected with either the wild-type (WT) eIF6 construct or each of the phosphorylation-defective eIF6 constructs (S174A, S175A, or S174,175A double mutant (DM)), and the relative distribution of the expressed Myc-His-tagged eIF6 was examined by immunofluorescence microscopy using anti-Myc antibody (Fig. 3C) and the results were quantified (Fig. 3D). Immunofluorescence analysis indicated that wild-type Myc-His-tagged eIF6 was predominantly cytosolic in the transfected cell population (∼65% of cells). Phosphorylation-defective Myc-His-tagged eIF6, however, was accumulated predominantly in the nucleus (Fig. 3, panels C and D). The effect was most pronounced for the S174,175A DM where nearly 80% of the transfected cell population showed the presence of eIF6 almost exclusively in their nuclei (Fig. 3D, subpanel DM). Similar subcellular distribution of the expressed Myc-His-tagged eIF6 was observed by Western blot analysis of the nuclear and the cytosolic fractions of the transfected cell lysates using anti-His antibody (Fig. 3, panels E and F). Taken together, the results presented in Fig. 3 suggest that phosphorylation at Ser-174 and Ser-175 by CK1 is required for the nuclear export of eIF6. It should be noted that this observation is in agreement with that reported previously from this laboratory (13) that lack of phosphorylation of yeast eIF6 (Tif6p) affects nuclear export of Tif6p and presumably of the pre-60 S ribosomal particles to which Tif6p is bound.
Ca2+-/Calmodulin-activated Calcineurin Phosphatase Associates with eIF6 in Vivo
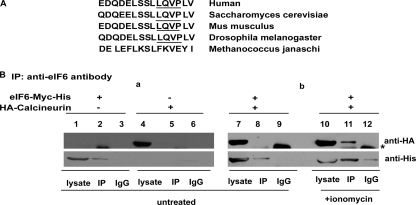
Results presented above showing that phosphorylation of eIF6 at Ser-174 and Ser-175 is required for eIF6 to be exported from the nucleus implies that the cytoplasmic eIF6 is the phosphorylated form of the protein. The question arises whether import of eIF6 from the cytoplasm to the nucleus is accompanied by dephosphorylation of the cytoplasmic eIF6. This hypothesis stems from the observations that nucleo-cytoplasmic shuttling of many mammalian and yeast proteins is regulated by signals that result in changes in their phosphorylation status. For example, nuclear accumulation of NFAT family of transcription factors in vertebrates and the stress-responsive transcription factor Crz1p in yeast is regulated by dephosphorylation mediated by the calcium/calmodulin-dependent calcineurin phosphatase (21–28). Indeed, examination of the amino acid sequence of eIF6 shows that, in addition to a conserved docking motif for CK1 (FXXXF, where X is any amino acid) (29) at amino acid positions 34–38, and well-characterized CK1 phosphorylation sites at Ser-174 and Ser-175, the protein also possesses a binding motif LXVP for the Ca2+-regulated protein phosphatase calcineurin (30). This putative calcineurin binding motif LQVP is present at amino acid positions 177–180, which is immediately adjacent to the CK1 phosphorylation sites at Ser-174 and Ser-175 (Fig. 4A). The presence of a calcineurin docking motif suggested that eIF6 might be regulated by calcineurin similar to the mammalian transcription factor NFAT.
FIGURE 4.
Ca2+/calmodulin-activated calcineurin associates with eIF6 in vivo. A, conserved amino acid residues surrounding the serine CK1 phosphorylation sites (Ser-174 and Ser-175) of eIF6 contains a putative binding site LxVP (30) for calcineurin in nucleated species. The putative calcineurin LQVP binding site is underlined. B, COS-7 cells were transiently transfected with combinations of Myc-His-tagged eIF6 construct and HA-tagged calcineurin A and untagged calcineurin B constructs as indicated. Transfected cells (48 h post-transfection), were either treated with 2 μm ionomycin or left untreated for 1 h, as indicated. eIF6 was immunoprecipitated using anti-eIF6 antibody (see “Experimental Procedures” for details). HA-tagged calcineurin A subunit was detected by Western blot analysis using anti-HA antibody. Myc-His-tagged eIF6 was also detected in Western blot analysis by anti-His antibody. Asterisk (*) denotes IgG heavy chain.
Calcineurin, a Ca2+/calmodulin-dependent protein phosphatase, is a heterodimer consisting of a catalytic subunit A and a regulatory subunit B. Although both yeast and mammalian cells contain many calcineurin substrates, most of our knowledge on the mechanism of calcineurin-mediated dephosphorylation reaction has been derived from extensive studies on NFAT family of transcription factors that are essential for cytokine gene expression necessary for T cell activation (21–25). These studies revealed the existence of two distinct binding motifs on NFAT that are used by calcineurin as its docking site on the substrate. The main docking site with a consensus motif PXIXIT is located near the N terminus of the regulatory region of NFAT (31). Interaction of calcineurin with this motif occurs through its catalytic domain that lies in the proximity to its active site (32). In addition to the PXIXIT binding motif, a second calcineurin binding motif with a moderately conserved consensus sequence, LXVP lies near the C terminus of the NFAT regulatory region (33). Calcineurin binds to this site at a hydrophobic pocket formed at the interface of its two subunits A and B (30). This hydrophobic pocket formed at the interface of the two calcineurin subunits is targeted by the immunosuppressant-immunophilin complexes (e.g. cyclosporin A (CsA)-cyclophilin or FK506-FK binding protein 12) (30, 33). Indeed, immunosuppressants CsA and FK506 have been widely used as specific potent inhibitors of calcineurin-mediated dephosphorylation reaction in vivo (23–28).
The presence of a putative calcineurin binding LXVP motif (Fig. 4, panel A) prompted us to investigate whether Ca2+-activated calcineurin interacts with the cytosolic eIF6, presumably causing its dephosphorylation and promoting its nuclear import. To test this hypothesis, we transiently transfected COS-7 cells with combinations of Myc-His-tagged eIF6 and HA-tagged calcineurin A and untagged calcineurin B cDNA constructs and analyzed cell lysates for eIF6-calcineurin A complexes by immunoprecipitating eIF6. Co-immunoprecipitation analysis showed that calcineurin interacted with eIF6-Myc-His under conditions when calcineurin was activated by calcium ionophore, ionomycin (Fig. 4B, subpanel b, lane 11). Calcineurin, however, did not interact with eIF6 under unstimulated condition (Fig. 4B, subpanel b, lane 8). These data indicate that eIF6 associates with Ca2+-activated calcineurin in vivo.
Nuclear Import of eIF6 from the Cytoplasm Requires Ca2+-dependent Serine-Threonine Protein Phosphatase Calcineurin
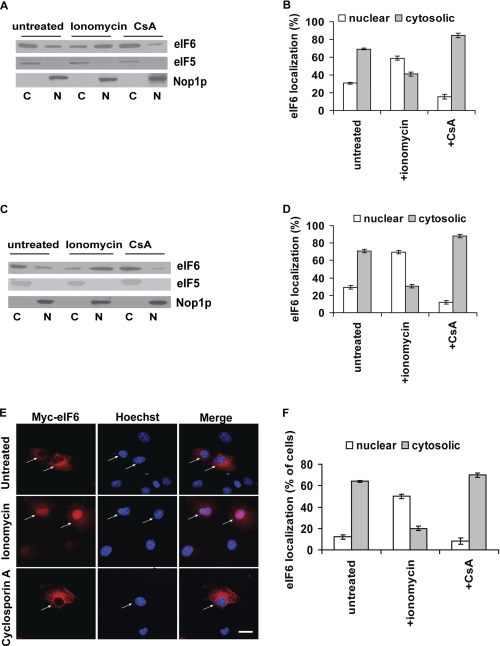
Dephosphorylation mediated by calcineurin might regulate the nuclear import of eIF6. The effect of calcium ionophore ionomycin or calcineurin inhibitor cyclosporin A (CsA) on intracellular localization of endogenous eIF6 in COS-7 cells was examined by Western blot analysis using rabbit polyclonal anti-eIF6 antibodies (Fig. 5, panels A and B). We also examined the effect of ionomycin and CsA on transiently transfected Myc-His-tagged eIF6 in COS-7 cells by Western blot analysis using rabbit polyclonal anti-eIF6 antibodies (Fig. 5, panels C and D) and by indirect immunofluorescence microscopy using anti-Myc antibody (Fig. 5, panels E and F). In all cases, we observed that whereas wild-type eIF6 was predominantly cytosolic in untreated cells, ionomycin promoted nuclear translocation of eIF6 within 30 min of treatment (Fig. 5, panels A–F). In contrast, treatment of cells with the calcineurin inhibitor cyclosporin A, led to nearly complete cytosolic localization of eIF6 (Fig. 5, panels A–F). The nuclear marker Nop1p and cytosolic marker eIF5 were not affected by ionomycin or cyclosporin A treatment (Fig. 5, panels A and C).
FIGURE 5.
Nuclear import of eIF6 from the cytoplasm is sensitive to calcineurin activator and inhibitor in vivo. A, COS-7 cells were treated either with 2 μm calcium ionophore ionomycin to activate calcineurin, or with 5 μm CsA, a specific calcineurin inhibitor. Nuclear (N) and cytoplasmic (C) fractions derived from extracts of these cells and also from control untreated cells, were subjected to Western blot analysis to detect endogenous eIF6 using anti-eIF6 antibody as a probe. B, relative distribution of endogenous eIF6 in the cytoplasmic and nuclear fractions of the COS-7 cells, as presented in panel A, was quantified. C, COS-7 cells, transiently transfected with the Myc-His-tagged eIF6 constructs, were treated either with 2 μm ionomycin, or with 5 μm cyclosporin A. Nuclear and cytoplasmic fractions derived from extracts of these cells and also from control untreated transfected cells, were subjected to Western blot analysis using anti-eIF6 antibody as a probe. D, data of panel C were quantified. E, COS-7 cells transiently transfected with the Myc-His tagged eIF6 constructs were treated either with 2 μm ionomycin, or with 5 μm cyclosporin A and Myc-His-tagged eIF6 was detected by indirect immunofluorescence staining. Bar, 10 μm. F, data of panel E were quantified. Each experiment depicted in this figure was performed three separate times, and data plotted are the average of the three independent experiments. Experiments of panels A–D were carried out and quantified as described under “Western blotting” (under “Experimental Procedures”), while those in panels E and F were carried out and quantified as described under “Immunofluorescence Microscopy” (under “Experimental Procedures”).
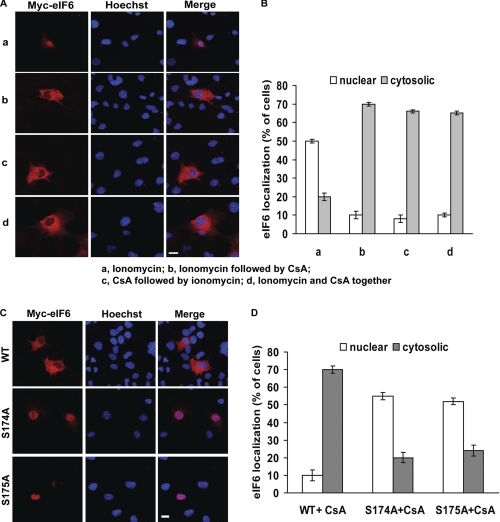
We also examined the kinetics of administration of ionomycin and CsA on the nuclear localization of eIF6 (Fig. 6A). While administration of ionomycin induced nuclear accumulation of Myc-His-tagged eIF6 (Fig. 6A, subpanel a), addition of cyclosporin A to ionomycin-stimulated transfected cells resulted in reappearance of the expressed eIF6 from the nucleus to the cytosol (Fig. 6A, subpanel b). Preincubation of the transfected cells with CsA prior to ionomycin stimulation inhibited nuclear translocation of the expressed eIF6 by ionomycin (Fig. 6A, subpanel c). Administration of ionomycin and cyclosporin A at the same time also resulted in cytosolic localization of the expressed eIF6 (Fig. 6A, subpanel d). The quantification of the results of Fig. 6A is presented in Fig. 6B.
FIGURE 6.
Nuclear export mediated by phosphorylation at Ser-174 and Ser-175 of eIF6 is sensitive to calcineurin inhibitor CsA. A, COS-7 cells were transiently transfected with wild-type Myc-His-tagged eIF6. In subpanel a, the cells were treated with 2 μm ionomycin for 30 min to induce nuclear localization of eIF6. In subpanel b, ionomycin-treated cells were further treated with 5 μm cyclosporin A (CsA) for 30 min before fixation. In subpanel c, cells were first treated with 5 μm cyclosporin A for 30 min followed by additional treatment with 2 μm ionomycin for another 30 min. In subpanel d, cells were simultaneously treated for 30 min with both ionomycin (2 μm) and cyclosporin A (5 μm). eIF6 localization was monitored under each condition by indirect immunofluorescence. (It should be noted that the immunofluorescence data of untreated control of panel A is not shown since the experiment presented in panel A was carried out together with that shown in Fig. 5E as a single set of experiments.) Bar, 10 μm. B, data of panel A were quantified. C, COS-7 cells were transiently transfected with wild-type Myc-His-tagged eIF6 or phosphorylation-defective eIF6 (S174A and S175A mutants). Transfected cells were treated with 5 μm cyclosporin A for 30 min. eIF6 localization was monitored under each condition by indirect immunofluorescence. Bar, 10 μm. D, data of panel C were quantified. (In each experiment depicted in this figure, indirect immunofluorescence was performed and analyzed as described under “Immunofluorescence Microscopy” under “Experimental Procedures”.)
Next, we tested the effect of CsA on the nuclear localization of phosphorylation defective eIF6 (S174A or S175A mutant). The prediction was if calcineurin regulates eIF6 through dephosphorylation of Ser-174 and/or Ser-175, administration of CsA would not affect the nuclear localization of phosphorylation-defective eIF6. While the phosphorylation-defective eIF6 was found mainly in the nucleus, their nuclear localization was no longer affected by CsA (Fig. 6, panels C and D). These data indicate that the effect of cyclosporin A on intracellular localization of eIF6 is abrogated by Ala replacement of Ser-174 or Ser-175 of eIF6.
eIF6 Shuttles between the Nucleus and the Cytoplasm
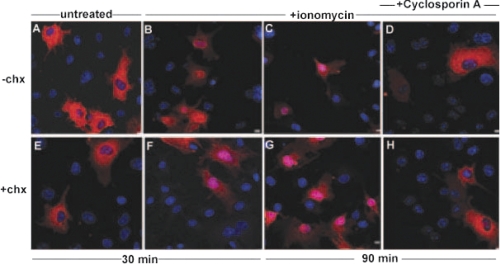
The reversible localization of eIF6 may be due to shuttling of pre-existing eIF6 molecules between the nucleus and the cytoplasm. Alternatively, eIF6 may be degraded following its entry and function in the nucleus and reappearance of eIF6 in the cytoplasm may be due to de novo eIF6 protein synthesis. To distinguish between these two possibilities, COS-7 cells, transiently transfected with the wild-type Myc-His-tagged eIF6 were treated for 1 h with a potent protein synthesis inhibitor cycloheximide (chx). Incubation of cycloheximide-treated cells with the calcineurin activator ionomycin resulted in the nuclear translocation of eIF6 similar to the results obtained with untreated cells (Fig. 7, compare panels E–G with panels A–C). This nuclear translocation of eIF6 could be reversed by the calcineurin inhibitors, cyclosporin A (Fig. 7, compare panels D and H) or FK520 (data not shown), even in the presence of cycloheximide. These results suggest that the same pool of eIF6 enters and exits the nucleus, which is regulated by calcineurin and CK1, respectively.
FIGURE 7.
Wild-type eIF6 shuttles between nucleus and cytoplasm independent of de novo protein synthesis. COS-7 cells, transiently transfected with the Myc-His-tagged wild-type eIF6 construct were pretreated (+chx, panels E–H) or not (−chx, panels A–D) with 10 μg/ml of cycloheximide (chx). Following incubation for 1 h, cells in all panels, except A and E, were treated with 2 μm ionomycin (panels B–D, F–H) as indicated. An aliquot of ionomycin-treated cells (panels B and F) as well as cells not treated with ionomycin (panels A and E) were monitored by immunofluorescence after 30 min of incubation. After an additional 30 min, another aliquot of ionomycin-treated cells were treated with 5 μm cyclosporin A, and cells were observed after an additional 30 min of incubation (panels D and H). Cells incubated with ionomycin for a total period of 90 min, but not treated with cyclosporin A, were also monitored (panels C and G). In each case, localization of eIF6 was monitored by its Myc tag (red) while the nuclei were stained with Hoechst (blue). Only the merged figures are shown here. Bar, 10 μm.
DISCUSSION
Many proteins shuttle continuously back and forth between the nucleus and cytoplasm of eukaryotic cells. The nucleo-cytoplasmic distribution of some of these proteins, particularly those involved in signaling, is known to be regulated. Some of the proteins undergo regulated binding and release from the receptors while the other group containing nuclear localization signals (NLSs) is regulated by masking and unmasking of NLSs. Both the release from the binding sites or exposure of NLS can be brought about by ligand binding, phosphorylation or dephosphorylation, or, in some cases, by proteolysis. Phosphorylation or dephosphorylation may cause conformational changes exposing a NLS or altering protein-protein interactions for proteins that do not contain any NLS (21, 22, 34).
eIF6, a highly conserved protein between yeast and mammals, does not appear to contain any NLS or nuclear export signal (NES). However, both the yeast and mammalian eIF6 are phosphorylated at Ser-174 and Ser-175 by the nuclear isoform of CK1 (10, 13) at a highly conserved amino acid sequence that also contains a putative calcineurin binding motif LXVP (30, 33) immediately adjacent to the CK1 phosphorylation sites (Fig. 4A). These observations prompted us to examine whether phosphorylation and dephosphorylation are involved in the nuclear export and import of eIF6.
In the present work, we present several lines of evidence which suggest that eIF6 shuttles back and forth between the nucleus and the cytoplasm of mammalian cells. This process is dependent on phosphorylation and dephosphorylation of eIF6 at Ser-174 and Ser-175 mediated by CK1 and the Ca2+/calmodulin-dependent protein phosphatase calcineurin, respectively. We show that the protein phosphatase calcineurin, following its activation by Ca2+, associates with eIF6 (Fig. 4) and thus appears to play an essential role in eIF6 nuclear import, as the localization of eIF6 changes from cytosolic to nuclear subsequent to calcineurin activation in vivo (Fig. 5). This event is blocked by the immunosuppressive drug cyclosporin A, that is known to be a specific calcineurin inhibitor (Figs. 5 and 6) suggesting that the dephosphorylated form of eIF6 is imported to the nucleus. The nuclear export of eIF6, on the other hand, requires rephosphorylation of Ser-174 and Ser-175. Failure to phosphorylate at these sites either by mutation of the serine residues to alanine or treatment of cells with a CK1 inhibitor causes a significant fraction of eIF6 to be retained in the nucleus (Fig. 3). These observations also suggest that it is the phosphorylated form of eIF6 that is exported to the cytoplasm. Additionally, nuclear import and export of eIF6 occur even in the presence of a potent protein synthesis inhibitor cycloheximide and is therefore a reflection of dynamics of import and export of pre-existing eIF6 molecules rather than de novo new eIF6 synthesis. It should be noted that under normal growth conditions of cells in culture, although eIF6 is mostly localized in the cytoplasm of mammalian cells, it, in fact, shuttles continuously between the nucleus and the cytoplasm. This is evident from our observations that inhibition of phosphorylation of eIF6 or addition of leptomycin B to the cell culture results in predominantly nuclear localization of eIF6, while inhibition of calcineurin activity by addition of cyclosporin A to the culture results in nearly total accumulation of eIF6 in the cytoplasm.
The question naturally arises why shuttling of eIF6 is required. eIF6 is essential for 60 S ribosome biogenesis, that is mostly a nucleolar function. Its association with the pre-60 S ribosomal particles is also required for the export of the pre-60 S particles from the nucleus to the cytoplasm where the final maturation of pre-60 S particles to mature 60 S ribosomes occurs. Because eIF6 is a limiting component, it is imperative that following its release from the pre-60 S ribosomal particles in the cytoplasm, it must recycle back to the nucleolus for continued 60 S ribosome biogenesis. Furthermore, the requirement of Ca2+-activated calcineurin phosphatase in the nuclear import of eIF6 suggests that calcineurin may also play a role in 60 S ribosome biogenesis. Additionally, the requirement of calcineurin, in principle, could also provide a means by which changes in intracellular Ca2+ level could modulate nuclear import of eIF6 and, consequently, nucleolar 60 S ribosome biogenesis. Clearly, additional studies will be necessary to answer these questions.
Nucleo-cytoplasmic shuttling of eIF6 described above is analogous to that observed for the mammalian NFAT family of transcription factors that regulate cytokine gene transcription during T-cell activation (21, 23–25) and the yeast stress-responsive transcription factor Crz1p (26–28). These transcription factors are present in mammalian or yeast cells, respectively, in an inactive state in the cytoplasm as phosphoproteins and appear to depend on nuclear import for their function in vivo. In response to signal-mediated elevated intracellular Ca2+ concentration, activated calcineurin dephosphorylates critical serine residues in these cytosolic transcription factors and allows these transcription factors to be imported to the nucleus for target gene activation. Cessation of Ca2+ signaling results in rephosphorylation of these transcription factors in the nucleus by the nuclear kinases (CK1 for Crz1p (28) and a combination of CK1 and GSK3 for NFAT transcription factor (29)) and allows these proteins to be exported to the cytosol as phosphoproteins (21–28). It should, however, be emphasized that the present work is the first to show nucleo-cytoplasmic shuttling of an essential ribosome biogenesis factor.
In view of our observation that eIF6 is phosphorylated by the nuclear isoform of CK1 and phosphorylation of eIF6 signals its nuclear export, there must exist a mechanism to prevent eIF6 phosphorylation in the nucleus before it has completed its function in the nucleus. In case of NFAT transcription factor, it has been shown that Ca2+ signaling induces an association between NFAT and calcineurin (23, 25) and these molecules are transported as a complex to the nucleus, where calcineurin continues to dephosphorylate NFAT4 in the nucleus antagonizing the effect of the nuclear kinase. We have shown that Ca2+-signaling (via ionomycin) also induces association between eIF6 and calcineurin (Fig. 4). It will be of interest to investigate whether these molecules are also transported as a complex to the nucleus. Alternatively, the possibility also exists that like the yeast system (9, 35), mammalian eIF6, following its entry to the nucleus, associates with the pre-66 S ribosomal RNP particles in the nucleolus to which a large number of other trans-acting proteins as well as many 60 S ribosomal proteins are also bound. These bound trans-acting proteins may mask direct interaction between bound eIF6 and the nuclear kinase. During subsequent maturation of the pre-66 S particles, most of these bound trans-acting factors are sequentially released (35). When the pre-60 S particles, containing bound eIF6, are close to be exported from the nucleus, eIF6 may then become accessible to the nuclear kinase for phosphorylation.
An important question that still awaits investigation is whether differences exist in the manner in which yeast eIF6 and mammalian eIF6 are recycled between the nucleus and the cytoplasm. This question arises because unlike mammalian eIF6, yeast eIF6 is predominantly localized to the nucleolus in exponentially growing cells (11, 12). However, like mammalian eIF6, the yeast eIF6 is also phosphorylated at Ser-174 and Ser-175 (10) in a highly conserved amino acid sequence that also contains a calcineurin binding motif LXVP (30) immediately adjacent to the CK1 phosphorylation site (Fig. 4A). Additionally, like mammalian eIF6, lack of phosphorylation of yeast eIF6 also affects its nuclear export (13). Thus the possibility exists that yeast eIF6 may also undergo nucleo-cytoplasmic shuttling similar to that described here for mammalian eIF6. The marked difference in the localization pattern of eIF6 in mammalian and yeast cells seems to suggest that fundamental differences may exist in the kinetics of nucleo-cytoplasmic shuttling of eIF6 between yeast and mammalian cells. Further work on yeast eIF6 is necessary to answer this question.
Is eIF6 phosphorylated only at Ser-174 and Ser-175 in mammalian cells? It has been reported that eIF6 is also phosphorylated by protein kinase C (PKC) in a reaction requiring Rack1 (17). PKC-mediated phosphorylation of eIF6 in the cytoplasm has been implicated to be required for the release of bound eIF6 from the 60 S ribosomal particles (17). However, the site of phosphorylation has not been directly determined. It has been inferred from deletion studies. Additionally, Rack1 is a 40 S ribosome-interacting protein (36, 37) while eIF6 is bound to the pre-60 S particles prior to its release in the cytoplasm. Thus, it is difficult to visualize a model in which a 60 S-bound eIF6 interacts with the Rack1-PKC complex. Furthermore, as far as the release of eIF6 from the pre-60 S particles is concerned, in yeast cells, the release is achieved by interaction with two cytoplasmic proteins Efl1p and Sdo1p (11, 12). Both these proteins have corresponding mammalian homologues, implying that these two proteins may also be involved in the release of eIF6 from the pre-60 S particles in the cytoplasm of mammalian cells. Clearly, further work is necessary to characterize the mechanism of release of eIF6 from the pre-60 S ribosomal particles in the cytoplasm of mammalian cells.
Acknowledgments
We thank Dr. Jayanta Chaudhuri of the Sloan-Kettering Cancer Research Center, New York and Dr. Romit Majumdar, formerly of the Albert Einstein College of Medicine, for critically reading the manuscript.
This work was supported, in whole or in part, by Grant GM15399 from the National Institutes of Health, by Cancer Core Support Grant P30CA13330 from the NCI, and institutional support from the Albert Einstein College of Medicine.
- eIF6
- eukaryotic (translation) initiation factor 6
- Tif6p
- yeast eIF6
- CK1
- casein kinase1
- CsA
- cyclosporin A
- rRNA
- ribosomal RNA
- Ni-NTA
- nickel-nitrilotriacetic acid
- DM
- double mutant at Ser-174 and Ser-175 of eIF6
- NLS
- nuclear localization signal.
REFERENCES
- 1. Si K., Chaudhuri J., Chevesich J., Maitra U. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14285–14290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Si K., Maitra U. (1999) Mol. Cell. Biol. 19, 1416–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Russell D. W., Spremulli L. L. (1979) J. Biol. Chem. 254, 8796–8800 [PubMed] [Google Scholar]
- 4. Russell D. W., Spremulli L. L. (1980) Arch. Biochem. Biophys. 201, 518–526 [DOI] [PubMed] [Google Scholar]
- 5. Valenzuela D. M., Chaudhuri A., Maitra U. (1982) J. Biol. Chem. 257, 7712–7719 [PubMed] [Google Scholar]
- 6. Raychaudhuri P., Stringer E. A., Valenzuela D. M., Maitra U. (1984) J. Biol. Chem. 259, 11930–11935 [PubMed] [Google Scholar]
- 7. Sanvito F., Piatti S., Villa A., Bossi M., Lucchini G., Marchisio P. C., Biffo S. (1999) J. Cell Biol. 144, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Basu U., Si K., Warner J. R., Maitra U. (2001) Mol. Cell. Biol. 21, 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C. J., Shabanowitz J., Hunt D. F., Woolford J. L., Jr. (2001) Mol. Cell 8, 505–515 [DOI] [PubMed] [Google Scholar]
- 10. Ray P., Basu U., Ray A., Majumdar R., Deng H., Maitra U. (2008) J. Biol. Chem. 283, 9681–9691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Senger B., Lafontaine D. L., Graindorge J. S., Gadal O., Camasses A., Sanni A., Garnier J. M., Breitenbach M., Hurt E., Fasiolo F. (2001) Mol. Cell 8, 1363–1373 [DOI] [PubMed] [Google Scholar]
- 12. Menne T. F., Goyenechea B., Sánchez-Puig N., Wong C. C., Tonkin L. M., Ancliff P. J., Brost R. L., Costanzo M., Boone C., Warren A. J. (2007) Nat. Genet. 39, 486–495 [DOI] [PubMed] [Google Scholar]
- 13. Basu U., Si K., Deng H., Maitra U. (2003) Mol. Cell. Biol. 23, 6187–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gross S. D., Anderson R. A. (1998) Cell Signal. 10, 699–711 [DOI] [PubMed] [Google Scholar]
- 15. Ho J. H., Kallstrom G., Johnson A. W. (2000) J. Cell Biol. 151, 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson A. W., Lund E., Dahlberg J. (2002) Trends Biochem. Sci. 27, 580–585 [DOI] [PubMed] [Google Scholar]
- 17. Ceci M., Gaviraghi C., Gorrini C., Sala L. A., Offenhäuser N., Marchisio P. C., Biffo S. (2003) Nature 426, 579–584 [DOI] [PubMed] [Google Scholar]
- 18. Perrino B. A., Fong Y. L., Brickey D. A., Saitoh Y., Ushio Y., Fukunaga K., Miyamoto E., Soderling T. R. (1992) J. Biol. Chem. 267, 15965–15969 [PubMed] [Google Scholar]
- 19. Mukherjee S., Chiu R., Leung S. M., Shields D. (2007) Traffic 8, 369–378 [DOI] [PubMed] [Google Scholar]
- 20. Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crabtree G. R., Olson E. N. (2002) Cell 109, (suppl.), S67–S79 [DOI] [PubMed] [Google Scholar]
- 22. Mattaj I. W., Englmeier L. (1998) Annu. Rev. Biochem. 67, 265–306 [DOI] [PubMed] [Google Scholar]
- 23. Shibasaki F., Price E. R., Milan D., McKeon F. (1996) Nature 382, 370–373 [DOI] [PubMed] [Google Scholar]
- 24. Timmerman L. A., Clipstone N. A., Ho S. N., Northrop J. P., Crabtree G. R. (1996) Nature 383, 837–840 [DOI] [PubMed] [Google Scholar]
- 25. Loh C., Shaw K. T., Carew J., Viola J. P., Luo C., Perrino B. A., Rao A. (1996) J. Biol. Chem. 271, 10884–10891 [DOI] [PubMed] [Google Scholar]
- 26. Stathopoulos A. M., Cyert M. S. (1997) Genes. Dev. 11, 3432–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stathopoulos-Gerontides A., Guo J. J., Cyert M. S. (1999) Genes. Dev. 13, 798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kafadar K. A., Zhu H., Snyder M., Cyert M. S. (2003) Genes. Dev. 17, 2698–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okamura H., Garcia-Rodriguez C., Martinson H., Qin J., Virshup D. M., Rao A. (2004) Mol. Cell. Biol. 24, 4184–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodríguez A., Roy J., Martínez-Martínez S., López-Maderuelo M. D., Niño-Moreno P., Ortí L., Pantoja-Uceda D., Pineda-Lucena A., Cyert M. S., Redondo J. M. (2009) Mol. Cell 33, 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aramburu J., Garcia-Cózar F., Raghavan A., Okamura H., Rao A., Hogan P. G. (1998) Mol. Cell 1, 627–637 [DOI] [PubMed] [Google Scholar]
- 32. Li H., Rao A., Hogan P. G. (2004) J. Mol. Biol. 342, 1659–1674 [DOI] [PubMed] [Google Scholar]
- 33. Liu J., Arai K., Arai N. (2001) J. Immunol. 167, 2677–2687 [DOI] [PubMed] [Google Scholar]
- 34. Nigg E. A. (1997) Nature 386, 779–787 [DOI] [PubMed] [Google Scholar]
- 35. Nissan T. A., Bassler J., Petfalski E., Tollervey D., Hurt E. (2002) EMBO J. 21, 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sengupta J., Nilsson J., Gursky R., Spahn C. M., Nissen P., Frank J. (2004) Nat. Struct. Mol. Biol. 11, 957–962 [DOI] [PubMed] [Google Scholar]
- 37. Nilsson J., Sengupta J., Frank J., Nissen P. (2004) EMBO Rep. 5, 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]