Abstract
Ricin A chain (RTA) inhibits protein synthesis by removing a specific adenine from the highly conserved α-sarcin/ricin loop in the large rRNA. Expression of RTA with its own signal sequence in yeast resulted in its translocation into the endoplasmic reticulum (ER) and subsequent glycosylation. Because RTA must unfold within the ER, it may be vulnerable to host defenses, such as the unfolded protein response (UPR). UPR was induced in cells expressing an active site mutant but not the wild type RTA, indicating that the active site of RTA played a role in perturbing the ER stress response. The inactive RTA without the signal sequence did not induce UPR, indicating that translocation into the ER was critical for induction of UPR. The wild type RTA inhibited activation of UPR not only due to ER stress induced by the protein itself but also by global effectors such as tunicamycin and dithiothreitol. Mature RTA without the signal sequence also inhibited UPR, providing evidence that inhibition of UPR occurred on the cytosolic face of the ER. RTA could not inhibit UPR when the spliced form of HAC1 mRNA was provided in trans, indicating that it had a direct effect on UPR upstream of HAC1-dependent transcriptional activation. Only the precursor form of HAC1 mRNA was detected in cells expressing RTA after exposure to ER stress, demonstrating that ricin inhibits activation of UPR by preventing HAC1 mRNA splicing. The RTA mutants that depurinated ribosomes but did not kill cells were not able to inhibit activation of UPR by tunicamycin, providing evidence that the inability to activate UPR in response to ER stress contributes to the cytotoxicity of ricin.
Ricin is a member of a class of molecules termed ribosome-inactivating proteins that remove a highly conserved adenine from the α-sarcin ricin loop of the large rRNA (1). The depurinated ribosome is no longer capable of fulfilling its role in protein synthesis (2). This rapidly lethal mechanism for debilitating and destroying cells has earned ricin the recent spotlight as a potential bioweapon. Ricin is produced in the seeds of castor bean, Ricinus communis. It is a heterodimeric toxin consisting of a catalytic A chain (RTA)3 covalently joined by a disulfide bond to a cell binding B chain (3, 4). Due to its potent cytotoxicity and wide availability, ricin has been classified as a category B priority by the Centers for Disease Control and Prevention. Currently there are no specific medical treatment options for ricin intoxication. To gain insight into the mechanism of ricin-induced cell death, we recently isolated a panel of nontoxic RTA mutants based on their inability to kill yeast cells (5). Analysis of these mutants provided evidence that ribosome depurination and translation inhibition are not sufficient for ricin-mediated cell death (5), suggesting that an extra-ribosomal activity of RTA may play a role in its cytotoxicity.
Ricin possesses several unique features to overcome the cellular defenses before reaching the ribosome as a single chain ribosome-inactivating protein. In mammalian cells the holotoxin binds initially to terminal galactose-containing cellular receptors via ricin B chain. After endocytosis, a small fraction of ricin navigates through the trans-Golgi network and enters the endoplasmic reticulum (ER) via retrograde transport (4, 6, 7). Upon entry into the ER, the disulfide bonds of holotoxin are reduced, allowing RTA to separate from the B chain. RTA then enters the cytosol from the ER to reach its target (6). The mechanism by which AB toxins enter the cytosol and how they evade degradation by the ubiquitin proteasome pathway are poorly understood. RTA is thought to enter the cytosol from the ER using the ER-associated degradation (ERAD) pathway (8, 9). A fraction of RTA escapes the ubiquitin mediated-degradation in the cytosol ultimately reaching ribosomes (10). Ricin undergoes conformational changes in the ER, including but not limited to reduction of disulfide bonds, partial unfolding, and transporter binding (4). During any of these manipulations, RTA may be vulnerable to host defenses designed to destroy foreign proteins. One of these defenses that RTA is likely to encounter is the ER-to-nucleus signaling pathway called the unfolded protein response (UPR).
UPR is triggered in cells upon accumulation of unfolded proteins within the ER. Various stresses, such as glucose starvation, inhibition of glycosylation, reducing conditions, alteration of calcium concentration, and viral infection can also trigger this defense. Several human diseases, are a result of perturbation in UPR, several human diseases, such as diabetes, neurodegenerative disorders, and cancer, are a result of perturbation in UPR (11). Accumulation of unfolded proteins in the ER in yeast is sensed by a transmembrane protein kinase/ribonuclease, Ire1p, that transmits a signal from the ER to the nucleus (12, 13). Ire1p is an ER resident transmembrane protein whose amino-terminal domain is located in the ER lumen and functions as a sensor for the accumulation of unfolded proteins (13). Its carboxyl-terminal cytosolic portion contains Ser/Thr protein kinase and endoribonuclease domains (12, 13). Accumulation of unfolded or misfolded proteins in the ER causes oligomerization and autophosphorylation of Ire1p (14). The cytosolic endoribonuclease domain of Ire1p is then activated and excises the intron in the unspliced HAC1 mRNA, Hac1u (u for uninduced), generating spliced HAC1 mRNA, Hac1i (i for induced) (15–17). The splicing event is so specific that Hac1u mRNA has been shown to be the only substrate for Ire1p in yeast (18). A tRNA ligase Rlg1p is then required to rejoin the cleaved ends (19). Hac1u is not a favorable mRNA for protein translation, but after splicing Hac1i is translated very efficiently, and its product, the basic leucine zipper (bZIP) transcription factor Hac1p activates transcription of the UPR target genes by binding to the unfolded protein response element (UPRE) (20). Activation of UPR reestablishes the protein folding capacity of the cell by increasing of genes encoding ER-resident chaperones and ERAD components that reduce ER stress by directing misfolded proteins from the ER to the cytosol for degradation by the 26 S proteasome (21–23).
The consequences of UPR induction in mammalian and yeast systems differ in some fundamental aspects. For example, the upstream modulator of UPR in yeast cells is limited to Ire1p and its activity on HAC1 mRNA. In animal cells, one modulator is Ire1α and its substrate is the XBP1 mRNA. However, the basic leucine-zipper transcription factor ATF6 and the PERK kinase also play equally important roles in the induction of UPR (11, 24). Because yeast lacks both ATF6 and PERK, it provides an ideal system to study the PERK- and ATF6-independent pathways of the ER stress response.
Considering RTA must unfold at least partially within the ER, it would seem likely that activation of UPR would occur. Because activation of UPR leads to expression of ERAD components, induction of this stress response may be deleterious to ricin. Ricin may have evolved to counteract host defenses within the ER, including UPR, in order to retrotranslocate from the ER to the cytosol. In this study we expressed RTA with and without its own amino-terminal signal sequence in yeast to investigate the events after entry of the toxin into the ER. We present the first evidence that ricin inhibits activation of UPR by preventing HAC1 mRNA splicing and Ire1p signaling. We further show that the ability to inhibit UPR contributes to the cytotoxicity of ricin.
EXPERIMENTAL PROCEDURES
Growth Conditions and Plasmids
The RTA plasmid contained the 35-residue signal sequence and the 267-residue mature RTA downstream of the GAL1 promoter (5). The RTA mutants contained the same cDNA with the point mutations. The mature RTA contained the 267-residue mature RTA without the signal sequence, and the mature E177K contained the 267-residue mature RTA with the E177K mutation at its active site. W303 yeast cells harboring RTA, E177K, mature RTA, mature E177K plasmids or the empty vector (LEU), and either the UPRE-lacZ reporter, pJC104 (URA), or the PGK1-lacZ reporter, pTI25 (TRP) (25) were grown overnight in the appropriate synthetic dropout media supplemented with 100 μg/ml myo-inositol, 2% glucose. Cells were pelleted and resuspended in the appropriate synthetic dropout media supplemented with 100 μg/ml myo-inositol, 2% galactose (for induction of RTA). A portion of cells was induced for UPR with either 1 μg/ml tunicamycin (Tm) or 2 mM dithiothreitol (DTT) simultaneously during the galactose induction of RTA. At each time point, 0.2 A600 units of cells were pelleted and frozen for subsequent lacZ reporter assays. The growth conditions were identical for BY4743 cells and Δhac1 cells. Spliced HAC1 was provided where indicated from pJC835 (HIS). Both pJC104 (12) and pJC835 (26) were gifts from Peter Walter. Ire1p-HA was over-expressed from the vector BG1805 (Open Biosystems).
lacZ Reporter Assay
The assay was conducted as described previously (25). Briefly, cells pelleted as described above were resuspended in 800 μl of Z-buffer (100 mM sodium phosphate, pH 7.0, 10 mM KCl, 1 mM MgSO4) + 40 mM β-mercaptoethanol. 100 μl of 0.1% SDS and 100 μl of chloroform were added and briefly vortexed. Cells were placed in a 30 °C water bath for at least 5 min to pre-equilibrate the lysates. After this incubation, 100 μl of orthonitrophenyl-D-galactopyranoside (4 mg/ml in Z-buffer) was added, samples were placed at 30 °C, and the timer was started. When samples appeared to be in the linear range for quantification, 250 μl of 1 M Na2CO3 was added to stop the reaction. Samples were vortexed briefly and centrifuged for phase separation. 100 μl of the supernatant was diluted into 200 μl of Z-buffer for quantification (in triplicate) along with a standard curve (in duplicate). lacZ activity was determined by calculating the A420 and dividing by the product of the A600 units and the length of incubation. This calculation was repeated on at least three individual transformants.
Analysis of Protein Expression
The total cell lysate from frozen yeast cells harvested during the time course of induction was fractionated into membrane and cytosolic fractions as previously described (27). Total protein (10 μg) from each time point was separated on 15% SDS-PAGE, transferred to nitro-cellulose, and probed with RTA polyclonal antibody (1:5000). RTA was visualized by chemiluminescence (PerkinElmer Life Sciences). The blots were then stripped for 30–45 min with 8 M guanidine hydrochloride and reprobed with antibody to dolichol phosphate mannose synthase (Dpm1p; Molecular Probes) (1:2000) and 3-phosphoglycerate kinase (Pgk1p; Molecular Probes) (1:5000). Hac1p levels were determined in cells co-transformed with pJC835 using monoclonal anti-HA (Covance) (1:5000).
RNA Analysis
Total RNA was extracted from yeast using hot phenol (25). Northern analysis was conducted using standard protocols. Blots were probed with PCR products corresponding to HAC1, ACT1, and KAR2 cDNA. For real-time RT-PCR analysis, cDNA was synthesized from 1 μg of total RNA in a 20-μl reaction containing 1× first-strand buffer (Invitrogen), 40 units/μl RNA Guard RNase inhibitor (Promega, Madison, WI), 0.5 μg of poly d(T) oligonucleotide (Promega), 40 mm dNTPs, and Superscript II (Invitrogen) reverse transcriptase. Quantification of transcript levels by real-time PCR analysis was performed using an ABI Prism 7000 Sequence Detection system using the manufacturer’s protocols.
For quantitative PCR, the primers used were as follows: KAR2, 5′-AGACTAAGCGCTGGCAAGCT-3′ and 5′-ACCAC-GAAAAGGGCGTACAG-3′; GCN4, 5′-CCAATTGTGCCCG-AATCC-3′ and 5′-CCTGGCGGCTTCAGTGTT-3′; DER1, 5′-GCAGCATCACTCGGTGTGTT-3′ and 5′-TTTCCGTTCT-TTTTCAGTTCGTAGT-3′; ACT1, 5′-TGGATTCCGGTGA-TGGTGTT-3′ and 5′-TCAAAATGGCGTGAGGTAGAGA-3′. For Northern blotting, a HAC1 probe was synthesized by first amplifying the first 717 nucleotides of HAC1 cDNA, sub-cloning into pYES2.1 (Invitrogen), and re-isolating the fragment after restriction digest followed by [32P]dCTP labeling (GE Healthcare). The primers used for HAC1 amplification were 5′-ATGGAAATGACTGATTTTGA-ACTA-3′ and 5′-TCATGAAGTG-ATGAAGAAATCATT-3′. Similarly, a probe for ACT1 was generated using the primers 5′-TGGATTCCG-GTGATGGTGTT-3′ and 5′-TTAG-AAACACTTGTGGTGAACG-3′.
RESULTS
Expression of RTA with Its Own Signal Sequence Results in Translocation into the Endoplasmic Reticulum and Glycosylation
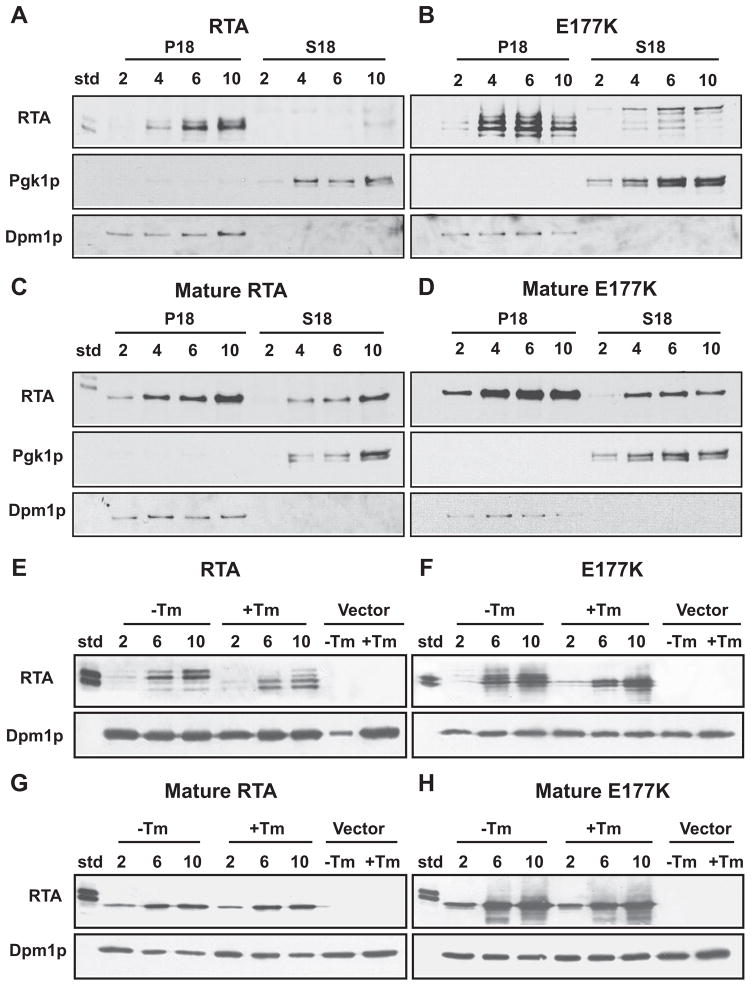
As shown in Fig. 1A, expression of RTA with its own 35-residue amino-terminal signal sequence from the GAL1 promoter in yeast resulted in the toxin accumulating in the membrane fraction (P18). To confirm that the membrane fraction was free of cytosolic contamination and comprised of the ER, the blots were stripped and reprobed with antibodies against the cytosolic marker, 3-phosphoglycerate kinase (Pgk1p), and the ER marker, dolichol phosphate mannose synthase (Dpm1p) (Fig. 1A). These results were repeated with an inactive form of RTA that was mutated at the active site. By changing residue 177 from a glutamic acid to a lysine (E177K), the toxicity of RTA was abolished (5). As shown in Fig. 1B, E177K also accumulated predominantly in the membrane fraction. The glycosylated RTA from castor beans contained two different forms (standard (std)). Unlike the wild type RTA, E177K expressed in yeast contained multiple forms (Fig. 1B). Expression of the mature RTA or mature E177K without the signal sequence resulted in a single form of the protein, which was more equally distributed in both the cytosolic and the membrane fractions (Fig. 1, C and D).
FIGURE 1. Immunoblot analysis of yeast expressing RTA or E177K either with (A and B) or without (C and D) its signal sequence.
Cells were grown for the hours indicated. 10 μg of fractionated supernatant (S18) and a corresponding volume of fractionated membranes (P18) were separated on SDS-PAGE. Blots were initially probed with polyclonal anti-RTA and subsequently stripped and reprobed with anti-Dpm1p and anti-Pgk1p. Cells expressing RTA (E), E177K (F), mature RTA (G), or mature E177K (H) were grown for the hours indicated in the presence or absence of 1 μg/ml tunicamycin (Tm). Unfractionated total extract (10 μg) was separated on a SDS-polyacrylamide gel. Blots were initially probed with polyclonal anti-RTA and subsequently stripped and reprobed with anti-Dpm1p. Cells harboring the empty vector were analyzed as indicated.
To determine whether the multiple bands observed with the RTA or E177K correspond to glycosylated forms, we carried out immunoblot analysis after treating cells with Tm, which inhibits glycosylation. As shown in Fig. 1E, the slower migrating forms of RTA disappeared after treatment with Tm, and new faster migrating forms appeared. Similarly, treatment of cells expressing the E177K with Tm led to the disappearance of the slower migrating forms of RTA and the appearance of faster migrating forms (Fig. 1F). In contrast, the mobility of the mature RTA or mature E177K did not change in the presence or absence of Tm (Fig. 1, G and H). These results indicated that expression RTA or E177K with the signal sequence resulted in translocation into the ER and glycosylation. In contrast, the mature RTA without the signal sequence was not translocated into the ER but was associated with the membrane fraction.
Expression of Inactive RTA That Can Translocate to the ER Induces UPR
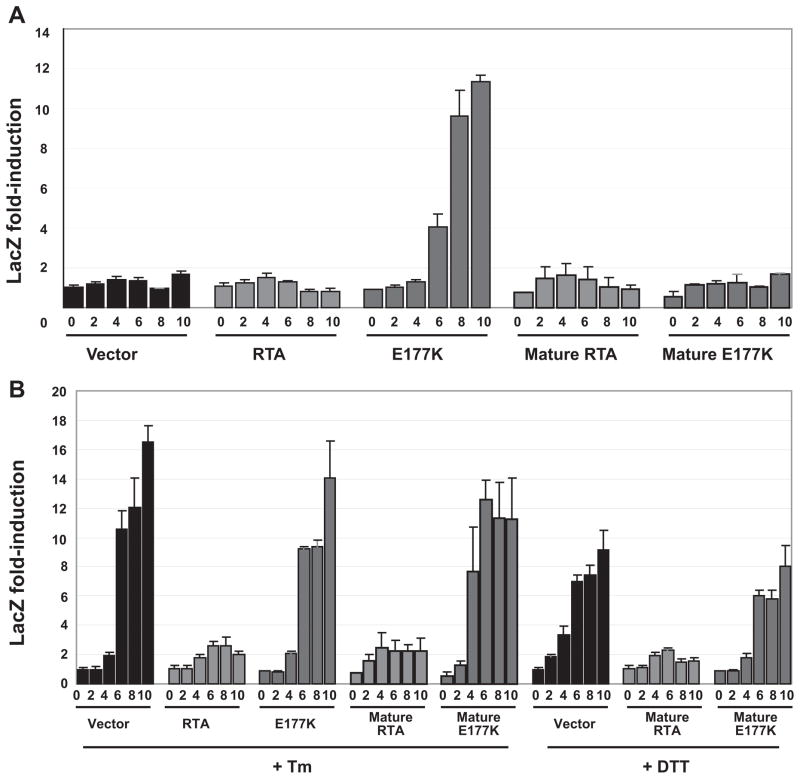
To determine whether RTA affects UPR, we expressed RTA in cells that also harbor the lacZ reporter containing several UPREs within the promoter region. This reporter system has been used to determine the presence and extent of UPR induction in yeast cells (16). β-Galactosidase is only produced during induction of UPR through binding of Hac1p to the UPRE in the promoter. Cells were induced for up to 10 h, and β-galactosidase activity was determined. As shown in Fig. 2A, compared with vector control cells, UPR was induced in yeast expressing E177K. Conversely, cells expressing RTA did not induce UPR. To determine whether inhibition of translation by RTA prevented induction of UPR, we co-transformed cells expressing RTA, E177K, or the vector with a plasmid containing lacZ downstream of the constitutively expressed phosphoglycerate kinase (PGK1) promoter. In cells harboring the vector, RTA, or E177K, lacZ expression from the PGK1 promoter was not significantly changed (Table 1). When β-galactosidase activity was expressed as the ratio of the UPRE/ PGK-driven lacZ, the activity was similar in cells expressing RTA or harboring the vector but higher in cells expressing E177K (Table 1). These results indicated that in cells expressing RTA, the inability to induce lacZ expression from the UPRE promoter was not due to inhibition of translation of the lacZ reporter.
FIGURE 2. β-Galactosidase activity in yeast cells (W303) expressing RTA or E177K either with or without its signal sequence co-transformed with the UPRE-lacZ reporter.
Cells were grown on galactose for the hours indicated in the absence (A) or presence of 1 μg/ml tunicamycin or 2 mM DTT (B). Equivalent amounts of cells were analyzed for lacZ activity using a colorimetric assay. Fold induction was normalized to the level observed in the vector control at time 0. The data are representative of three independent assays.
TABLE 1.
Data are millliunits of β-galactosidase activity upon induction
| Vector control |
RTA |
E177K |
||||
|---|---|---|---|---|---|---|
| 6 h of induction | 10 h of induction | 6 h of induction | 10 h of induction | 6 h of induction | 10 h of induction | |
| Millliunits | Millliunits | Millliunits | ||||
| β-Galactosidase from UPRE-driven lacZ | ||||||
| Galactose | 111 ± 15a (1.0)b | 139 ± 12 (1.3) | 105 ± 8 (1.0) | 68 ± 13 (0.6) | 333 ± 54 (3.0) | 937 ± 26 (8.5) |
| +1 μg/ml tunicamycin | 874 ± 102 (7.9) | 1360 ± 94 (12.3) | 212 ± 28 (1.9) | 165 ± 17 (1.5) | 763 ± 11 (6.9) | 1160 ± 209 (10.5) |
| +2 mM DTT | 579 ± 34 (5.2) | 757 ± 111 (6.8) | 189 ± 14 (1.7) | 127 ± 18 (1.1) | 496 ± 29 (4.5) | 662 ± 119 (6.0) |
| β-Galactosidase from PGK1-driven lacZ | ||||||
| Galactosec | 1500 ± 115 (1.0) | 972 ± 115 (0.6) | 1330 ± 107 (0.9) | 1100 ± 75 (0.7) | 1420 ± 113 (0.9) | 1020 ± 73 (0.7) |
| Ratio of UPRE/PGK1-driven reporter expressionb | ||||||
| Galactose | 1.0 | 1.9 | 1.1 | 0.8 | 3.2 | 12.4 |
| +1 μg/ml tunicamycin | 7.9 | 19.0 | 2.2 | 2.0 | 7.3 | 15.4 |
| +2 mM DTT | 5.2 | 10.6 | 1.9 | 1.6 | 4.7 | 8.8 |
S.D.
Fold above vector control at 6 h on galactose alone.
No significant variation in PGK1-driven lacZ expression was observed between vector control, RTA, and E177K in the presence of tunicamycin or DTT.
Because E177K seemed to strongly activate UPR, we next determined whether translocation to the ER was responsible for the induction. The mature RTA and the mature E177K were transformed into yeast cells bearing the UPRE-driven lacZ reporter plasmid. As expected, expression of either mature RTA or mature E177K without the signal sequence was no longer capable of causing UPR activation through ER stress (Fig. 2A). We conclude from these data that RTA can induce UPR only under conditions when the signal sequence is present but, as in E177K, when the activity of the protein is attenuated.
RTA Inhibits Induction of UPR by Either Tunicamycin or DTT
One explanation for the lack of UPR induction in cells expressing wild type RTA is that the toxin inhibits induction of the UPR pathway. To test this hypothesis, we examined the effect of RTA on UPR that is activated by Tm or DTT. Both Tm and DTT can induce UPR via related but distinct mechanisms. Tm inhibits protein glycosylation, whereas DTT prevents disulfide bond formation. As expected, both agents caused an up-regulation of UPRE-driven lacZ in vector control cells (Table 1 and Fig. 2B). An enhancement of UPR was observed in cells expressing E177K after treatment with Tm or DTT (Table 1). In contrast, RTA inhibited induction of UPR by both Tm and DTT (Table 1 and Fig. 2B). Therefore, the wild type RTA appears to have a direct effect on UPR upstream of UPRE-dependant transcriptional activation.
Because RTA inhibited induction of UPR upon ER stress, we next determined whether translocation into the ER was necessary for inhibition of UPR. This would allow us to discriminate between RTA targets within the lumen of the ER and targets on the cytosolic side of the ER. We treated cells expressing both mature RTA and mature E177K without the signal sequence with Tm and DTT and showed that only with mature RTA, but not with mature E177K, is there continued inhibition of Tm-and DTT-promoted UPR (Fig. 2B). These results demonstrated that RTA could inhibit activation of UPR regardless of whether it can translocate to the ER.
Expression of KAR2 or DER1 Is Not Induced in Cells Expressing RTA
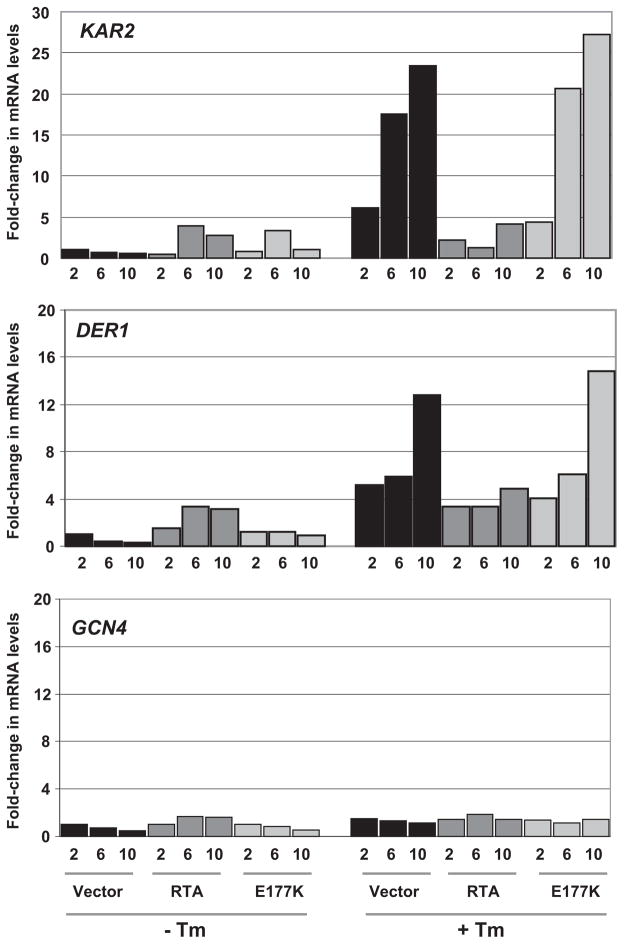
The expression of the KAR2 gene, encoding Kar2p the yeast ortholog of mammalian Bip, is up-regulated by UPR (20). The mRNA levels corresponding to DER1, a component of the ERAD pathway, also increase during UPR (20). In contrast, GCN4 expression is not induced by UPR (20). To determine whether RTA affected expression of genes induced during UPR, we examined the mRNA levels corresponding to KAR2, DER1, and GCN4. As shown in Fig. 3, real time quantitative PCR analysis indicated that in the absence of Tm, KAR2 and DER1 mRNA levels were slightly higher in cells expressing RTA or E177K compared with cells harboring the vector. However, at 10 h after Tm treatment, KAR2 and DER1 mRNA levels increased by more than 20 and 12-fold, respectively, in cells harboring the vector or expressing E177K but not in cells expressing RTA. In contrast, we did not detect any increase in GCN4 mRNA levels in cells expressing RTA or E177K or harboring the vector after treatment with Tm. The expression of actin mRNA as an internal control did not change in these cells. Similar results were obtained by Northern blot analysis (data not shown). These results demonstrated that RTA impairs the induction of expression of KAR2 and DER1 mRNA in response to stress.
FIGURE 3. Real-time PCR analysis of mRNA levels in yeast cells (W303) expressing RTA or E177K in the absence or presence of 1 μg/ml tunicamycin.
Cells were grown on galactose for the hours indicated. The mRNA levels for the genes indicated were normalized to actin mRNA using the ΔΔCT method from Applied Biosystems.
HAC1 mRNA Is Not Spliced in Cells Expressing RTA
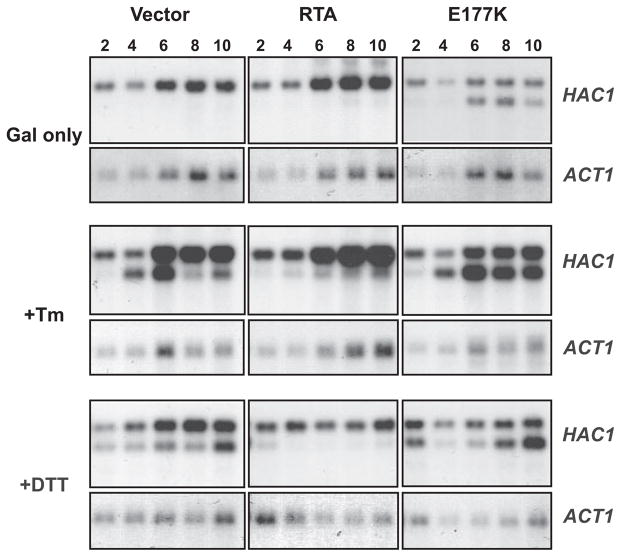
Previous results indicated that translocation of RTA into the ER was not necessary for inhibition of UPR. A potential explanation for the inhibitory activity of RTA could then be explained by interference with one of the events occurring on the cytosolic face of the ER. These include trans-phosphorylation of Ire1p and splicing of HAC1 mRNA, translation of HAC1 mRNA, Hac1p binding, and transcriptional activation of the UPRE-containing genes. To determine more precisely the step inhibited in the UPR signaling pathway, we examined the splicing of HAC1 mRNA. To address the status of HAC1 mRNA, Northern blot analysis was conducted on cells expressing RTA or E177K or harboring the vector. As shown in Fig. 4, HAC1 mRNA was normally unspliced in the absence of ER stress in cells harboring the empty vector (VC). However, HAC1 mRNA was spliced when E177K was expressed in yeast. HAC1 mRNA was rapidly spliced in vector control cells after treatment with Tm or DTT (Fig. 4). In contrast, HAC1 mRNA was not spliced in cells expressing RTA, even after treatment with Tm or DTT. These results indicated that the inhibition of UPR by ricin occurs through a mechanism that specifically prevents HAC1 mRNA splicing.
FIGURE 4. HAC1 mRNA splicing in yeast cells (W303) expressing RTA or E177K.
Cells were grown on galactose (Gal) ± 1 μg/ml tunicamycin or ±2 mM DTT for the hours indicated. 10 μg of total RNA was separated on a 1.8% denaturing agarose gel followed by Northern blotting. The blots were first probed for HAC1 mRNA and later stripped and reprobed for actin mRNA.
UPR Is Not Inhibited by Ricin after Splicing of HAC1 mRNA
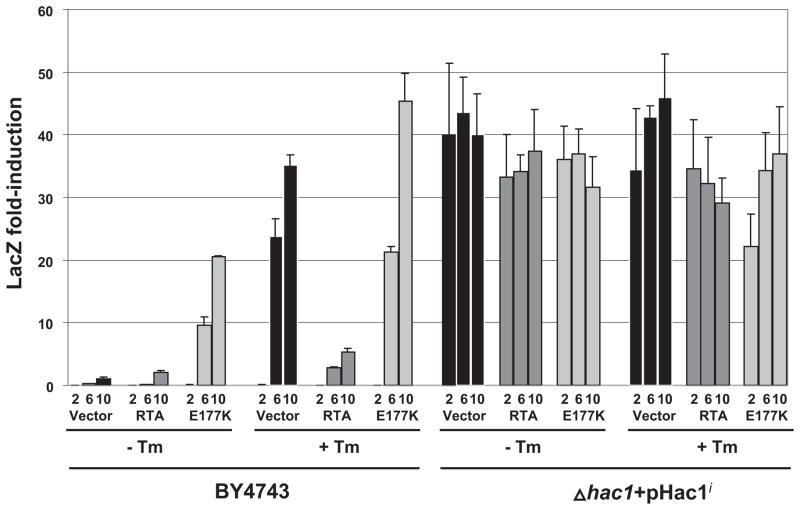
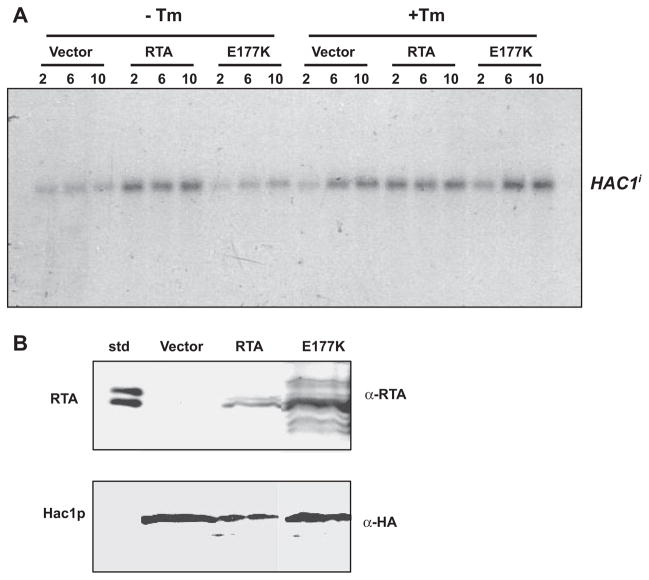
Previous results indicated that the mature form of HAC1 mRNA is markedly decreased in cells expressing RTA. The RTA might inhibit HAC1 message either by increasing its turnover, sequestering it, or by inhibiting processing of the unspliced form, Hac1u. To determine whether this inhibition occurs upstream of the generation of HAC1i, Δhac1 cells, which contain a deletion of the endogenous HAC1, and the isogenic wild type BY4743 cells were co-transformed with RTA, E177K, or the vector and the UPRE-driven lacZ reporter plasmid, pJC104. By assaying UPR in yeast in which the endogenous HAC1 was deleted, the effects of UPR induction could accurately be attributed to only the spliced HAC1 mRNA provided in trans. Predictably, there was no detectable induction of UPR even in the presence of Tm in Δhac1 cells (data not shown). We then co-transformed these cells with the Hac1i. By providing the spliced form of HAC1 mRNA in trans, we were able to observe the effect of RTA at points downstream of the appearance of Hac1i in the cytosol. As measured by lacZ activity, constitutive up-regulation of UPR was observed in cells expressing RTA or E177K or harboring the vector when Hac1i was provided in trans (Fig. 5). Northern blot analysis demonstrated no alterations in the levels of spliced HAC1 mRNA when provided in trans (Fig. 6A), ruling out the possible effects of RTA on the stability of Hac1i. If RTA inhibited UPR via direct inhibition of Hac1p binding to the UPRE, we would have expected a decrease in the reporter activity. These results indicated that the RTA does not affect the stability of Hac1i, translation of Hac1i, or trans activation of the UPRE-containing genes by Hac1i. To confirm that Hac1p can be translated in cells expressing RTA, we carried out immunoblot analysis of Hac1p in the BY4743 cells expressing RTA or E177K or harboring the vector. As shown in Fig. 6B, Hac1p accumulated in cells expressing RTA or E177K or containing the vector. We conclude from these results that that RTA inhibits UPR at a point before HAC1 mRNA splicing and not via inhibition of translation or modulation of Hac1p activity.
FIGURE 5. β-Galactosidase activity in yeast cells expressing RTA or E177K in the presence or absence of tunicamycin.
Wild type (BY4743) or Δhac1 cells harboring a plasmid containing the Hac1i (Δhac1 + pHac1i) were grown on galactose ± 1 μg/ml tunicamycin for the hours indicated. Equivalent amounts of cells were analyzed for β-galactosidase activity using a colorimetric assay. Fold induction was normalized to the level observed in the vector control at 10 h. The data are representative of three independent assays.
FIGURE 6. HAC1 mRNA and protein expression in yeast cells expressing RTA or E177K.
A, Δhac1 cells harboring a plasmid containing the Hac1i (Δhac1 + pHac1i) were grown on galactose ± 1 μg/ml tunicamycin for the hours indicated. Total RNA (10 μg) was separated on a 1.8% denaturing aga-rose gel followed by Northern blotting for HAC1 mRNA. B, immunoblot analysis of Hac1p protein levels. Wild type (BY4743) cells harboring a plasmid containing the Hac1i were grown for 10 h, and total protein (10 μg) was separated on a SDS-polyacrylamide gel. Blots were initially probed with anti-RTA and subsequently with anti-HA to detect Hac1p. std, RTA standard.
Role of Inhibition of UPR in the Cytotoxicity of RTA
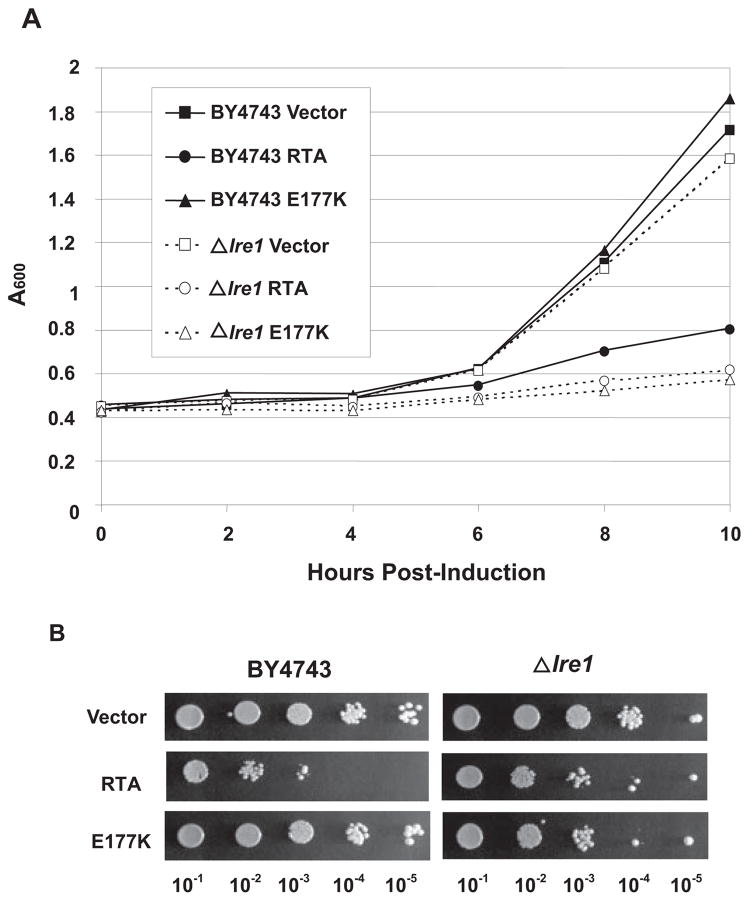
To determine whether inhibition of UPR plays a role in the cytotoxicity of RTA, we examined the growth and the viability of Δire1 cells, which contain a deletion of the endogenous IRE1, and the isogenic wild type BY4743 cells expressing RTA or E177K or harboring the vector. As shown in Fig. 7A, growth of Δire1 cells expressing E177K was reduced to similar levels as cells expressing the wild type RTA. Viability of Δire1 cells expressing E177K was also reduced to a similar level as the Δire1 cells expressing the wild type RTA (Fig. 7B), indicating that a nontoxic RTA mutant reduced the viability of cells that were unable to induce UPR. Because an RTA mutant that was not able to depurinate ribosomes or inhibit translation reduced the growth and the viability of Δire1 cells, these results provided evidence that the Ire1p function was critical for the survival of cells expressing E177K.
FIGURE 7. Growth and viability of Δire1 cells expressing RTA and E177K.
A, expression of RTA and E177K was induced in Δire1cells and isogenic BY4743 cells by growing cells on medium containing galactose. An A600 reading was taken for growth measurement at the indicated times after induction. B, cells were induced for 10 h in SD-Leu media containing galactose, and serial dilutions were plated on noninducing SD-Leu plates containing glucose.
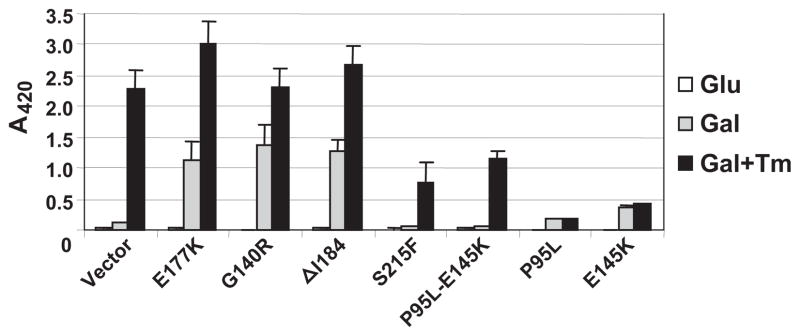
To obtain further evidence that the inability to activate UPR plays a role in the cytotoxicity of RTA, we examined UPR in yeast expressing the nontoxic RTA mutants G140R and ΔI184 that did not inhibit translation, the nontoxic RTA mutants S215F and P95L/E145K that inhibited translation, and the toxic RTA mutants P95L and E145K (5). As shown in Fig. 8, UPR was induced in cells expressing the nontoxic RTA mutants G140R and ΔI184 that did not inhibit translation as in cells expressing the E177K but not in cells expressing the nontoxic mutants S215F and P95L/E145K that depurinated ribosomes and inhibited translation to the same level as wild type RTA (5). The UPR was not induced in cells expressing the toxic mutants P95L or E145K in the absence of Tm. After Tm treatment, UPR was induced to a higher level in cells expressing the inactive RTA mutants G140R and ΔI184 but not in cells expressing the toxic RTA mutants P95L and E145K. UPR was induced in cells expressing S215F or P95L/E145K after Tm treatment but not to the same level as in cells expressing the inactive RTA mutants. These results indicated that the nontoxic RTA mutants that inhibited translation but did not kill cells were able to induce UPR in response to ER stress, suggesting that the ability to activate UPR might contribute to their survival.
FIGURE 8. β-Galactosidase activity in yeast cells expressing RTA mutants co-transformed with the UPRE-lacZ reporter.
Cells were grown overnight in media containing glucose (Glu), and RTA expression was induced by transferring cells to galactose for 8 h in the absence (Gal) or presence of 1 μg/ml tunicamycin (Gal + Tm). Equivalent amounts of cells were analyzed for lacZ activity using a colorimetric assay. The data represent the analysis of three individual colonies containing each construct and are expressed as the mean ± S.E.
DISCUSSION
Through a series of genetic and molecular studies, we demonstrate here the inhibition of UPR by ricin. Specifically, we showed that expression of RTA with its own signal sequence resulted in its translocation to the ER and subsequent glycosylation. Interestingly, expression of the inactive form that could translocate to the ER-induced UPR, but not the wild type RTA, providing evidence that the active site of ricin, played a role in perturbing the ER stress response. The inhibition of UPR was not unique to ER stress induced by the wild type protein but also by global effectors with different mechanisms of action, such as tunicamycin and DTT. Analysis of the nontoxic RTA mutants provided a causal link between the ability of ricin to inhibit ER stress induced UPR and its cytotoxicity.
For ricin to be successful in reaching its cytosolic target, the ribosome, it must be retrotranslocated from the ER to the cytosol. Considering RTA must unfold within the ER, it would seem likely that activation of UPR would occur. Induction of UPR would lead to activation of transcription of ERAD components (21–23). Because ERAD is normally used to retrotranslocate misfolded proteins from the ER to the cytosol for destruction by the ubiquitin proteasome pathway, ricin must evade complete degradation by this pathway. Evidence for this was provided from recent studies, which showed that overexpression of an ERAD-related protein, the α-mannosidase I-like protein (EDEM) strongly protected against the cytotoxicity of ricin by decreasing its retrotranslocation to the cytosol (28). However, when EDEM was available for ricin and the interaction between EDEM and misfolded proteins was inhibited, it promoted retrotranslocation of ricin from the ER to the cytosol (28). Therefore, for RTA to retrotranslocate to the cytosol, this ER stress response must either be inhibited greatly or abolished altogether. Otherwise, induction of UPR might successfully disable the toxin.
Two well characterized activities of ricin are to inhibit translation and subsequently reduce cell viability. It is important to verify that the inhibition of UPR observed is not confounded by inhibition of translation by ricin. To confirm that translation inhibition was not responsible for inhibition of lacZ expression from the UPRE promoter, we demonstrated that expression of lacZ from a PGK promoter was unaffected in the presence of RTA. Additionally, RTA did not inhibit UPR in cells expressing the spliced form of HAC1, indicating that it does not completely inhibit Hac1p translation (Fig. 5). Immunoblot analysis confirmed that Hac1p accumulated in cells expressing RTA (Fig. 6B). Third, we presented evidence that the nontoxic RTA mutants that depurinated ribosomes and inhibited translation were not able to inhibit activation of UPR in response to ER stress, separating the inhibitory activity of RTA on translation from its activity on UPR. Last, a nontoxic RTA mutant that did not depurinate ribosomes reduced the viability of cells that could not induce UPR, indicating that Ire1p function was critical for survival. The northern data provided the most definitive evidence of a defect at the level of splicing, independent of translation. It has recently been shown that the pro-apoptotic protein Bax is required for efficient UPR in mammalian cells (29). We have observed that overexpression of Bax can also induce UPR in yeast.4 Using this finding as a guide, we speculate that the decrease in viability seen in cells expressing RTA, if due to apoptosis, would not be expected to promote inhibition of UPR.
RTA expressed in yeast with its signal sequence as well as the mature RTA without the signal sequence that reduced the viability of yeast cells were both associated with the membrane fraction. However, only RTA with the signal sequence was translocated into the ER. In cells expressing either protein we did not observe induction of β-galactosidase expression or splicing of HAC1 mRNA after treatment with ER stress inducers, indicating that inhibition of UPR occurred on the cytosolic face of the ER. In contrast, UPR was induced in cells expressing the active site mutant with the signal sequence, but not the mature E177K, demonstrating that the inactive RTA could modulate UPR only when it could translocate into the ER. All nontoxic RTA mutants that did not depurinate ribosomes induced UPR, possibly by accumulating in the ER, since activation of UPR would lead to overexpression of ERAD components, which would inhibit retrotranslocation of these mutants to the cytosol (28). In contrast, all toxic mutants that depurinated ribosomes, inhibited activation of UPR. These results provided evidence that an intact active site was needed for inhibition of UPR by the wild type protein, possibly to allow the protein to retrotranslocate to the cytosol. We have previously shown that ribosome depurination and translation inhibition are not sufficient for ricin-mediated cell death (5), suggesting that the protein must attenuate another target to enhance its cytotoxicity. The results presented here demonstrate that the active site of RTA appears to be critical not only for ribosome depurination but also for inhibition of UPR.
Only the precursor form of the HAC1 mRNA was detected in cells expressing RTA after ER stress, indicating that ricin inhibits UPR by preventing splicing of the HAC1 mRNA. The unspliced HAC1 mRNA is localized to the cytoplasm and is associated with functional polyribosomes, which are stalled on the mRNA due to a direct 16-nucleotide-long base pairing interaction between the HAC1 5′-untranslated region and the intron (16, 30, 31). Because inhibition of UPR occurred on the cytosolic face of the ER and required an intact active site, RTA may inhibit UPR by targeting the unspliced HAC1 mRNA. This is consistent with evidence indicating that ribosome-inactivating proteins target other RNAs besides the rRNA (32). A single chain ribosome-inactivating protein, pokeweed antiviral protein, binds to the cap structure on mRNA and depurinates the mRNA downstream of the cap structure in vitro (33). Pokeweed antiviral protein autoregulates its own mRNA (34) and can target other RNAs in vivo (35, 36). By analogy with pokeweed antiviral protein, RTA may bind to the unspliced form of HAC1 mRNA and depurinate it, preventing long distance base-pairing between the 5′-untranslated region and the intron. Disruption of the interaction between HAC1 5′-untranslated region and intron has been shown to lead to accumulation of the unspliced HAC1 mRNA and reduce the efficiency of splicing (31). Alternatively, RTA may attenuate the enzymatic activity of Ire1p either through a direct interaction with Ire1p or through interactions with lumenal proteins.
We present evidence here that ricin inhibits adaptation responses to ER stress by preventing HAC1 mRNA splicing and Ire1p signaling to downstream mediators of UPR, such as KAR2 and DER1. If cells are subjected to continuous ER stress and cannot respond by inducing UPR, they commit to apoptosis (20). Viability of yeast cells expressing the precursor or the mature form of RTA is decreased significantly (5). A nontoxic RTA mutant that could not depurinate ribosomes reduced the viability of Δire1 cells, providing evidence that Ire1p function was critical for survival in the presence of RTA. The Δire1 cells expressing RTA were slightly more viable than the isogenic BY4743 cells expressing RTA (Fig. 7B), suggesting that the Ire1p function was critical for the cytotoxicity of RTA. Taken together, these results suggest that there is an ideal balance between the presence of Ire1p, so that some of the toxin is allowed into the cytosol, and its complete absence, which causes so much ER stress that the cell inevitably dies. The RTA mutants that depurinated ribosomes but did not kill cells were not able to inhibit activation of UPR after ER stress, providing further evidence that the inability to activate UPR in response to ER stress contributes to ricin-mediated cell death. Consistent with these results, recent evidence in mammalian cells suggested a link between UPR signaling and cell survival after ER stress by demonstrating that termination of IRE1 activity is an important factor in allowing cell death after UPR activation (37). Other ribosome-inactivating toxins that need to enter the cytosol from the ER to carry out their enzymatic action may also inhibit UPR to avoid destruction by the ubiquitin proteasome pathway. Therefore, the results described here for ricin may be broadly applicable to other toxins. Several viruses have been shown to induce ER stress and activate UPR (38–41). Cancer cells need UPR for their survival (42). Modulation of UPR by ricin may represent a unique opportunity to inhibit a pathway required by viruses to replicate and cancer cells to grow. Additionally, activation of UPR is required for effective antibody production (43). Through the knowledge gained by studying the effect of RTA on UPR, protection strategies against ricin to counter the threat of its use as a potential bioweapon can be developed so that the beneficial immune response is not dampened.
Acknowledgments
We thank Drs. Rong Di and John McLaughlin for valuable discussions and helpful comments on the manuscript and Dr. Peter Walter for the kind gifts of pJC104 and pJC835.
Footnotes
This work was supported in part by National Institutes of Health Grant AI072425 (to N. E. T.) and National Science Foundation Grant MCB0348299 (to N. E. T.).
The abbreviations used are: ER, endoplasmic reticulum; UPR, unfolded protein response; UPRE, UPR element; RTA, ricin A chain; ERAD, ER-associated degradation; Tm, tunicamycin; DTT, dithiothreitol; PGK, 3-phosphoglycer-ate kinase; HA, hemagglutinin.
B. A. Parikh and N. E. Tumer, unpublished information.
References
- 1.Endo Y, Tsurugi K. J Biol Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 2.Lord JM, Roberts LM, Robertus JD. FASEB J. 1994;8:201–208. [PubMed] [Google Scholar]
- 3.Stirpe F, Battelli MG. Cell Mol Life Sci. 2006;63:1850–1866. doi: 10.1007/s00018-006-6078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartley MR, Lord JM. Biochim Biophys Acta. 2004;1701:1–14. doi: 10.1016/j.bbapap.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Li XP, Baricevic M, Saidasan H, Tumer NE. Infect Immun. 2007;75:417–428. doi: 10.1128/IAI.01295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapak A, Falnes PO, Olsnes S. Proc Natl Acad Sci U S A. 1997;94:3783–3788. doi: 10.1073/pnas.94.8.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wesche J. Int J Med Microbiol. 2002;291:517–521. doi: 10.1078/1438-4221-00161. [DOI] [PubMed] [Google Scholar]
- 8.Wesche J, Rapak A, Olsnes S. J Biol Chem. 1999;274:34443–34449. doi: 10.1074/jbc.274.48.34443. [DOI] [PubMed] [Google Scholar]
- 9.Simpson JC, Roberts LM, Romisch K, Davey J, Wolf DH, Lord JM. FEBS Lett. 1999;459:80–84. doi: 10.1016/s0014-5793(99)01222-3. [DOI] [PubMed] [Google Scholar]
- 10.Deeks ED, Cook JP, Day PJ, Smith DC, Roberts LM, Lord JM. Biochemistry. 2002;41:3405–3413. doi: 10.1021/bi011580v. [DOI] [PubMed] [Google Scholar]
- 11.Zhao L, Ackerman SL. Curr Opin Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Cox JS, Shamu CE, Walter P. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 13.Mori K, Ma W, Gething MJ, Sambrook J. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 14.Welihinda AA, Kaufman RJ. J Biol Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- 15.Sidrauski C, Walter P. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 16.Cox JS, Walter P. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 17.Kawahara T, Yanagi H, Yura T, Mori K. Mol Biol Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niwa M, Patil CK, DeRisi J, Walter P. Genome Biology. 2005;6:R3. doi: 10.1186/gb-2004-6-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidrauski C, Cox JS, Walter P. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 20.Bernales S, Papa FR, Walter P. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 21.McCracken AA, Brodsky JL. Curr Top Microbiol Immunol. 2005;300:17–40. doi: 10.1007/3-540-28007-3_2. [DOI] [PubMed] [Google Scholar]
- 22.Meusser B, Hirsch C, Jarosch E, Sommer T. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 23.Romisch K. Annu Rev Cell Dev Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- 24.Kaufman RJ. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 25.Tumer NE, Parikh BA, Li P, Dinman JD. J Virol. 1998;72:1036–1042. doi: 10.1128/jvi.72.2.1036-1042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng DT, Spear ED, Walter P. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parikh BA, Baykal U, Di R, Tumer NE. Biochemistry. 2005;44:2478–2490. doi: 10.1021/bi048188c. [DOI] [PubMed] [Google Scholar]
- 28.Slominska-Wojewodzka M, Gregers TF, Walchli S, Sandvig K. Mol Biol Cell. 2006;17:1664–1675. doi: 10.1091/mbc.E05-10-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- 30.Chapman RE, Walter P. Curr Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- 31.Ruegsegger U, Leber JH, Walter P. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- 32.Barbieri L, Valbonesi P, Bonora E, Gorini P, Bolognesi A, Stirpe F. Nucleic Acids Res. 1997;25:518–522. doi: 10.1093/nar/25.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudak KA, Bauman JD, Tumer NE. RNA. 2002;8:1148–1159. doi: 10.1017/s1355838202026638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parikh BA, Coetzer C, Tumer NE. J Biol Chem. 2002;277:41428–41437. doi: 10.1074/jbc.M205463200. [DOI] [PubMed] [Google Scholar]
- 35.Di R, Tumer NE. Mol Plant-Microbe Interact. 2005;18:762–770. doi: 10.1094/MPMI-18-0762. [DOI] [PubMed] [Google Scholar]
- 36.Picard D, Kao CC, Hudak KA. J Biol Chem. 2005;280:20069–20075. doi: 10.1074/jbc.M413452200. [DOI] [PubMed] [Google Scholar]
- 37.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isler JA, Maguire TG, Alwine JC. J Virol. 2005;79:15388–15397. doi: 10.1128/JVI.79.24.15388-15397.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isler JA, Skalet AH, Alwine JC. J Virol. 2005;79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jordan R, Wang L, Graczyk TM, Block TM, Romano PR. J Virol. 2002;76:9588–9599. doi: 10.1128/JVI.76.19.9588-9599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su HL, Liao CL, Lin YL. J Virol. 2002;76:4162–4171. doi: 10.1128/JVI.76.9.4162-4171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee AS, Hendershot LM. Cancer Biol Ther. 2006;5:721–722. doi: 10.4161/cbt.5.7.3120. [DOI] [PubMed] [Google Scholar]
- 43.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. J Clin Investig. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]