1. Introduction
“Thus, principles of evolution are dramatically exhibited by studying strains of Proteus on a solid medium, which contains a source of nutrients and oxidizable organic compounds. If the descendants of an individual are to compete successfully in this defined habitat, they must have evolved: (i) highly efficient enzyme catalysts to release and trap free energy, (ii) mobile mechanisms to access the site where more free energy is available, and (iii) mechanisms to stake out a claim on free energy—territoriality, one of the fundamental principles of ecology. Microbes did it first.”
---Ralph S. Wolfe1
Members of the bacterial species Proteus mirabilis are capable of rapid swarming over a hard agar surface. Swarming colonies display a striking phenotype in which a visible boundary forms between swarms of different strains. In contrast, boundaries do not arise between two swarms of a single strain (Figure 1). This behavior is a demonstration that P. mirabilis swarms are capable of territoriality and self versus non-self recognition. Though first described over 70 years ago, the molecular mechanisms underlying boundary formation in P. mirabilis have remained stubbornly elusive despite the combined efforts of several research groups. In this article we attempt to give a concise review of the historical and current models for Proteus boundary formation. We begin with a brief background on the genus Proteus and specifically the species Proteus mirabilis, followed by a review of swarm boundary formation. We then analyze the two prominent models for boundary formation: (1) preemptive antagonism and (2) recognition. We conclude with the broader implications of P. mirabilis territoriality and self vs non-self recognition.
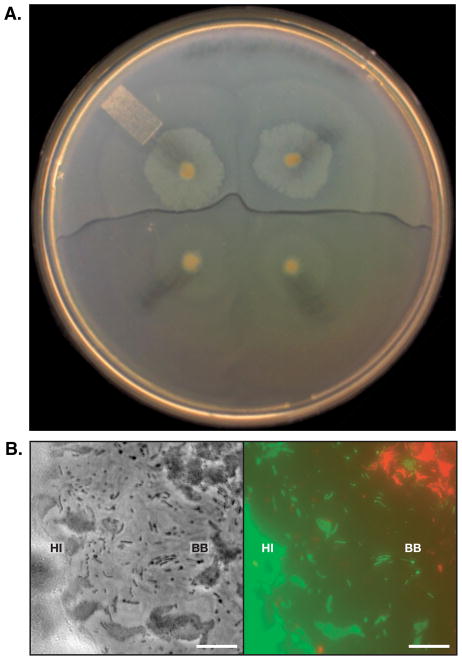
Figure 1. Boundary between P. mirabilis strains.
(A) A boundary between two different wild-type strains of P. mirabilis, HI4320 (HI) and BB2000 (BB). Top, two swarms of HI that have merged with each other; bottom, two swarms of BB that have merged with each other. Swarms of the same strain merge. (B) Higher resolution phase contrast and fluorescent microscope image of a boundary zone between HI (green) and BB (red). Phase on left, pseudocolored overlay on right. Bar, 10 μm. Reprinted from Reference 31. Copyright 2008 American Association for the Advancement of Science; reprinted with permission from AAAS.
2. The genus Proteus
2.1 General information about the ecology of Proteus
The genus Proteus is a member of the Enterobacteriaceae family. Proteus can be found in soil and contaminated water, and for some Proteus species, in the intestinal tract of animals. In soil and water, and perhaps healthy animals, Proteus species are likely involved in the degradation of organic material. These bacteria are noted for their ability to utilize urea as a source of nitrogen and energy2. The two Proteus species most commonly found in humans are P. mirabilis and to a lesser extent P. vulgaris3. Jacobsen et al. and Manos et al. recently reviewed Proteus pathogenesis in detail2,3. In brief, P. mirabilis is an opportunistic pathogen that causes complicated urinary tract infections, especially in people requiring long-term indwelling catheterization4. In hospital and nursing home settings, Proteus species can also cause infections in the ear, nose, throat, and skin, among others, especially immunocompromised individuals. Infections at multiple sites in a single patient are caused by single P. mirabilis strains; P. mirabilis strains isolated from the catheter, urine, feces and kidney stones of a single patient are often the same strain, suggesting that P. mirabilis infections are clonal5–7.
2.2 Proteus mirabilis virulence factors
P. mirabilis employs a wide variety of virulence factors during colonization and subsequent infection. One major virulence factor is urease, an enzyme that converts urea to ammonia and carbon dioxide. Activity of urease results in an increase of the local alkalinity and eventually the crystallization of magnesium ammonium phosphate and calcium phosphate8. The alkaline conditions can lead to the formation of kidney stones and of crystalline biofilms that encrust the catheter tubing8. Biofilms often result from the attachment of cells to catheter walls via adhesins2. Other virulence factors include a hemolysin, an IgA-degrading protease, mannose-resistant Proteus-like fimbriae (MR/P), mannose-resistant Klebsiella-like hemagglutinin (MR/K), P. mirabilis fimbriae (PMF), nonagglutinating fimbriae (NAF), and ambient-temperature fimbriae (ATF)4,9–11. Production of these virulence factors, including urease is associated with surface-attached growth12.
2.3 The Proteus mirabilis life cycle
P. mirabilis cells undergo a morphologically distinct developmental cycle upon contact with a surface or in viscous liquid environments (Figure 2). In normal liquid growth medium, P. mirabilis exists as short (1 to 2 μ.m in length), rod-shaped swimmer cells that have six to ten peritrichous flagella. Upon contact with a surface, swimmer cells differentiate into elongated (10 to 80 μ.m), hyper-flagellated swarmer cells3. The main component of the flagella, the flagellar filament protein FlaA, is the same for both the swimmer and swarmer morphotypes3. Swarmer cells have multiple copies of the chromosome yet appear to be a single cell without a division plane or septum. Some of the developmental cues leading to swarmer cell differentiation include increased expression of flagellar genes as directed by the flagellar regulator, FlhDC13–16 inhibition of flagellar rotation17, and the accumulation of putrescine18. Expression of virulence genes is coincident with the developmental switch from a swimmer cell to the swarmer cell morphotype17,19–22. The initiation of P. mirabilis swarmer cell differentiation has been thoroughly reviewed recently3,23.
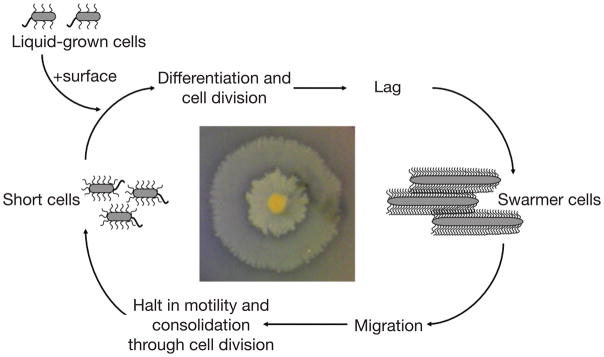
Figure 2. Diagram of the developmental cycle of surface-attached P. mirabilis.
Swarming colonies form bull’s eye patterns (photograph at center) because of the repeated cycles of migration followed by consolidation. This figure was adapted from Reference 57.
Upon initial surface contact there is a lag period, and then swarmer cells migrate rapidly across the surface in a coordinated cell-to-cell behavior. The swarming cells migrate as rafts of cells connected by flagella. Cells in rafts are aligned in a parallel manner as rafts move24. Occasionally an individual cell moves alone near the front of an advancing swarm. Swarming rafts and cells leave visible tracks on the agar surface (Figure 3). Following an initial period of migration, the swarmer cells arrest movement and divide into shorter cells. This results in a macroscopic consolidation ring. Interestingly, the consolidation phase appears to be independent of nutrient availability and metabolite changes25,26. This arrest in migration is puzzling because the medium still provides an excess of nutrients and energy sources. Perhaps an internal (and unknown) factor in swarmer cells dictates the consolidation. After a brief delay the new short cells differentiate into swarmer cells and population migration resumes. The repeated cycles of migration followed by consolidation result in the characteristic bull’s eye pattern of P. mirabilis swarm colonies (Figure 2). The ability to swarm has been implicated in pathogenesis, perhaps for migration across for catheters or initial colonization, as evidenced by the fact that non-swarming P. mirabilis strains are not able to ascend to the urinary tract in mouse models and exhibit significantly reduced virulence20; however swarmer cells are not observed in tissues isolated from infected mice, and non-flagellated P. mirabilis strains do not exhibit decreased infectivity when inoculated intravenously into mice27,28. It is likely that P. mirabilis cells utilize the swarming behavior to rapidly migrate across catheters and perhaps ascend the urinary tract against the flow of urine4.
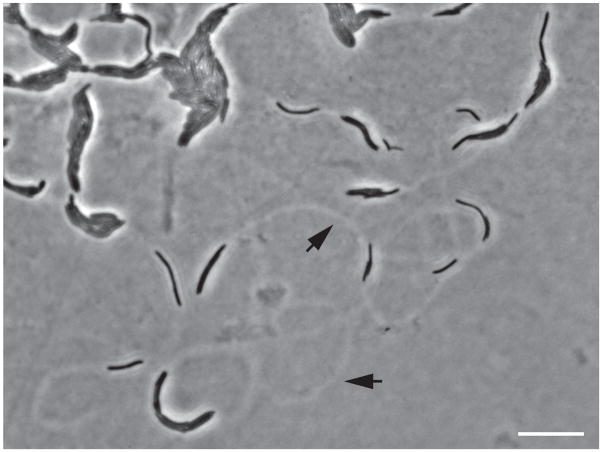
Figure 3. Phase contrast micrograph of the leading edge of a P. mirabilis swarm.
The cells are black, and the agar is gray. Rafts of cells and individual cells are evident as are tracks (black arrows) on the agar surface. Bar, 10μm.
3. Swarm boundary formation and territoriality
3.1 Discovery of swarm boundaries
In 1946, Louis Dienes described the formation of a boundary between clinical isolates of P. mirabilis from two patients29. Dienes observed that colonies of the same isolate merge at the juncture where two swarming populations approach. However, a boundary, visible to the naked eye, forms at the intersection between two colonies of two different Proteus strains:
“Halos starting from colonies of different Proteus strains do not penetrate into each other. Even after several days of incubation a sharp line of demarcation persists between the areas covered by different strains. The halos sometimes stop a short distance from each other without coming into contact, indicating the diffusion of antagonistic substances into the medium. Meanwhile extension proceeds in other directions.”29
As such, the boundary is often referred to as a Dienes line and the occurrence as the Dienes phenomenon. P. vulgaris exhibits similar territorial behavior29. Strains that merge are considered members of the same identity group, while strains that form boundaries are of different identity groups. Typing by Dienes line formation is a rapid method for discriminating amongst clinical isolates of P. mirabilis, and its discriminatory effectiveness has been confirmed by ribotyping and pulsed-field gel electrophoresis7,30.
3.2 Description of swarm boundaries
The Dienes line boundary is marked by the arrest in forward migration of approaching P. mirabilis swarms. The boundary can range in diameter from 0.5 up to 3.0 mm. After the initial interaction of swarm fronts, subsequent layers of swarming cells continue to approach the boundary but do not cross, forming a metaphoric canyon in which the stacked layers of cells at the swarm front amass like cliff walls and the boundary lays sparse like a dry riverbed. When observed at higher resolution, boundaries appear to consist of unoccupied regions, cell debris, and round bodies (Figure 1)31. It is likely that the debris is due in part to production of Proteus-specific antibacterial peptides called proticines32,33. The round bodies in the boundaries are sometimes associated with the filamentous swarmer cells29,34. Dienes described it is as if the swarmer cells were emptying their cytoplasmic contents into the round bodies29. These large, round bodies also form when P. mirabilis is refrigerated, transferred to tap water, or exposed to the poison mercuric chloride, HgCl229,34, indicating that P. mirabilis cells may form round bodies under stress conditions. There are suggestions that the round bodies are viable29. However, it is more likely that the round bodies represent cells that have died35. Reports have suggested that boundary formation only occurs when P. mirabilis cells are in the swarming phase of the developmental cycle; however, the round bodies are produced even when the cells are mixed in liquid culture, i.e. are short swimmer cells29,36.
4. Territoriality
4.1 Territorial boundaries between Proteus mirabilis swarms
Boundary formation between P. mirabilis strains is a vivid example of intra-specific territoriality in bacteria. Territoriality, generally defined as the behavior to establish and maintain an area that the organism occupies exclusively and defends, is commonly associated with larger organisms and is likely based on complex communication strategies, and even perhaps division of labor37,38. In the case of P. mirabilis, one strain is able to hold off and inhibit colonization by a competing different strain. It is an act of mutual exclusion. Similar territorial behavior has more recently been reported in other bacteria, for example, Escherichia coli and Myxoccocus xanthus39,40. Territorial behavior in larger organisms is generally considered to result from overt aggression or advertisement38. Dienes initially hypothesized that the boundary formation could be due to antagonistic substances. Since then, two hypotheses have emerged as the dominant explanations for boundary formation in Proteus: (1) preemptive antagonism and (2) recognition (Figure 4). We recently provided evidence that recognition is a key element of boundary formation (discussed below)31.
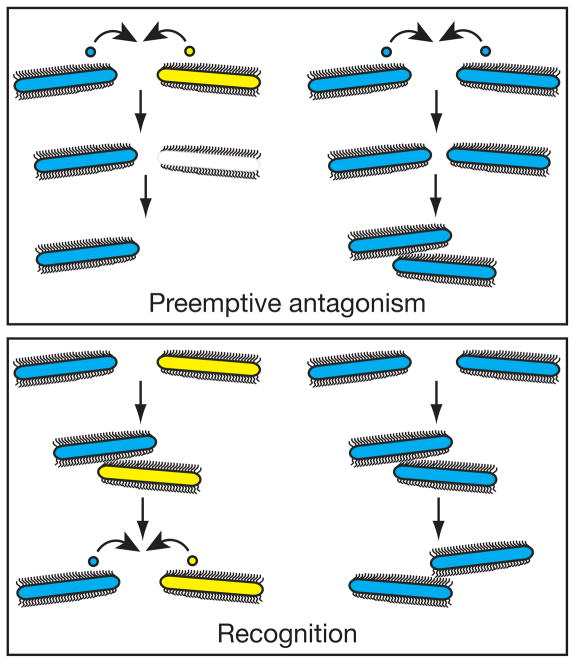
Figure 4. Models for Proteus boundary formation.
Top. The preemptive antagonism model, which postulates that secretion of proticines, or other toxins, by the dominant Proteus strain into the zone between strains causes cell death in the inferior strain, ultimately resulting in boundary formation. Bottom. The recognition model, which postulates that upon interaction with the approaching swarm, P. mirabilis cells can recognize and identify identical strains as self and different strains as non-self. This may or may not result in killing.
4.2 The preemptive antagonism model
The preemptive antagonism hypothesis postulates that boundary formation results from an antagonistic signal that is conveyed from one strain to the other. Preemptive antagonism is essentially overt aggression. This is opposed to the recognition hypothesis (discussed in a later section), which postulates that P. mirabilis strains are able to recognize and differentiate themselves from each other without aggression or prior to an aggressive reaction through a mechanism that enables advertisement. These hypotheses are not mutually exclusive. Boundary formation might involve a combination of recognition and aggression.
The preemptive antagonism model infers death of cells in the boundary zone as a result of direct killing mechanisms, or at least growth inhibition. This model predicts that swarming Proteus cells secrete toxins that are specific for other Proteus strains. The toxins may work as a toxin-antitoxin system and are highly specific to the surface composition of other Proteus strains. The preemptive antagonism hypothesis posits that secretion of strain-specific toxins and the resultant cell death caused by the toxins are primarily responsible for boundary formation between P. mirabilis strains32,34,35. Interestingly when an Anapore membrane, which allows the relatively free diffusion of small molecules and prohibits the migration of cells, is placed between swarms of two different P. mirabilis strains, a boundary will fail to form; in fact both swarms will move directly to the edge of the membrane before arresting movement, suggesting that a freely diffusible small molecule is unlikely the source of boundary formation34.
As mentioned above, round bodies are evident in Dienes lines29. Though still debated, cells of each of the strains in approaching swarms appear to form round bodies. Dienes believed that there was a viable subpopulation of round bodies emerging primarily from one of the strains41; more recent studies suggest that round bodies are primarily formed by one strain and not the other. Evidence suggests that close cell proximity, if not direct cell-cell contact, is necessary for round body formation34,35, but the data really only support the idea that there is not a diffusible substance inducing round body formation. We favor the idea that round bodies reflect cell death caused by the interactions between incompatible strains.
It is likely that the activity of proticines contributes to the formation of round bodies. As discussed above, proticines are Proteus-produced bacteriocins, proteinaceous antibacterial toxins of approximately 10 components that can inhibit growth and cause cell death of sensitive Proteus strains33,42. Though the mechanisms of proticine production are not completely understood, it appears that these toxins function by binding specific cell surface receptors in a manner similar to the action of colicins; different Proteus strains probably display different receptors 33,42. Bernard Senior published an extensive investigation of the link between proticines and boundary formation in P. mirabilis and P. vulgaris isolates32. Strains were placed in identity groups based on the formation of boundaries. Those strains that did not form boundaries with each other were placed in the same group. If a boundary formed between two strains they were placed in different groups. Senior also examined proticine production by each strain and the proticines to which each strain was sensitive, resulting in a proticine production/sensitivity (P/S) type for each strain. Out of 204 Proteus isolates, 98 distinct groupings emerged32,43. There were 14 subclasses for proticine production and separately for proticine sensitivity, as well as a classification called atypical boundary formation. For the vast majority but not all strains, boundary formation correlated with proticine P/S types32.
Of note, there were four strains that neither produced nor were sensitive to proticines. In this subset of isolates, one strain formed a boundary with all of the others, even though none of the strains produced (or were sensitive to) proticines32. These exceptions indicate that proticine production and sensitivity do not define the basis of boundary formation32. Proticines likely contribute to the cell death observed in the boundary zones but they are not required for boundary formation. Evidence indicates that boundary formation does not involve a diffusible substance, and perhaps cell-to-cell contact is needed for boundaries to develop31,34. Yet proticines are diffusible and they function to inhibit cell growth in liquid media33.
4.3. The recognition model
Central to territoriality is the ability for an organism to recognize self, or identity, and discriminate self from non-self. According to the recognition hypothesis, Proteus cells at the edge of an advancing swarm can identify cells in an approaching swarm as self or non-self, after which there is an appropriate reaction that leads to a merger of the swarms or boundary formation. Boundary formation might or might not involve or require killing. This sort of recognition behavior is similar to allorecognition in eukaryotic cells. The capability for self vs non-self recognition is a common theme in biology39,44–46. We recently identified a genetic locus in P. mirabilis that encodes components sufficient for self vs non-self recognition and for the definition of strain-specific identity31.
5. The ids self-identity locus
5.1 The ids genes
We identified genetic determinants of self vs non-self recognition in a screen for P. mirabilis mutants that formed a boundary with the wild-type parent31. This was a screen for recognition defects. We isolated one mutant and mapped the mutation to a six-gene locus termed idsABCDEF for identification of self. Swarms of a mutant in which the entire idsA-F region was deleted formed a boundary with the parent, and with other wild-type P. mirabilis strains31. The recognition defect could be complemented by ectopic expression of the ids genes; complemented strains merged with the parent and formed boundaries with the idsA-F deletion strain31. These results demonstrate that the idsA-F genes are required and sufficient for self-non-self recognition in P. mirabilis swarms.
5.2 Parent and ids mutants do not kill each other
Unlike the debris-containing boundary between two wild-type P. mirabilis strains, there is little visible debris contained in the boundaries between ids mutants and the parent. Furthermore, when mixed in broth an idsA-F deletion strain and the parent grow equally well31. Therefore the idsA-F locus does not appear to be a toxin-antitoxin system. Rather, functional copies of the ids genes are needed for self or non-self recognition.
5.3. Analysis of ids gene products
There is essentially nothing known about the biochemistry of the ids gene products, but sequence comparisons and genetic analyses have provided insights about possible Ids functions. IdsA shows sequence similarity to the Hcp protein31. Hcp is secreted and forms hexameric rings that can polymerize into tubular structures47,48. IdsB shows sequence similarity to VgrG family members31. Hcp and VgrG are components of Type VI secretion systems, which are widely distributed on Proteobacteria. It is believed that secretion of other proteins is mediated by a surface structure involving Hcp and VgrG 49–52. Though P. mirabilis appears to have a type VI secretion system, the other necessary components of a complete type VI secretion system, such as the clpV ATPase and icmF, are not located near the ids genes on the P. mirabilis genome53. IdsC, IdsD, and IdsE are each unique to P. mirabilis, and IdsF is a conserved hypothetical bacterial protein of unknown function31.
The idsABCDEF locus was present in each of seven P. mirabilis isolates examined. These seven isolates all formed boundaries with each other. Among the seven strains, the polypeptides encoded by idsA, idsB, idsC, and idsF were over 96% identical31. In contrast, IdsD and IdsE contained regions of high variability (72 - 84% pair-wise identity and 32 to 80% pair-wise identity, respectively) flanked by regions of over 96% identity31. This sequence pattern of variable regions flanked by conserved regions is reminiscent of alleles encoding antigenic variation in other bacteria54.
Genetic analyses have led to a model describing functional roles for the ids gene products. First, idsABCDEF constitute an operon (unpublished). The idsBCDEF genes are required for self vs non-self recognition, and IdsF has a distinct function from the functions of IdsBCDE31. Specific IdsA mutants behave like the parent. IdsA is not required for self vs non-self identification. There are three other hcp homologs in the P. mirabilis genome and we believe one or more of them may be capable of substituting for IdsA. The, idsB, idsC, or idsF genes code for required self vs non-self recognition functions, but idsB, idsC, and idsF are general functions that are interchangeable between strains. In contrast, idsD and idsE encode strain-specific (or self) identity and are sufficient to confer a new self identity when introduced into a heterologous strain31.
5.4. The bar code model for ids function
The informatics and genetic analyses described above have led to our current bar code and scanner model for ids function. IdsD and IdsE function as unique identifiers, or bar codes, for each P. mirabilis strain. IdsA, IdsB, IdsC, and IdsF function to display or interpret the information conveyed by IdsD and IdsE; they act as scanners or display equipment. Although we know that the idsABCDE operon confers self vs non-self identity, we believe that there may be other such determinants31. The available genetic data support a view in which the initial stage of P. mirabilis boundary formation encompasses social recognition and signaling between the approaching swarms.
6. The cell to cell contact hypothesis and an alternative hypothesis
A simple and elegant experiment showing that non-self recognition is not mediated by a diffusible factor was published recently34. Two different isolates of P. mirabilis were spotted on a plate and separated by a highly porous Anapore membrane. Both isolates swarmed up to the membrane and a territorial boundary did not form as it did in control experiments where the isolates were not separated by a membrane. This is strong evidence that non-self recognition is not mediated by a factor that can diffuse through the agar and through the dialysis membrane. It was also taken as evidence that cell-to-cell contact is required for non-self recognition. But could there be other explanations for the results, an explanation that does not invoke a cell-to-cell contact requirement? We will present one interesting possibility. At the leading edge of swarms, cells in rafts as well as individual cells exist as a thin layer on the agar surface, and they leave visible trails (Figure 3). It could very well be that materials in the trails, perhaps IdsD and E serve as signals for the scanning devices and cell-to-cell contact is not required for non-self identification. There is as yet no evidence to discriminate between the cell-to-cell contact model and a model whereby a non-diffusible signal is laid down on the surface as cells move. Further, the physical mechanisms of boundary formation are still poorly understood: is it that the populations migrate, interact, and then one or both population(s) retreats; is it that one or both population(s) dies after interacting with the other population; is it that after initial cells meet, a signal is secreted that discourages future migration into the same region? All of these possibilities support a model in which recognition precedes antagonism in P. mirabilis boundary formation.
7. Clinical implications
The prevalence of P. mirabilis infections is likely to rise as the U.S. population ages and the number of patients requiring indwelling catheters continues to increase. Clinical isolates of P. mirabilis with resistance to a broad range of antibiotics are emerging55. Identical P. mirabilis strains are vertically transferred from mother to newborn and in rare cases, can cause neonatal meningitis and subsequent brain abscesses56. Further, P. mirabilis infections are usually clonal5,6. P. mirabilis may use self vs non-self recognition and territoriality to ensure dominance of a single strain, perhaps during colonization of catheters or the urethra. The increased presence of antibiotic-resistant strains and the ability of clones to establish dominance could have implications for increased occurrences of antibiotic resistance in urinary tract infections of immunocompromised individuals.
8. Biological significance
At the cellular level self vs non-self recognition is a behavior conserved throughout biology, from bacteria to animals and to vertebrate immune systems. The puzzle of how cells or organisms recognize and differentiate self from non-self is fundamental to fields as far from each other as immunology and ecology. The boundary formation between P. mirabilis is clearly a complex interplay of cell-to-cell communication and population development. In boundary formation, there is not only social recognition but also territorial competition between approaching Proteus strains. It seems likely that recognition and antagonism both play a role in the overall boundary formation process, but antagonism is not required for boundary formation. Molecular details of the dance leading to boundary formation remain to be determined. How do Ids proteins function to convey strain-specific identity? How is self-identity maintained? How did territoriality emerge in Proteus? What are the costs and benefits of this social activity? Proteus provides a wonderful tool to dissect the costs and benefits of territoriality. Where else in the bacterial world does territoriality occur?
Acknowledgments
The authors’ work described was supported in part by the following grants from the NIH: AI55396 and GM59026. In addition the W. M. Keck Foundation provided support to initiate our work on P. mirabilis territoriality.
Biographies

Dr. Gibbs received her Bachelor’s degree in Biochemical Sciences from Harvard University and earned her Ph.D. in Microbiology and Immunology with Dr. Julie Theriot at Stanford University. After receiving her doctorate, she trained as a postdoctoral fellow at the University of Washington in Seattle with Dr. E. Peter Greenberg where she analyzed the genetic components of territorial behavior and self-identity in bacteria. She recently joined Harvard University as an Assistant Professor in the Department of Molecular and Cellular Biology, where she is building a research group that focuses on the social behaviors of microbes, specifically investigating the molecular mechanisms that underlie the ability of bacterial cells to discriminate self from non-self.

Dr. Greenberg received his Bachelor’s degree from Western Washington University, a Master’s from the University of lowa, PhD from the University of Massachusetts. After a postdoctoral at Harvard he joined the faculty at Cornell University in 1977, eventually moved back to the University of lowa (1987) and finally returned to the Pacific Northwest as a member of the UW Medicine Microbiology faculty in 2005. Dr. Greenberg has spent his scientific career uncovering aspects of microbial social behavior. Due in part to his efforts we now understand that many bacteria communicate by using chemical signals. Bacterial communication controls virulence in a variety of pathogenic bacteria, and it has thus become a target for development of new therapeutic strategies. Bacteria have also become models for studies of selection for and evolution of cooperative behavior.
References
- 1.Wolfe RJ. [(accessed February 15, 2010).];Microbiol Biol Ed [Online] 2009 14:6 . http://archive.microbeHbrary.org/edzine/details.asp?id=2jys.
- 2.Jacobsen SM, Stickler DJ, Mobley HL, Shirtliff ME. Clin Microbiol Rev. 2008;21:26. doi: 10.1128/CMR.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manos J, Belas R. Prokaryotes. 2006;6:245. [Google Scholar]
- 4.Mobley HL, Belas R. Trends Microbiol. 1995;3:280. doi: 10.1016/s0966-842x(00)88945-3. [DOI] [PubMed] [Google Scholar]
- 5.Sabbuba NA, Mahenthiralingam E, Stickler DJ. J Clin Microbiol. 2003;41:4961. doi: 10.1128/JCM.41.11.4961-4965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabbuba NA, Stickler DJ, Mahenthiralingam E, Painter DJ, Parkin J, Feneley RCL. J Urol. 2004;171:1925. doi: 10.1097/01.ju.0000123062.26461.f9. [DOI] [PubMed] [Google Scholar]
- 7.Mathur S, Sabbuba NA, Suller MTE, Stickler DJ, Feneley RCL. Eur J Clin Microbiol Infect Dis. 2005;24:643. doi: 10.1007/s10096-005-0003-0. [DOI] [PubMed] [Google Scholar]
- 8.Jones BV, Mahenthiralingam E, Sabbuba NA, Stickler DJ. J Med Microbiol. 2005;54:807. doi: 10.1099/jmm.0.46123-0. [DOI] [PubMed] [Google Scholar]
- 9.Kimberly EW, Sasan M-J, Lockatell CV, David J, Robert B. Mol Microbiol. 1999;32:825. [Google Scholar]
- 10.Walker KE, Moghaddame-Jafari S, Lockatell CV, Johnson D, Belas R. Mol Microbiol. 1999;32:825. doi: 10.1046/j.1365-2958.1999.01401.x. [DOI] [PubMed] [Google Scholar]
- 11.Phan V, Belas R, Gilmore BF, Ceri H. Infect Immun. 2008;76:4859. doi: 10.1128/IAI.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rather PN. Environ Microbiol. 2005;7:1065. doi: 10.1111/j.1462-2920.2005.00806.x. [DOI] [PubMed] [Google Scholar]
- 13.Allison C, Lai HC, Gygi D, Hughes C. Mol Microbiol. 1993;8:53. doi: 10.1111/j.1365-2958.1993.tb01202.x. [DOI] [PubMed] [Google Scholar]
- 14.Furness RB, Fraser GM, Hay NA, Hughes C. J Bacteriol. 1997;179:5585. doi: 10.1128/jb.179.17.5585-5588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemmer KM, Rather PN. Res Microbiol. 2007;158:295. doi: 10.1016/j.resmic.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Clemmer KM, Rather PN. J Med Microbiol. 2008;57:931. doi: 10.1099/jmm.0.47778-0. [DOI] [PubMed] [Google Scholar]
- 17.Belas R, Suvanasuthi R. J Bacteriol. 2005;187:6789. doi: 10.1128/JB.187.19.6789-6803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sturgill G, Rather PN. Mol Microbiol. 2004;51:437. doi: 10.1046/j.1365-2958.2003.03835.x. [DOI] [PubMed] [Google Scholar]
- 19.Allison C, Lai HC, Hughes C. Mol Microbiol. 1992;6:1583. doi: 10.1111/j.1365-2958.1992.tb00883.x. [DOI] [PubMed] [Google Scholar]
- 20.Allison C, Emödy L, Coleman N, Hughes C. J Infect Dis. 1994;169:1155. doi: 10.1093/infdis/169.5.1155. [DOI] [PubMed] [Google Scholar]
- 21.Burall LS, Harro JM, Li X, Lockatell CV, Himpsl SD, Hebel JR, Johnson DE, Mobley HLT. Infect Immun. 2004;72:2922. doi: 10.1128/IAI.72.5.2922-2938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coker C, Poore CA, Li X, Mobley HL. Microbes Infect. 2000;2:1497. doi: 10.1016/s1286-4579(00)01304-6. [DOI] [PubMed] [Google Scholar]
- 23.Fraser GM, Hughes C. Curr Opin Microbiol. 1999;2:630. doi: 10.1016/s1369-5274(99)00033-8. [DOI] [PubMed] [Google Scholar]
- 24.Jones BV, Young R, Mahenthiralingam E, Stickler DJ. Infect Immun. 2004;72:3941. doi: 10.1128/IAI.72.7.3941-3950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauprich O, Matsushita M, Weijer CJ, Siegert F, Esipov SE, Shapiro JA. J Bacteriol. 1996;178:6525. doi: 10.1128/jb.178.22.6525-6538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuyama T, Takagi Y, Nakagawa Y, Itoh H, Wakita J, Matsushita M. J Bacteriol. 2000;182:385. doi: 10.1128/jb.182.2.385-393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen AM, Lockatell CV, Johnson DE, Mobley HL. Infect Immun. 2003;71:3607. doi: 10.1128/IAI.71.6.3607-3613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zunino P, Piccini C, Legnani-Fajardo C. Microb Pathog. 1994;16:379. doi: 10.1006/mpat.1994.1038. [DOI] [PubMed] [Google Scholar]
- 29.Dienes L. Proc Soc Exp Biol Med. 1946;63:265. doi: 10.3181/00379727-63-15570. [DOI] [PubMed] [Google Scholar]
- 30.Pfaller MA, Mujeeb I, Hollis RJ, Jones RN, Doern GV. J Clin Microbiol. 2000;38:1077. doi: 10.1128/jcm.38.3.1077-1080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibbs KA, Urbanowski ML, Greenberg EP. Science. 2008;321:256. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senior BW. J Gen Microbiol. 1977;102:235. doi: 10.1099/00221287-102-2-235. [DOI] [PubMed] [Google Scholar]
- 33.Senior BW. J Med Microbiol. 1983;16:323. doi: 10.1099/00222615-16-3-323. [DOI] [PubMed] [Google Scholar]
- 34.Budding AE, Ingham CJ, Bitter W, Vandenbroucke-Grauls CM, Schneeberger PM. J Bacteriol. 2009;191:3892. doi: 10.1128/JB.00975-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolstenholme J. J Gen Microbiol. 1973;74:343. doi: 10.1099/00221287-74-2-343. [DOI] [PubMed] [Google Scholar]
- 36.Tracy O, Thomson EJ. J Clin Pathol. 1972;25:69. doi: 10.1136/jcp.25.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hölldobler B, Lumsden CJ. Science. 1980;210:732. doi: 10.1126/science.210.4471.732. [DOI] [PubMed] [Google Scholar]
- 38.Wilson E. Sociobiology: The New Synthesis. 25. Harvard University Press; Cambridge: 2000. [Google Scholar]
- 39.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Science. 2005;309:1245. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 40.Vos M, Velicer GJ. Curr Biol. 2009;19:1763. doi: 10.1016/j.cub.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dienes L. Proc Soc Exp Biol Med. 1947;66:97. doi: 10.3181/00379727-66-15994. [DOI] [PubMed] [Google Scholar]
- 42.Coetzee HL, De Klerk HC, Coetzee JN, Smit JA. J Gen Virol. 1968;2:29. doi: 10.1099/0022-1317-2-1-29. [DOI] [PubMed] [Google Scholar]
- 43.Senior BW. J Med Microbiol. 1977;10:7. doi: 10.1099/00222615-10-1-7. [DOI] [PubMed] [Google Scholar]
- 44.De Tomaso AW, Nyholm SV, Palmeri KJ, Ishizuka KJ, Ludington WB, Mitchel K, Weissman IL. Nature. 2005;438:454. [Google Scholar]
- 45.Dudley SA, File AL. Biol Lett. 2007;3:435. doi: 10.1098/rsbl.2007.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reece SE, Drew DR, Gardner A. Nature. 2008;453:609. doi: 10.1038/nature06954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. Science. 2006;312:1526. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng J, Leung K. Mol Microbiol. 2007;66:1192. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- 49.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Proc Natl Acad Sci USA. 2007;104:15508. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cascales E. EMBO Rep. 2008;9:735. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filloux A, Hachani A, Bleves S. Microbiology. 2008;154:1570. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 52.Aschtgen M-S, Gavioli M, Dessen A, Lloubès R, Cascales E. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2009.07028.x. [DOI] [PubMed] [Google Scholar]
- 53.Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Walker D, Whithead S, Thomson NR, Rather PN, Parkhill J, Mobley HLT. J Bacteriol. 2008;190:4027. doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Woude MW, Bäumler AJ. Clin Microbiol Rev. 2004;17:581. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones ME, Draghi DC, Thornsberry C, Karlowsky JA, Sahm DF, Wenzel RP. Ann Clin Microbiol Antimicrob. 2004;3:14. doi: 10.1186/1476-0711-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bingen E, Boissinot C, Desjardins P, Cave H, Brahimi N, Lambert-Zechovsky N, Denamur E, Blot P, Elion J. J Clin Microbiol. 1993;31:1055. doi: 10.1128/jcm.31.5.1055-1059.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider R, Lockatell CV, Johnson D, Belas R. Microbiology. 2002;148:773. doi: 10.1099/00221287-148-3-773. [DOI] [PubMed] [Google Scholar]