Abstract
The mode of viral hemorrhagic septicemia virus (VHSV) transmission in the Great Lakes basin is largely unknown. In order to assess the potential role of macroinvertebrates in VHSV transmission, Diporeia spp., a group of amphipods that are preyed upon by a number of susceptible Great Lakes fishes, were collected from seven locations in four of the Great Lakes and analyzed for the presence of VHSV. It was demonstrated that VHSV is present in some Diporeia spp. samples collected from lakes Ontario, Huron, and Michigan, but not from Lake Superior. Phylogenetic comparison of partial nucleoprotein (N) gene sequences (737 base pairs) of the five isolates to sequences of 13 other VHSV strains showed the clustering of Diporeia spp. isolates with the VHSV genotype IVb. This study reports the first incidence of a fish-pathogenic rhabdovirus being isolated from Diporeia, or any other crustacean and underscores the role macroinvertebrates may play in VHSV ecology.
Findings
The viral hemorrhagic septicemia virus (VHSV), genotype IVb, is a recent invader to the Laurentian Great Lakes basin and has been associated with mortalities in a number of resident freshwater fish species [1], [2], [3], [4]. While laboratory studies demonstrated that the virus can be transmitted to naïve fish by both immersion and injection [5], [6], [7], the mode of VHSV transmission in the Great Lakes basin is largely unknown. In a previous study, it was concluded the pisocolid intermittent leech Myzobdella lugubris harbors VHSV [8]. Whether other macroinvertebrates can act as a vector or reservoir for VHSV remains to be elucidated.
In the Great Lakes foodweb, amphipods of the genera Diporeia, Gammarus, and Hyalella occupy a central position as they transform energy from lower to higher trophic levels [9]. Unfortunately, Diporeia spp. have experienced a sharp decline sharp decline in abundance over the last two decades [10]; the cause(s) of which puzzle scientists. To tackle this enigma, a study was designed that involved comprehensive parasitological and microbiological analysis of Diporeia spp. collected from lakes Ontario, Huron, Michigan and Superior [Faisal and Winters: Pathogens impacting Diporeia spp. in the Great Lakes, submitted].
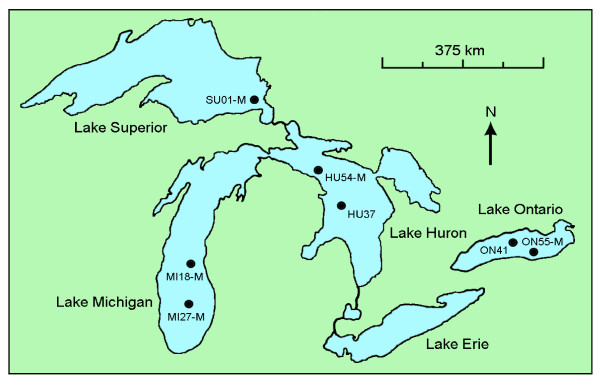
Diporeia spp. were collected between August 2007 and April 2008 by taking Ponar grabs from seven locations in the Great Lakes basin at depths between 74-190 meters. The approximate locations of collections of Diporeia spp. are shown in Figure 1. Collected Diporeia spp. were pooled (five amphipods/pool), immersed briefly in absolute ethanol for surface disinfection, and then rinsed several times in sterile water. Samples (~100 μg) were homogenized with a sterile mortar and pestle and then diluted with 1 ml Earle's salt-based minimal essential medium (MEM, INVITROGEN). Homogenized Diporeia contents were removed with a sterile transfer pipette, dispensed into a sterile 1.5 ml centrifuge tube, and centrifuged at 5500 rcf for 20 min and supernatants were immediately used for virus isolation.
Figure 1.
Map of the Laurentian Great Lakes showing where Diporeia spp. were collected for this study. The solid circles denote sampling locations.
Since there is currently no amphipod cell line that could be used to aid in the isolation of amphipod-pathogenic viruses, virus isolation was performed according to the standard protocols detailed in the American Fisheries Society Blue Book [11] and the Office International des Epizooties [12], using the Epithelioma papulosum cyprinii (EPC) cell line [13]. Inoculated 96-well plates containing EPC cells grown with MEM (5% fetal bovine serum) were incubated at 15°C for 21 days, and were observed for the formation of cytopathic effects (CPE). Second and third blind passages were performed and assessed for the presence of CPE. All cell culture positive samples of Diporeia homogenates (ON41, ON55-M, HU54-M, MI18-M, and MI27-M) caused CPE on EPC in the form of focal areas of rounded, refractile cells which progressed to full lysis of the cell monolayer.
Reverse transcriptase polymerase chain reaction (RT-PCR) was then performed on all samples (Table 1) Total RNA was extracted from inoculated cell culture supernatant using a QIAamp® Viral RNA Mini Kit (QIAGEN). Reverse transcription was accomplished by a two-step protocol using the Affinity Script Multiple Temperature Reverse Transcriptase RT-PCR™ (AGILENT TECHNOLOGIES). The primer set used in this assay was recommended by the Office de International Epizootics for the detection of a 811 base pair sequence of the VHSV nucleocapsid (N) gene: 5'-GGG GAC CCC AGA CTG T-3' (forward primer) and 5'-TCT CTG TCA CCT TGA TCC-3' (reverse primer). Amplicons of 811 base pairs were amplified in all cell culture positive samples.
Table 1.
Locations in the Laurentian Great Lakes from which Diporeia spp. were collected for this study (CPE = formation of cytopathic effect; RT-PCR = results for amplification of the viral hemorrhagic septicemia virus nucleoprotein gene).
| Lake | Station | Depth (m) | Latitude | Longitude | Date | CPE | RT-PCR |
|---|---|---|---|---|---|---|---|
| Ontario | ON41 | 129 | 43°43.0000 N | 078°01.6299 W | 4/25/08 | + | + |
| ON55-M | 190 | 43°26.6000 N | 077°26.2903 W | 4/25/08 | + | + | |
| Huron | HU37 | 74 | 44°45.7001 N | 082°47.0103 W | 8/7/07 | - | - |
| HU54-M | 124 | 45°31.0000 N | 083°25.0301 W | 8/7/07 | + | + | |
| Michigan | MI18-M | 160 | 42°44.0601 N | 086°59.9803 W | 4/16/08 | + | + |
| MI27-M | 103 | 43°36.0101 N | 086°55.0002 W | 4/18/08 | + | + | |
| Superior | SU01-M | 95 | 46°59.5601 N | 085°09.6300 W | 8/18/07 | - | - |
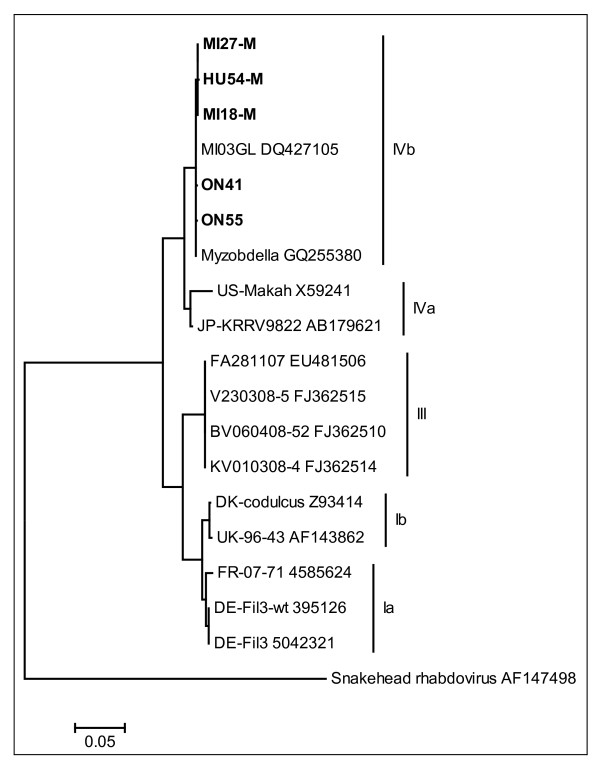
The RT-PCR yielded five samples (ON41, ON55-M, HU54-M, MI18-M, and MI27-M) with the characteristic VHSV 811 bp band. The amplicons were further purified with the Wizard® SV Gel and PCR Clean-up System (PROMEGA) and then sequenced from both directions. Overlapping sequences for each isolate were aligned using the BioEdit contig assembly program version 7.0.9.0 [14] and the aligned contigs were used for multiple alignments performed by ClustalW [15]. Phylogenetic analysis of the VHSV Diporeia strain with 14 nucleoprotein encoding genes from other species of rhabdovirus was done by generating the phylogenetic dendrogram (Figure 2) using MEGA 4 [16] and the Neighbor-Joining algorithm [17]. Phylogenetic analysis of the five Diporeia isolate sequences (737 bp) base pairs [GenBank: HQ214133-HQ214135, HQ415762-HQ415763] showed that the sequences clustered with the VHSV IVb-MI03 strain, the index strain of the Great Lakes VHSV [GenBank: DQ427105]. In comparison to the Great Lakes VHSV strain, two nucleotide substitutions were observed in Diporeia isolates from both lakes Huron and Michigan: a transition from cytosine to thymine at nucleotide position 408 which caused a silent mutation and a transversion from guanine to thymine at nucleotide position 907 which caused a mutation from glycine to cysteine.
Figure 2.
Distance tree constructed for phylogenetic comparison of isolates obtained in this study. The tree generated using the Neighbor-joining algorithm and maximum likelihood method shows high phylogenetic similarity between the five viral hemorrhagic septicemia virus (VHSV) isolates obtained in this study (ON41, ON55-M, HU54-M, MI18-M, and MI27-M) and other isolates belonging to the VHSV genotype IVb. The alignment file used to produce the tree contained partial VHSV nucleoprotein (N) gene sequences (737 nucleotide positions). Snakehead rhabdovirus was used as the outgroup. The scale bar indicates the number of substitutions per nucleotide site.
Our findings provided evidence that VHSV can exist within Diporeia spp. Whether VHSV propagates in the cells of these amphipods or just existed in their viscera or gills is currently unknown and deserves further investigation. Indeed, the presence of VHSV in Diporeia spp. in three of the four Great Lakes sampled is surprising since these amphipods were collected from depths that ranged from 74-190 meters where none of the susceptible fish are known to reside. Diporeia spp. feed on detritus and planktonic organisms and is known to scavenge for food items in lower depths. Such a feeding habit has the potential to transfer VHSV from the benthos to the pelagic zone through their excreta or by being preyed upon by susceptible fish; lake whitefish for example. Moreover, based exclusively on data generated in this study, one cannot rule out that VHSV is a pathogen of Diporeia spp. or that Diporeia spp. can be a reservoir for VHSV in the Great Lakes. Regardless of these currently unanswered questions, this study reports the first incidence of a fish-pathogenic rhabdovirus being isolated from Diporeia, or other crustacean. This finding underscores the dire need to better understand the role of macroinvertebrates in disease ecology.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AW conducted field collection. AW and MF, designed the study, performed all laboratory assays, and drafted the manuscript. Both authors reviewed and approved of the final manuscript.
Contributor Information
Mohamed Faisal, Email: faisal@cvm.msu.edu.
Andrew D Winters, Email: winter28@msu.edu.
Acknowledgements
The authors thank the crew of the R/V Lake Guardian for their assistance with sample collection. In order to conduct the study, we are indebted to the generous funding provided by the United States Environmental Protection Agency - Great Lakes National Protection Office (Grant #: GL00E36101) and the United States Department of Agriculture - Animal and Plant Health Inspection Service (Grant#: 10-9100-1293-GR).
References
- Elsayed E, Faisal M, Thomas M, Whelan G, Batts W, Winton J. Isolation of viral haemorrhagic septicaemia virus from muskellunge, Esox masquinongy (Mitchill), in Lake St. Clair, Michigan, USA reveals a new sublineage of the North American genotype. J Fish Dis. 2006;29:611–619. doi: 10.1111/j.1365-2761.2006.00755.x. [DOI] [PubMed] [Google Scholar]
- Gagné N, MacKinnon AM, Boston L, Souter B, Cook-Versloot M, Griffiths S, Olivier G. Isolation of viral hemorrhagic septicemia virus from mummichog, stickleback, striped bass and brown trout in eastern Canada. J Fish Dis. 2007;30:213–223. doi: 10.1111/j.1365-2761.2007.00802.x. [DOI] [PubMed] [Google Scholar]
- Groocock GH, Getchell RG, Wooster GA, Britt KL, Batts WN, Winton JR, Casey RN, Casey JW, Bowser PR. Detection of viral hemorrhagic septicemia in round gobies in New York State (USA) waters of Lake Ontario and the St. Lawrence River. Dis Aquat Org. 2007;76(3):187–192. doi: 10.3354/dao076187. [DOI] [PubMed] [Google Scholar]
- Lumsden JS, Morrison B, Yason C, Russell S, Young K, Yazdanpanah A, Huber P, Al-Hussinee L, Stone D, Way K. Mortality event in freshwater drum Aplodinotus grunniens from Lake Ontario, Canada, associated with viral haemorrhagic septicemia virus, Type IV. Dis Aquat Org. 2007;76(2):99–111. doi: 10.3354/dao076099. [DOI] [PubMed] [Google Scholar]
- Kim R, Faisal M. Comparative susceptibility of representative Great Lakes fish species to the North American viral hemorrhagic septicemia virus Sublineage IVb. Dis Aquat Org. 2010;91:23–34. doi: 10.3354/dao02217. [DOI] [PubMed] [Google Scholar]
- Kim R, Faisal M. Experimental studies confirm the wide host range of the Great Lakes viral haemorrhagic septicaemia virus genotype IVb. J Fish Dis. 2010;33:83–88. doi: 10.1111/j.1365-2761.2009.01093.x. [DOI] [PubMed] [Google Scholar]
- Kim R, Faisal M. The Laurentian Great Lakes strain (MI03) of the viral haemorrhagic septicaemia virus is highly pathogenic for juvenile muskellunge, Esox masquinongy (Mitchill) J Fish Dis. 2010;33(6):513–527. doi: 10.1111/j.1365-2761.2010.01150.x. [DOI] [PubMed] [Google Scholar]
- Faisal M, Schulz C. Detection of Viral Hemorrhagic Septicemia virus (VHSV) from the leech Myzobdella lugubris Leidy, 1851. Paras Vect. 2009;2:45. doi: 10.1186/1756-3305-2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley MA, Vanderploeg HA. Ingestion of live filamentous diatoms by the Great Lakes amphipod, Diporeia sp.: a case study of the limited value of gut contents analysis. Hydrobiol. 1991;223:141–148. doi: 10.1007/BF00047635. [DOI] [Google Scholar]
- Nalepa TF, Fanslow DL, Lang GA. Transformation of the offshore benthic community in Lake Michigan: recent shift from the native amphipod Diporeia spp. to the invasive mussel Dreissena rostriformis bugensis. Freshwater Bio. 2009;54:466–479. doi: 10.1111/j.1365-2427.2008.02123.x. [DOI] [Google Scholar]
- American Fisheries Society-Fish Health Section. Suggested procedures for the detection and identification of certain finfish and shellfish pathogens. American Fisheries Society: Bethesda, MD; 2007. [Google Scholar]
- Office International des Epizooties (OIE) Manual of Diagnostic Tests for Aquatic Animals. 4. World Animal Health Organization, Paris; 2006. Chapter 2.1.5 Viral Hemorrhagic Septicemia. [Google Scholar]
- Fijan N, Sulimanovic D, Bearotti M, Musinie D, Zwillenberg LD, Chilmonczyk S, Vantherot JF, deKinkilin P. Some properties of the Epithelioma papulosum cyprinii (EPC) cell line from carp Cypnnus carpio. Ann Virol. 1983;134:207–220. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biol and Evolution. 1987. pp. 406–425. [DOI] [PubMed]