Abstract
Background
Ash (Fraxinus spp.) is a dominant tree species throughout urban and forested landscapes of North America (NA). The rapid invasion of NA by emerald ash borer (Agrilus planipennis), a wood-boring beetle endemic to Eastern Asia, has resulted in the death of millions of ash trees and threatens billions more. Larvae feed primarily on phloem tissue, which girdles and kills the tree. While NA ash species including black (F. nigra), green (F. pennsylvannica) and white (F. americana) are highly susceptible, the Asian species Manchurian ash (F. mandshurica) is resistant to A. planipennis perhaps due to their co-evolutionary history. Little is known about the molecular genetics of ash. Hence, we undertook a functional genomics approach to identify the repertoire of genes expressed in ash phloem.
Methodology and Principal Findings
Using 454 pyrosequencing we obtained 58,673 high quality ash sequences from pooled phloem samples of green, white, black, blue and Manchurian ash. Intriguingly, 45% of the deduced proteins were not significantly similar to any sequences in the GenBank non-redundant database. KEGG analysis of the ash sequences revealed a high occurrence of defense related genes. Expression analysis of early regulators potentially involved in plant defense (i.e. transcription factors, calcium dependent protein kinases and a lipoxygenase 3) revealed higher mRNA levels in resistant ash compared to susceptible ash species. Lastly, we predicted a total of 1,272 single nucleotide polymorphisms and 980 microsatellite loci, among which seven microsatellite loci showed polymorphism between different ash species.
Conclusions and Significance
The current transcriptomic data provide an invaluable resource for understanding the genetic make-up of ash phloem, the target tissue of A. planipennis. These data along with future functional studies could lead to the identification/characterization of defense genes involved in resistance of ash to A. planipennis, and in future ash breeding programs for marker development.
Introduction
Ash (Fraxinus spp.) is a dominant tree species in many urban and forest landscapes of North America (NA) [1], [2]. The emerald ash borer (Agrilus planipennis Fairmaire, EAB), which is indigenous to Eastern Asia has killed millions of ash trees since its accidental introduction to NA, primarily in the Midwestern United States and Southeastern Ontario [3], [4]. Larvae feed on phloem and outer xylem of trees of all sizes, girdling the tree and ultimately killing it within 1–4 years after symptoms become apparent [3], [4]. Black (F. nigra Marshall), green (F. pennsylvanica Marshall), and white ash (F. americana L.) are known to be highly susceptible, while blue ash (F. quadrangulata Michx) appears to be less preferred [5], [6]. If the pattern of invasion continues, A. planipennis has the potential to decimate ash throughout NA with substantial economic and ecological impact [3], [7]–[8].
Conversely, A. planipennis is not reported to be a major pest in Asia, where Manchurian ash (F. mandshurica Rupr) is a primary host [9]. In a common garden experiment, Manchurian ash was found to be much more resistant to A. planipennis than were NA green and white ash, perhaps by virtue of the co-evolutionary history shared by A. planipennis and Manchurian ash [10]. Phloem tissue of Manchurian ash was found to have high constitutive concentrations of phenolic-based hydroxycoumarins, phenylethanoids and calceloariosides, which may contribute to its resistance to A. planipennis [11].
Second generation sequencing technologies have been applied to a wide variety of studies such as transcriptome sequencing, single nucleotide polymorphism (SNP) discovery, mutation mapping, alternative splicing identification etc. [12]–[15]. In particular, gene discovery via transcriptome analysis has greatly helped in genomic analysis of several non-model organisms including plants viz., Cucumis sativus [16], Eucalyptus grandis [13], Castanea dentate and C. mollisima [17] and Pinus contorta [18]. Roche® 454 GS FLX Titanium is a high throughput sequencing platform that makes it possible to generate massive amounts of information in a short period of time with unprecedented high sequencing depth and low cost [19]. The generated expressed sequenced tags (ESTs) databases are invaluable for gene mining and annotation [20]–[24], phylogenetic analysis [25], molecular markers [26] and expression analysis [27].
Given the status of A. planipennis as an aggressive killer of NA ash trees, we undertook a functional genomics approach to identify the repertoire of genes expressed in phloem tissue of different ash species including green, white, black, blue, and Manchurian ash. This study will enable us to identify genes that are potentially involved in A. planipennis resistance of Manchurian ash, and to characterize the genetic makeup of ash phloem for future studies. Results stemming from this study could be used in future ash targeted breeding programs and increase fundamental understanding of interaction between ash trees and wood-borers such as A. planipennis.
Results and Discussion
Transcriptome analysis
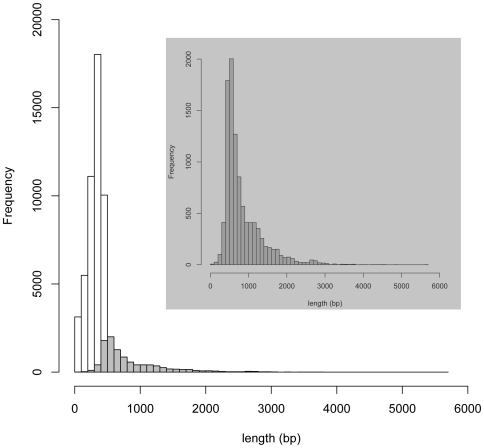
So far, one cDNA library has been developed for F. excelsior (European ash), and no EST sequences were available for NA and Manchurian ash as of October, 2010. The 454 pyrosequencing has made possible genomic studies in non-model organisms because it overcomes the limitations of conventional Sanger sequencing [18]. Our pyrosequencing study contributes a significant number of ESTs for future functional genomic studies in ash, yielding 203,718 total reads and 63,096,022 bases from which 79% and 60% were aligned respectively with an inferred read error of 2.21%. Assembled contigs had an average size of 649 bp with the largest contig being 5,662 bp. The singletons had an average size of 329 bp with the largest being 964 bp. Overall, we obtained 58,673 high quality ash sequences totaling 23,580,430 bp (Figure 1). To our knowledge, this is the first comprehensive study on the transcriptome of ash phloem.
Figure 1. Summary of Fraxinus spp. transcriptomic sequences.
The singleton sequences are represented by clear bars and the contig sequences by shaded bars (insert).
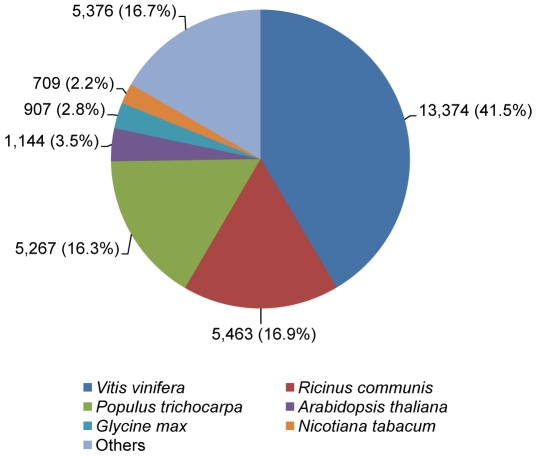
A sequence similarity search was done using BLASTx algorithm. This analysis revealed that 45% of the ash transcriptomic sequences to be potentially ash-specific i.e., no significant matches (E value cutoff of 1e-5) to protein sequences in the GenBank nr database. The vast majority (>99%) of the sequences with significant matches matched to plant sequences (Figure 2), out of which 41.5% matched with Vitis vinifera L., 16.3% with Populus trichocarpa (Torr. & Gray) and 16.9% with Ricinus communis L. In our dataset, 9 sequences matched to viral sequences, 18 to artificial sequences, and 42 to bacterial sequences. Although some of these (viral, artificial and bacterial) sequences may be derived from organisms that are naturally associated with ash trees, they were excluded from further analysis due to possible contamination.
Figure 2. A pie chart showing species distribution of the top BLAST hits of the Fraxinus spp. sequences to various plant species.
Comparative analysis
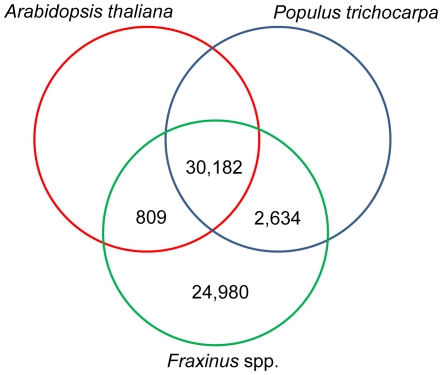
The ash transcriptomic sequences were compared to protein sequences of the model plant Arabidopsis thaliana (Brassicaceae) and the black cottonwood P. trichocarpa (Salicaceae) (also known as western balsam poplar or California poplar). These two species were chosen as they represent model plant systems whose genomes have been sequenced. Of the total 58,604 ash sequences, 24,980 (42.6%) had no significant similarity to any protein identified within the genomes of A. thaliana or P. trichocarpa (Figure 3). Similar observations of species specific transcript sequences were observed in other transcriptomic studies and were attributed to the presence of novel sequences or transcripts of 5′ and 3′ untranslated regions or genes with homologs in other species whose biological functions are not yet assigned. [28]–[30]. About 2,634 (4.5%) sequences were shared between P. trichocarpa and Fraxinus spp., but not with A. thaliana, suggesting that they are potential tree-specific sequences. Only 809 (1.4%) sequences were shared between A. thaliana and Fraxinus spp (Table S1). There were 30,182 sequences (51.5%) that were shared among all three plant species under comparison. Comparative genomics explore similarity with transcriptomes of other species, reveals species specific details and define genes which are conserved or diverging in plant species [31], [32]. Genome sequencing in non-model organisms, particularly forest trees is still in its infancy. For such species functional and comparative genomics is possible upon obtaining a good EST database. These studies seem to be the best source for deciphering the putative function of novel genes [33].
Figure 3. A venn diagram showing the comparisons of the sequences from Fraxinus spp. with the genomes sequences of Arabidopsis thaliana and Populus trichocarpa.
Gene Ontology
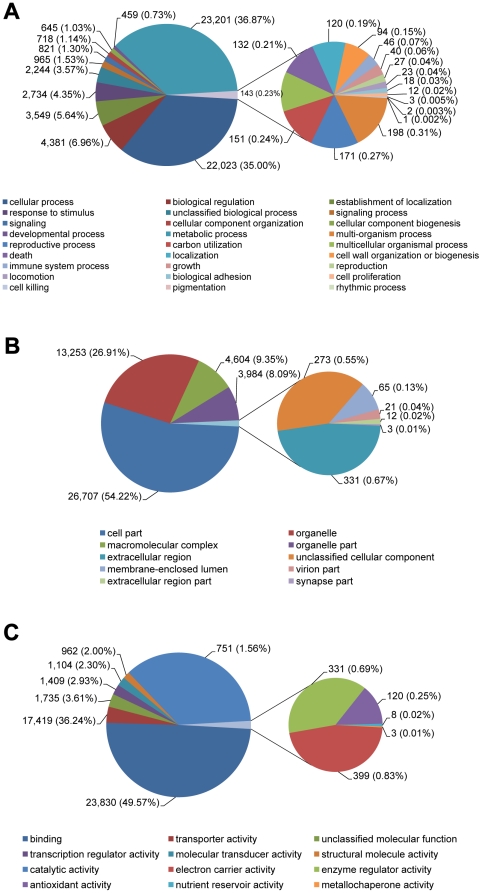
The derived Fraxinus phloem transcripts were assigned to three functional groups based on Gene Ontology (GO) terminology: Biological Process, Molecular Function and Cellular Component (Table S2). The software assigned 1,248 biological process terms to 19,958 transcripts (Figure 4A), 396 cellular component terms to 17,977 transcripts (Figure 4B), and 1,386 molecular function terms to 24,294 transcripts (Figure 4C). The most represented biological process terms were related to development (36.87%) and carbon utilization (35%). For those assigned to the cellular component, 54.22% were presumably associated with the cell part and 26.91% of the transcripts were representative for the organelle. Finally, the majority of terms represented in molecular function were for binding (49.57%) and catalytic activity (36.24%) suggesting a high degree of basal metabolic activity. It is well documented that a higher percentage of transcripts are involved in binding and catalytic activity in phloem compared to other parts of the plant [34]. We identified 417 Fraxinus transcripts encoding for proteins that are potentially involved in stress responses, including 120 transcripts encoding for heat-shock proteins. These proteins could be involved in responding to external stimuli including biotic and abiotic factors [35]. Functional annotation is a prerequisite to better understand transcriptomic data (especially of non-model systems). The GO facilitates functional characterization of genes, transcripts and proteins of any organisms with respect to cellular component, biological process and molecular function in a species-independent manner as reported in several other studies [36]–[38].
Figure 4. Depiction of Gene ontology (GO) terms for the transcriptomic sequences of Fraxinus spp.
(A) Biological Process, (B) Cellular Component and (C) Molecular Function.
Metabolic pathways
Overall 4,667 sequences were assigned to 142 KEGG metabolic pathways and the number of transcripts in the different pathways ranged from 1 to 1,422 (Table S3). The highest number of transcripts (1,422) was found in the secondary metabolites biosynthesis pathway (Table 1). In total, 788 transcripts that are involved in 7 biosynthesis pathways for alkaloids including indole alkaloid, isoquinoline alkaloid, tropane, piperidine and pyridine alkaloids were predicted in the ash sequences. Alkaloids are important components of the plant defense system against insect herbivory and so far 12,000 different alkaloids have been reported in plants which are involved in plant defense, growth and development [39], [40].
Table 1. Putative defense pathways identified in Fraxinus spp.
| Pathway | # of ESTs |
| Biosynthesis of secondary metabolites | 1422 |
| Biosynthesis of plant hormones | 668 |
| Biosynthesis of phenylpropanoids | 531 |
| Thiamine metabolism | 232 |
| Arginine and proline metabolism | 191 |
| Cysteine and methionine metabolism | 169 |
| Valine, leucine and isoleucine degradation | 151 |
| Phenylalanine metabolism | 127 |
| Lysine degradation | 123 |
| Tryptophan metabolism | 123 |
| Tyrosine metabolism | 112 |
| Flavonoid biosynthesis | 96 |
| Drug metabolism (cyt P450) | 93 |
| Metabolism of xenobiotic by cyt P450 | 72 |
| Anthocyanin biosynthesis | 22 |
| Isoflavonoid biosynthesis | 10 |
In this study, we recovered a high number of transcripts (531) that were mapped to the phenylpropanoid biosynthesis pathway. This pathway leads to the production of several phenolic compounds (flavonoids, tannins, coumarins etc.,) that plays an important role in plant defense against herbivores, microbes, and wounding [41]–[44]. Although not all of the major genes reported in the pathway were found in this study, this information provides a good base for further analysis and to better understand the potential role of phenylpropanoids in ash defense against biotic stress.
Protein domains
A domain search using HMMER3 software identified 2,534 distinct domains in 19,291 ash transcriptomic sequences (Table S4). Among the top Pfam domains, the most abundant ones were protein kinase domains (588) and protein tyrosine kinase domains (464). Protein kinases are primarily involved in plant signal transduction pathways [45], [46] and also participate in plant defense responses wherein they play an important role in signaling during pathogen recognition and activation of other plant defense mechanisms [47]–[49]. On the other hand, proteins containing tyrosine kinase domains and protein tyrosine phosphatases (PTPs) regulate abscisic acid (ABA) transduction pathways in plants [50]. The role of PTPs has been largely ignored; however a few tyrosine specific phosphatases were reported in A. thaliana [51]–[52]. PTPs are well documented in Daucus carota, Mimosa pudica, Arabidopsis hypocotyls and suspension cells [53]–[56]. Other abundant domains included metallothionein (263) and RNA recognition motif (RRM, 231). While metallothioneins are primarily involved in copper detoxification [57], RRM (also known as RNA binding domain or Ribonucleoprotein domain) plays an important role in post transcriptional events and in particular is involved in the 3′ end processing of chloroplast mRNA, [58], [59]. In a recent study, RRMs have emerged as key players in plant morphogenesis and RNA metabolism in chloroplast and mitochondria [60]. Further, we identified 154 RAS family members, which constitute RAS, RHO, RAB/YPT, ARF and RAN. RAS and RHO GTPases are considered to be important components in signaling cascades [61].
Interestingly, 153 cytochrome P450 domains were predicted in the derived ash sequences. Plant cytochrome P450 monoxygenases are thought to be involved in many biochemical pathways including the biosynthesis of secondary metabolites (e.g. phenylpropanoids, alkaloids, terpenoids, glucosinolates etc.), which have been well studied in plant-insect interactions [62]. However, plant cytochrome P450s are also involved in the biosynthesis of brassinosteroids and plant growth regulators [63], [64].
In total, 92 PPR (pentatricopeptide repeat) domains were identified in the ash sequences. The PPR repeat domain of ∼35 amino acids are well-known protein family members of both prokaryotes and eukaryotes [65] and appeared to function as sequence-specific RNA-binding proteins involved in post-transcriptional processes within organelles and translation initiation [66]–[71]. We also identified the PIWI domain in 18 transcripts and the PAZ domain in 6 transcripts. These domains are reported to be up regulated in the egg of A. thaliana and the presence of these domains suggests their role in epigenetic regulations through small RNA pathways [72].
Genes of Interest
Plants being sessile overcome various biotic and abiotic stress conditions through controlled gene expression. Immediate recognition of the biotic or abiotic factors/stimuli (i.e. in the early stages) is one of the key factors in plant defense [73]. Of the potential genes of interest listed in Table 2, we are particularly interested in those genes that participate in the early stages of plant defense including calcium dependent protein kinases (CDPKs); the transcription factors (TFs) WRKYs, MYBs and ethylene response factor (ERF); and a lipoxygenase (LOX3).
Table 2. Genes of interest recovered from the Fraxinus spp. transcriptomic database.
| Candidate genes | Number of occurrence |
| Proteases | 282 |
| cytochrome P450 | 192 |
| Lipase | 94 |
| WRKYS * | 47 |
| CDPKS | 43 |
| MYB | 37 |
| Hydroxyproline-rich glycoprotein | 29 |
| Protease/proteinase inhibitors | 16 |
| Phytoalexin deficient 4 (PAD4) | 11 |
| Hypersensitive-induced response protein | 9 |
| DREB | 07 |
| Myrosinase | 7 |
| Lipoxygenase | 6 |
| Jasmonic acid-amino conjugating enzyme | 3 |
| Pathogen-related protein | 3 |
| ERF | 02 |
*Candidate genes assayed in this study (in bold).
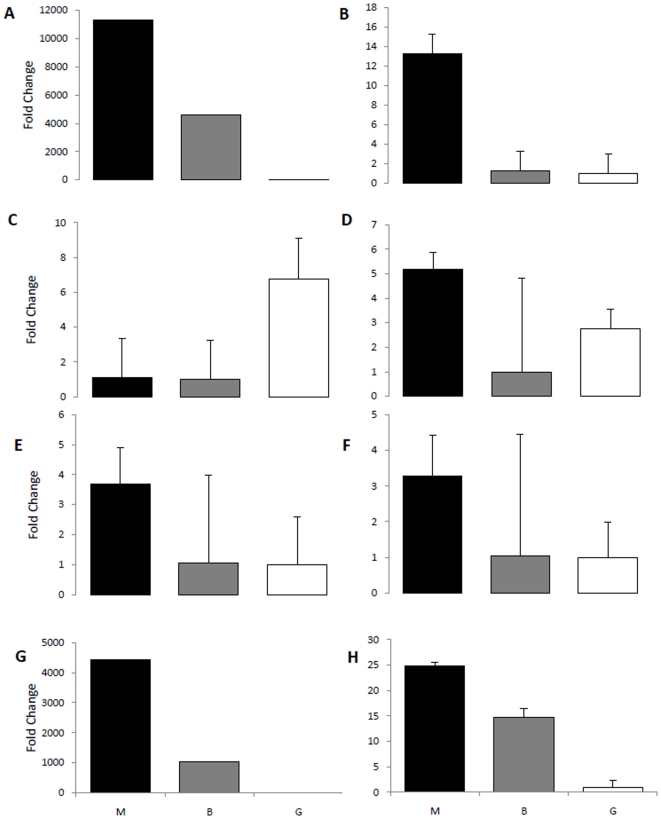
In the current study, both CDPKs (CDPK 349 and CDPK 361) showed the highest mRNA levels in Manchurian ash followed by black ash and green ash (Figure 5A and 5B). Interestingly, both CDPKs of ash significantly matched with CDPK3 of Nicotiana tabacum and P. trichocarpa (1e-14 and 6e-53). In a recent study, it was reported that CDPK3 and CDPK13 are involved in herbivory-induced signaling network via the regulation of defense related transcriptional machinery in A. thaliana [74]. Usually a dramatic change in cytosolic Ca2+ is observed through signaling pathways mediated by CDPKs in plants upon biotic and abiotic stress [75]–[77]. These findings along with the expression analysis could suggest that the recovered Fraxinus CDPKs (349 and 361) may regulate the transcriptional machinery involved in defense response. We posit that, the observed (constitutive) high mRNA levels for both CDPKs in Manchurian ash (compared to the susceptible black and green ash) may represent an enhanced capability to defend against A. planipennis. However, further studies need to be performed to confirm these observations.
Figure 5. Quantitative PCR analysis of candidate early regulators in different Fraxinus spp.
Includes Manchurian ash (M, black bars), black ash (B, grey bars) and green ash (G, white bars). Expression levels are shown for two CDPKs (A & B); two WRKYs (C & D), two MYBs (E & F), an ethylene response factor (G), and a lipoxygenase (H). An ash glucose-6-phosphate dehydrogenase (G6PD) was used as the internal reference gene. Standard error of the mean for two biological replicates (nested with two technical replicates) is represented by the error bars.
Upon physiological and environmental stimuli TFs (sequence specific DNA binding proteins) modulate transcription of specific target genes by binding to cis-elements located in gene promoters and/or introns [78]–[80]. TFs represent potential candidate genes for developing novel traits in crop plants [81]. In the current transcriptomic study, we found high occurrence of WRKYs, which are key regulators in higher plants represent the top ten largest families of transcription factors and are found throughout the green plants [82]. These early regulators are involved in modulating defense responses, abiotic stress and biosynthesis of secondary metabolites [83]–[86]. Expression analysis of two WRKYs (WRKY 7 and WRKY21) in ash revealed higher mRNA levels for WRKY 7 in Manchurian ash and for WRKY21 in green ash compared to the other ash species assayed (Figure 5C and 5D). These results may suggest that the profiled WRKYs respond to different external stimuli including biotic and abiotic stress. As reviewed in many previous studies, overexpression of OsWRKY resulted in enhanced salt and drought tolerance and the AtWRKY 25 mutants exhibited increased thermosensitivity [82], [87]. Besides these biotic and abiotic stress responses, WRKY proteins are reported to be involved in sugar signaling and seed development [88]–[91].
Both the MYBs (MYB8679 and MYB10337) assayed in ash were highly expressed in Manchurian ash compared to the mRNA levels observed for green and black ash species (Figure 5E and 5F). MYBs are reported to be involved in several physiological and biochemical processes including defense and stress response, regulation of secondary metabolism and signaling pathways [92]–[96].
In the current study, we found an ethylene response factor (ERF) that showed higher mRNA levels in Manchurian ash compared to green and black ash (Figure 5G). ERFs are important TFs that bind to the GCC motif of promoter region of ethylene-regulated genes [97]. In response to insect attack, plants usually show an ethylene burst which eventually activates polyphenol oxidase, peroxidase and proteinase inhibitor activities [98]. Ethylene is known to regulate a large number of genes related to defensive proteins and secondary metabolites [99], [100]. In a recent study it was shown that several genes involved in ethylene signaling were upregulated during forest tent caterpillar (Malacosma disstria) feeding on hybrid poplar leaves [101].
Expression analysis of an ash lipoxygenase 3 (LOX3) revealed the highest mRNA levels in Manchurian ash compared to black and green ash (Figure 5H). LOXs are versatile catalysts that participate in various physiological processes and are ubiquitous in nature [102]. In particular, LOXs in higher plants play a vital role in lipid peroxidation processes during plant defense responses and represent precursors for the biosynthesis of jasmonic acid related products, growth and development, senescence and during abiotic stress [102]–[104]. Although it is thought that LOX genes are up-regulated upon insect attack and/or wounding, perhaps the higher constitute levels of LOX3 in Manchurian ash could represent a primed response to A. planipennis.
The current study reports a number of transcription factors and other early regulators that are potentially involved in resistance to A. planipennis. Further studies need to be performed to learn the molecular functions of these reported genes which were observed to be expressed more abundantly in resistant Manchurian ash compared to the susceptible NA ash species.
Molecular markers
We have identified 1,272 single nucleotide polymorphisms (SNPs) in 410 ash transcriptome sequences (Table 3 and Table S5). Among them, 823 were transitions, i.e., changes from one purine to another purine or one pyrimidine to another pyrimidine, and 449 were transversions, changes between purines or pyrimidines (Table 3). This ratio of transitions to transversions (2∶1) of SNP occurrence in ash correlates well with other systems [105]. About 94% of the microsatellite loci predicted were dinucleotide (389) and tri-nucleotide repeats (532) followed by quad-nucleotide (37), hexa-nucleotide (17) and penta-nucleotide (5) repeats (Table 4 and Table S6). In general, EST-derived microsatellites are shorter than the genomic microsatellites [106], however, long dinucleotide microsatellites (CT)22 were predicted in the current study. Primers were designed for 25 microsatellites (10 primers for dinucleotide repeats and 15 primers for trinucleotide repeats) from the above predicted microsatellites. Seventeen of the 25 primers showed single band amplification in a PCR run, out of which seven were genotyped to check for polymorphism among three ash species (white, green, and Manchurian). Results indicate all seven loci to be polymorphic among the three species studied, providing a valuable resource of molecular markers for ash (Table 5). Similar observations were reported in transcriptomic studies of C. sativus and E. grandis, wherein 454 pyrosequencing was shown to be an excellent method for large scale prediction of molecular markers for future genetic linkage and QTL analysis in non-model organisms [13], [16]. Given that these microsatellite and SNP markers were predicted from transcriptomic sequences, they are likely linked to protein-coding genes, and therefore might have substantial physiological implications.
Table 3. Summary of putative SNPs in Fraxinus spp. transcriptomic sequences.
| SNP types | Number |
| Transition | |
| A-G | 411 |
| C-T | 412 |
| Transversion | |
| A-C | 123 |
| A-T | 137 |
| C-G | 80 |
| G-T | 109 |
| Total | 1,272 |
Table 4. Summary of microsatellite loci predicted in Fraxinus spp. transcriptomic sequences.
| Number of repeats | Di-nucleotide repeats | Tri-nucleotide repeats | Quad-nucleotide repeats | Penta-nucleotide repeats | Hexa-nucleotide repeats |
| 5 | 312 | 22 | 4 | 14 | |
| 6 | 128 | 9 | 3 | ||
| 7 | 52 | 2 | 1 | ||
| 8 | 129 | 14 | 1 | ||
| 9 | 86 | 10 | |||
| 10 | 46 | 3 | 1 | ||
| 11 | 34 | 2 | 2 | ||
| 12 | 29 | 8 | |||
| 13 | 10 | 1 | |||
| 14 | 12 | ||||
| 15 | 8 | 1 | |||
| 16 | 8 | ||||
| 17 | 3 | 1 | |||
| 18 | 6 | ||||
| 19 | 3 | ||||
| 20 | 9 | ||||
| 21 | 1 | ||||
| 22 | 2 | ||||
| 23 | 1 | ||||
| 24 | 2 | ||||
| Subtotal | 389 | 532 | 37 | 5 | 17 |
Table 5. Number of alleles in seven loci of three ash species (Fraxinus americana- White, F. mandshurica- Manchurian and F. pennsylvanica-Green) respectively.
The basic understanding of host resistance mechanisms in angiospermous trees including ash against woodborers is very limited. While NA ash species (green, black and white ash) are highly susceptible to A. planipennis, the incidence of attacks against Asian ash species including Manchurian and Chinese has been historically low, suggesting host resistance in the latter [107]. To date there is no evidence that any native NA ash species possesses resistance to EAB attack, which makes the entire ash population highly vulnerable to EAB invasion. Thus, results stemming from this functional genomics study to discover host resistance factors in ash, could feed into future ash breeding/genetic improvement programs.
Conclusions
The utilization of second generation sequencing for ash species has revealed various metabolic pathways that are of high interest with respect to ash resistance to A. planipennis. Data pertaining to the constitutive expression levels of early gene regulators in different ash species, revealed higher levels in Manchurian ash compared to NA ash. Results obtained in this study will lay the foundation for future differential gene expression analysis among different ash species and in deciphering the pathways of secondary metabolism which is related to plant defense. Molecular markers predicted in the current study will further help in population genomics and gene based association studies. These studies will provide critical insights to develop NA ash species that are resistant to A. planipennis through breeding programs and/or the application of transgenic technology.
Materials and Methods
Sample collection and RNA extraction
Two 10 mm in diameter phloem plugs from un-infested (by A. planipennis) green (F. pennsylvanica), white (F. americana), black (F. nigra), blue (F. quadrangulata) and Manchurian ash (F. mandshurica) were collected in February 2009 from a common garden established in 2003 at Novi, MI. The trees sampled did not have D-shaped exit holes and/or vertical splits on the trunk which are indicators of EAB infestation [3]. At least three different trees were sampled per species and the phloem plugs were immediately wrapped in aluminum foil and stored in liquid nitrogen. Approximately 70 mg of phloem tissue (phloem plugs homogenized in liquid nitrogen) per species was used for RNA extraction. Total RNA was extracted using Trizol® Reagent (Invitrogen, Carlsberg, CA) following manufacturer's protocol and stored at −80°C until further use.
cDNA library construction
The RNA extracted from the five Fraxinus species described above was aliquoted and pooled to construct a cDNA library. RNA isolated from different ash species was pooled in order to capture a diverse population of transcripts and potential species specific transcripts. Further, pooling of the RNA samples represents a cost effective transcriptomic approach to build an EST database for closely related species of a non-model organism. A SMART cDNA library construction kit (Clontech, Mountain View, CA) was used following manufacturer's protocol with modifications: i) A modified CDSIII/3′ primer (5′-TAG AGG CCG AGG CGG CCG ACA TGT TTT GTT TTT TTT TCT TTT TTT TTT VN-3′; PAGE purified) and SuperScript II reverse transcriptase (Invitrogen, Carlsberg, CA) were used for first-strand cDNA synthesis, ii) cDNA size fractionation was excluded and final products were cleaned and eluted using a QIAquick PCR purification kit (Qiagen, Valencia, CA).
Roche 454 sequencing
cDNA was sheared by nebulization and DNA fragments of approximately 500–800 bp were isolated by agarose gel electrophoresis and subsequent extraction. The isolated DNA was blunt ended, ligated to adapters and immobilized on beads. Single stranded DNA was later isolated from these beads. The isolated library was subjected to Quality Control using RNA 6000 (Agilent Technologies). Concentration and ligation of adapters were estimated using quantitative real-time PCR (qPCR). The emPCR reactions were performed to amplify a single template onto a single sequencing bead. One-quarter of a pico-titer plate was sequenced at the Purdue Genomics Core Facility (West Lafayette, IN) using the GS FLX Titanium chemistry (Roche Diagnostics, Indianapolis, IN).
Bioinformatic analysis
The 454 transcriptome reads were assembled using Newbler software package (Roche Diagnostics) after the removal of adapter sequences. To achieve better consistency, the contigs and singletons were renamed in the format of “ASH454ONE000001” where “ASH” stands for the ash genus, “454” for 454 sequencing technology, “ONE” for the first trial, and “000001” for an arbitrarily assigned number. The ash transcriptome sequences were annotated by searching against GenBank non-redundant database using the BLASTx algorithm [108]. Also, the sequences were compared to the protein sequences of A. thaliana in TAIR9 release from The Arabidopsis Information Resource (http://www.arabidopsis.org/) and P.trichocarpa v1.1 (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) using BLASTx algorithm. Protein domains were identified by searching against the Pfam database release 24.0 [109] using HMMER v3 program [110]. The Blast2GO software [111], [112] was used to predict the functions of the sequences, assign Gene Ontology terms, and predict the metabolic pathways in Kyoto Encyclopedia of Genes and Genome [113]–[115]. Microsatellite markers were identified using the Msatfinder version 2.0.9 program [116]. SNPs in the library were predicted using gsMapper software (Roche Diagnostics) with an arbitrary criterion of at least 4 reads supporting the consensus or variant.
Gene mining and quantitative real-time PCR
The ash transcriptome database was mined for genes potentially involved in plant defense. The constitutive gene expression profiles of potential early regulators: CDPK349, CDPK361, MYB, LOX, WRKY and ERF were analyzed using qPCR. cDNA was synthesized from green, black and Manchurian ash using a SuperScript™ First-Strand synthesis kit (Invitrogen, Carlsberg, CA) following manufacturer's protocol. We selected these three ash species to include one genetically close to Manchurian (black ash) and a species distantly related (green ash) which show different levels of susceptibility to A. planipennis [5], [6], [117]. Primers were designed using Beacon Designer 7 software (Table S7). The cycling parameters were 95°C for 5 min followed by 39 cycles of 95°C for 10 s and 60°C for 30 s ending with a melting curve analysis (65°C to 95°C in increments of 0.5°C every 5 s) to check for nonspecific product amplification. Relative gene expression was analyzed by the 2-ΔΔCT method [118]. An ash glucose-6-phosphate dehydrogenase (G6PD) was used as the internal reference gene, which has been previously shown to serve as a good internal control in plants [119].
Microsatellites analysis
Samples from eight individual trees of green, white and Manchurian ash were collected from the U.S. Forest Service, Northern Research station experimental plot Delaware, OH. Genomic DNA was extracted using E.Z.N.A. DNA kit (Omega Bio-Tek, Northcross, GA). Primers were designed for 25 of the predicted microsatellite markers of which only seven were used for genotyping (Table S8). Amplifications were performed in 10 µl reactions. Each reaction contained 5 µl of 2X-Failsafe PCR mix (Epicentre Biotech, Madison, WI), 0.5 U Taq polymerase, 2 pmol reverse primer, 4 pmol modified forward primer (M13 sequence at 5′ end) and ∼10 ng of DNA. M13-tagging protocol was followed using 4pmol of M13 fluorescently-tagged primer (5′-CACGACGTTGTAAAACGAC-3′) [120]. Thermocycling conditions were as follows: 94°C for 5 min, 35 cycles of 94°C for 20 s, 59°C for 20 s, 72°C for 30 s followed by eight cycles of 94°C for 30 s, 53°C for 15 s and 72°C for 30 s with a final extension at 72°C for 10 minutes [121]. PCR products were genotyped using Beckman-Coulter CEQ8800XL (Fullerton, CA) at the Molecular and Cellular Imaging Center (OARDC, Wooster, OH). Alleles were determined using CEQ Fragment Analysis software.
Data Deposition
The Roche 454 reads of Fraxinus species were submitted to NCBI Sequence Read Archive under the accession number of SRA020745.3
Supporting Information
Comparison of Fraxinus spp. sequences with the model plants Arabidopsis thaliana and Populus trichocarpa.
(XLSX)
Gene Ontology annotation results of Fraxinus spp. sequences.
(XLSX)
KEGG summary of Fraxinus spp. sequences.
(XLSX)
Pfam domain search of Fraxinus spp. sequences.
(XLSX)
Predicted SNPs in Fraxinus spp. sequences.
(XLSX)
Microsatellites (SSRs) loci in Fraxinus spp. sequences.
(XLSX)
List of primers for Fraxinus qPCR analysis
(DOC)
List of primers for SSRs identified from Fraxinus spp.
(DOC)
Acknowledgments
Help provided by the Purdue Genomics Core Facility (Phillip San Miguel and Rick Westerman) in performing the high throughput 454 sequencing and initial analysis of the raw sequencing data is much appreciated. Technical help provided by Binny Bhandary and Bryant Chambers (Department of Entomology, The Ohio State University/OARDC, Wooster, OH) is highly acknowledged. We thank Drs. Jennifer Koch and Mary Mason, U.S. Forest Service, Northern Research Station, Delaware, OH for allowing us to collect ash samples for validation of microsatellites. We also appreciate the help provided by Dr. Andy Michel (Department of Entomology, The Ohio State University/OARDC, Wooster, OH) with regards to the microsatellite analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The research was supported by a grant from the United States Department of Agriculture APHIS Accelerated Emerald Ash Borer Research Program (GRT00011769/60016270) and by State and Federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.MacFarlane DW, Meyer SP. Characteristics and distribution of potential ash tree hosts for emerald ash borer. For Ecol Manage. 2005;213:15–24. [Google Scholar]
- 2.Raupp MJ, Cumming AB, Raupp EC. Street tree diversity in eastern North America and its potential for tree loss to exotic borers. Arbor Urban For. 2006;32:297–304. [Google Scholar]
- 3.Herms DA, Stone AK, Chatfield JA. Emerald ash borer: the beginning of the end of Ash in North America? 2004, Ohio Agriculture Research and Development Center, Ohio State University Extension Special Circular 1932003. 2004:62–71. In: Chatfield JA, Draper EA, Mathers HM, Dyke DE, Bennett PJ, and Boggs JF. Ornamental Plants: Annual Reports and Research Reviews eds. [Google Scholar]
- 4.Poland T, McCullough D. Emerald ash borer: invasion of the urban forest and the threat to North America's ash resource. J Forestry. 2006;104:118–124. [Google Scholar]
- 5.Anulewicz AC, Mccullough DG, Cappaert DL, Poland TM. Host range of the emerald ash borer (Agrilus planipennis Fairmaire) (Coleoptera: Buprestidae) in North America: results of multiple-choice field experiments. Environ Entomol. 2008;37:230–241. doi: 10.1603/0046-225x(2008)37[230:hrotea]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Smith A. MS thesis. The Ohio State University, Columbus, , OH: 2006. Effects of community structure on forest susceptibility and response to the emerald ash borer invasion of the Huron River watershed in southeast Michigan. [Google Scholar]
- 7.Kovacs KF, Haight RG, McCullough DG, Mercader RJ, Seigert NW, et al. Cost of potential emerald ash borer damage in U.S. communities. Ecol Econ. 2009:2009–2019. [Google Scholar]
- 8.Gandhi KJK, Herms DA. Direct and indirect effects of alien insect herbivores on ecological processes and interactions in forests of eastern North America. Biol Invasions. 2010;12:389–405. [Google Scholar]
- 9.Yu C. Beijing: China Forestry Publishing House; 1992. Agrilus arcopoli Obenberger. In: Xiao GR (ed) Forest insects of China, 2nd edn. pp. 400–401. [Google Scholar]
- 10.Rebek EJ, Herms DA, Smitley DR. Interspecific variation in resistance to emerald ash borer (Coleoptera: Buprestidae) among North American and Asian Ash (Fraxinus spp.). Environ Entomol. 2008;37:242–246. doi: 10.1603/0046-225X(2008)37[242:IVIRTE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 11.Eyles A, Jones W, Riedl K, Cipollini D, Schwartz S, et al. Comparative phloem chemistry of Manchurian (Fraxinus mandshurica) and two North American Ash species (F. americana and F. pennsylvanica). J Chem Ecol. 2007;33:1430–1448. doi: 10.1007/s10886-007-9312-3. [DOI] [PubMed] [Google Scholar]
- 12.Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Novaes E, Drost DR, Farmerie WG, Pappas GJ, Grattapaglia D, et al. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics. 2008;9:312. doi: 10.1186/1471-2164-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber A, Weber K, Carr K, Wilkerson C, Ohlrogge J. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr Opin Plant Biol. 2009;12:107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo S, Zheng Y, Joung J, Liu S, Zhang Z, et al. Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genomics. 2010;11:384–396. doi: 10.1186/1471-2164-11-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barakat A, DiLoreto DS, Zhang Y, Smith C, Baier K, et al. Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol. 2009;9:51. doi: 10.1186/1471-2229-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parchman TL, Geist KS, Grahnen JA, Benkman CW, Buerkle A. Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery. BMC Genomics. 2010;11:180. doi: 10.1186/1471-2164-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore MJ, Dhingra A, Soltis PS, Shaw R, Farmerie WG, et al. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wicker T, Schlagenhauf E, Graner A, Close T, Keller B, et al. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung F, Win J, Lang JM, Hamilton J, Vuong H, et al. Analysis of the Pythium ultimum transcriptome using Sanger and pyrosequencing approaches. BMC Genomics. 2008;9:542. doi: 10.1186/1471-2164-9-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 23.Emrich S, Barbazuk W, Li L, Schnable P. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao C, Evans C, Jensen R, Sobral B. Identification of new genes in Sinorhizobium meliloti using the Genome Sequencer FLX system. BMC Microbiol. 2008;8:72. doi: 10.1186/1471-2180-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishiyama T, Fujita T, Shin-I T, Seki M, Nishide H, et al. Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: Implication for land plant evolution. Proc Natl Acad Sci U S A. 2003;100:8007–8012. doi: 10.1073/pnas.0932694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalo MJ, Oliver M, Garcia-Mas J, Monfort A, Dolcet-Sanjuan R, et al. Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor Appl Genet. 2005;110:802–811. doi: 10.1007/s00122-004-1814-6. [DOI] [PubMed] [Google Scholar]
- 27.Barbazuk B, Emrich S, Chen H, Li L, Schnable P. SNP discovery via 454 transcriptome sequencing. Plant J. 2007;51:910–918. doi: 10.1111/j.1365-313X.2007.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittapalli O, Bai X, Mamidala P, Rajarapu SP, Bonello P, et al. Tissue-specific transcriptomics of the exotic invasive insect pest emerald ash borer. PLoS One. 2010;5:e13708. doi: 10.1371/journal.pone.0013708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang H, Carlson JE, Leebens-Mack JH, Wall PK, Mueller LA, et al. An EST database for Liriodendron tulipifera L. floral buds: the first EST resource for functional and comparative genomics in Liriodendron. Tree Genet Genomes. 2008;4:419–433. [Google Scholar]
- 30.Falara V, Fotopoulos V, Margaritis T, Anastasaki T, Pateraki I, et al. Transcriptome analysis approaches for the isolation of trichome-specific genes from the medicinal plant Cistus creticus subsp. creticus. Plant Mol Biol. 2008;68:633–651. doi: 10.1007/s11103-008-9399-0. [DOI] [PubMed] [Google Scholar]
- 31.Quesada T, Li Z, Dervinis C, Li Y, Bocock PN, et al. Comparative analysis of the transcriptomes of Populus trichocarpa and Arabidopsis thaliana suggests extensive evolution of gene expression regulation in angiosperms. New Phytol. 2008;180:408–420. doi: 10.1111/j.1469-8137.2008.02586.x. [DOI] [PubMed] [Google Scholar]
- 32.Caicedo AL, Purugganan MD. Comparative plant genomics. Frontiers and prospects. Plant Physiol. 2005;138:545–547. doi: 10.1104/pp.104.900148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Luo HM, Sun C, Song JY, Wu Q, et al. EST analysis reveals putative genes involved in glycyrrhizin biosynthesis. BMC Genomics. 2010;11:268. doi: 10.1186/1471-2164-11-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omid A, Keilin T, Glass A, Leshkowitz D, Wolf S. Characterization of phloem-sap transcription profile in melon plants. J Exp Bot. 2007;58:3645–3656. doi: 10.1093/jxb/erm214. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Eyles A, Mandich D, Bonello P. Systemic aspects of host-pathogen interactions in Austrian pine (Pinus nigra): a proteomics approach. Physiol and Mol Plant Pathol. 2006;68:149–157. [Google Scholar]
- 36.Conesa A, Gotz S. Blast2GO: a Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genomics. 2008;28:619–632. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Botton A, Galla G, Conesa A, Bachem C, Ramina A, et al. Large-scale Gene Ontology analysis of plant transcriptome-derived sequences retrieved by AFLP technology. BMC Genomics. 2008;9:347. doi: 10.1186/1471-2164-9-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sathiyamoorthy S, In JG, Gayathri S, Kim Y, Yang DC. Gene ontology study of methyl jasmonate-treated and non-treated hairy roots of Panax ginseng to identify genes involved in secondary metabolic pathway. Russ J Genet. 2010;7:828–835. [PubMed] [Google Scholar]
- 39.Adler LS, Karban R, Strauss SY. Direct and indirect effects of alkaloids on plant fitness via herbivory and pollination. Ecology. 2001;82:2032–2044. [Google Scholar]
- 40.Ziegler J, Facchini PJ. Alkaloid biosynthesis: metabolism and trafficking. Ann Rev Plant Biol. 2008;59:735–69. doi: 10.1146/annurev.arplant.59.032607.092730. [DOI] [PubMed] [Google Scholar]
- 41.Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Ann Rev Plant Physiol and Plant Mol Biol. 1989;40:347–369. [Google Scholar]
- 42.Treutter D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 2005;7:581–591. doi: 10.1055/s-2005-873009. [DOI] [PubMed] [Google Scholar]
- 43.Bernays EA. Plant tannins and insect herbivores: an appraisal. Eco Entomol. 1981;6:353–60. [Google Scholar]
- 44.Dixon RA, Achnine L, Kota P, Liu C-J, Reddy MS, et al. The phenylpropanoid pathway and plant defense: a genomics perspective. Mol Plant Pathol. 2002;3:371–390. doi: 10.1046/j.1364-3703.2002.00131.x. [DOI] [PubMed] [Google Scholar]
- 45.Hirt H, Scheel D. MAP Kinases in Plant Signal Transduction In Hirt, H: Results and Problems in Cell Differentiation: (Springer, Heidelberg) 2000. pp. 85–93. [DOI] [PubMed]
- 46.Tena G, Asai T, Chiu WL, Sheen J. Plant MAP kinase signaling cascades. Curr. Opin. Plant Biol. 2001;4:392–400. doi: 10.1016/s1369-5266(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 47.Zhang S, Klessig DF. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001;6:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]
- 48.Romeis T. Protein kinases in the plant defence response. Curr. Opin. Plant Biol. 2001;4:407–414. doi: 10.1016/s1369-5266(00)00193-x. [DOI] [PubMed] [Google Scholar]
- 49.Nurnberger T, Scheel D. Signal transmission in plant immune response. Trends Plant Sci. 2001;6:372–379. doi: 10.1016/s1360-1385(01)02019-2. [DOI] [PubMed] [Google Scholar]
- 50.Ghelis T, Bolbach G, Clodic G, Habricot Y, Miginiac E, et al. Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol. 2008;148:1668–1680. doi: 10.1104/pp.108.124594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kerk D, Templeton G, Moorhead GB. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008;146:351–367. doi: 10.1104/pp.107.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rayapureddi JP, Kattamuri C, Chan FH, Hegde RS. Characterization of a plant, tyrosine-speciWc phosphatase of the aspartyl class. Biochemistry. 2005;44:751–758. doi: 10.1021/bi0481794. [DOI] [PubMed] [Google Scholar]
- 53.Barizza E, Sciavo FL, Terzi M, Filippini F. Evidence suggesting protein tyrosine phosphorylation in plants depends on the developmental conditions. FEBS Lett. 1999;447:191–194. doi: 10.1016/s0014-5793(99)00272-0. [DOI] [PubMed] [Google Scholar]
- 54.Kameyama K, Kishi Y, Yoshimura M, Kanzawa N, Sameshima M, et al. Tyrosine phosphorylation in plant bending. Nature. 2000;407:37. doi: 10.1038/35024149. [DOI] [PubMed] [Google Scholar]
- 55.Huang HJ, Lin YM, Huang DD, Takahashi T, Sugiyama M. Protein tyrosine phosphorylation during phytohormone-stimulated cell proliferation in Arabidopsis hypocotyls. Plant Cell Physiol. 2003;44:770–775. doi: 10.1093/pcp/pcg082. [DOI] [PubMed] [Google Scholar]
- 56.Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, et al. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol. 2008;4:193. doi: 10.1038/msb.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roosens NH, Bernard C, Leplae R, Verbruggen N. Evidence for copper homeostasis function of metallothionein (MT3) in the hyperaccumulator Thlaspi caerulescens. FEBS Lett. 2004;577:9–16. doi: 10.1016/j.febslet.2004.08.084. [DOI] [PubMed] [Google Scholar]
- 58.Schuster G, Gruissem W. Chloroplast mRNA-3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 1991;10:1493–1502. doi: 10.1002/j.1460-2075.1991.tb07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maris C, Dominguez C, Allain F. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 60.Kroeger TS, Watkins KP, Friso G, Van Wijk KJ, Barkan, A A plant-specific RNA binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl Acad. Sci. U S A. 2009;106:4537–4542. doi: 10.1073/pnas.0812503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu G, Li H, Yang Z. Arabidopsis RopGAPs are a novel family of Rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for Rop-specific GTPase stimulation Plant Physiol. 2000;124:1625–1636. doi: 10.1104/pp.124.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuler MA. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 1996;112:1411–1419. doi: 10.1104/pp.112.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell. 2005;17:776–790. doi: 10.1105/tpc.104.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chapple C. Molecular genetics analysis of plant cytochrome P450-dependent monooxygenases. Ann Rev Plant Physiol Mol Biol. 1998;49:311–343. doi: 10.1146/annurev.arplant.49.1.311. [DOI] [PubMed] [Google Scholar]
- 65.Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 66.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 67.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mili S, Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol Cell Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 70.Nakamura T, Schuster G, Sugiura M, Sugita M. Chloroplast RNA-binding and pentatricopeptide repeat proteins. Biochem Soc Trans. 2004;32:571–574. doi: 10.1042/BST0320571. [DOI] [PubMed] [Google Scholar]
- 71.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, et al. Arabidopsis Female Gametophyte Gene Expression Map Reveals Similarities between Plant and Animal Gametes. Curr Biol. 2010;20:506–512. doi: 10.1016/j.cub.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 73.Maffei ME, Mithofer A, Boland W. Before gene expression: early events in plant insect interaction. Trends Plant Sci. 2007;12:310–16. doi: 10.1016/j.tplants.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 74.Kanchiswamy CN, Takahashi H, Quadro S, Maffei ME, Bossi S, et al. : Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 2010;26:10. doi: 10.1186/1471-2229-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pandey S, Tiwari SB, Upadhyaya KC, Sopory SK. Calcium signaling: linking environmental signals to cellular functions. Crit Rev Plant Sci. 2000;19:291–318. [Google Scholar]
- 76.Ludwig AA, Saitoh H, Felix G, Freymark G, Miersch O, et al. Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc Natl Acad Sci U S A. 2005;102:10736–10741. doi: 10.1073/pnas.0502954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romeis T, Ludwig AA, Martin R, Jones JDG. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001;20:5556–5567. doi: 10.1093/emboj/20.20.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Ann Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 79.Martinez E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol. 2002;50:925–947. doi: 10.1023/a:1021258713850. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol. 2005;5:1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Century K, Reuber TL, Ratcliffe OJ. Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 2008;147:20–29. doi: 10.1104/pp.108.117887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rushton PJ, Somssich IE, Ringler P Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 83.Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 84.Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol. 2003;51:21–37. doi: 10.1023/a:1020780022549. [DOI] [PubMed] [Google Scholar]
- 85.Mare C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM. Hv WRKY38: a new transcription factor involved in cold and drought response in barley. Plant Mol Biol. 2004;55:399–416. doi: 10.1007/s11103-004-0906-7. [DOI] [PubMed] [Google Scholar]
- 86.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. TheWRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 87.Li S, Fu Q, Huang W, Yu D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009;28:683–693. doi: 10.1007/s00299-008-0666-y. [DOI] [PubMed] [Google Scholar]
- 88.Sun CX, Hoglund AS, Olsson H, Mangelsen E, Jansson C. Antisense oligodeoxynucleotide inhibition as a potent strategy in plant biology: identification of SUSIBA2 as a transcriptional activator in plant sugar signalling. Plant Journal. 2005;44:128–138. doi: 10.1111/j.1365-313X.2005.02515.x. [DOI] [PubMed] [Google Scholar]
- 89.Sun CX, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, et al. A novel WRKY transcription factor, SUSIBA2, participates in sugar signalling in barley by binding to the sugar responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou Y, Zhang X, Kang X, Zhao X, Ni M. SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell. 2009;21:106–117. doi: 10.1105/tpc.108.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell. 2007;19:2531–43. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, et al. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gocal GFW, Poole AT, Gubler F, Watts RJ, Blundell C, et al. Long day up-regulation of a GAMYB gene during Lolium temulentum inflorescence formation. Plant Physiol. 1999;119:1271–1278. doi: 10.1104/pp.119.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 96.Newman LJ, Perazza DE, Juda L, Campbell MM. Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 2004;37:239–250. doi: 10.1046/j.1365-313x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- 97.Pirrello J, Jaimes-Miranda F, Sanchez-Ballesta MT, Tournier B, Khalil-Ahmad Q, et al. Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol. 2006;47:1195–1205. doi: 10.1093/pcp/pcj084. [DOI] [PubMed] [Google Scholar]
- 98.von Dahl CC, Baldwin IT. Deciphering the role of ethylene in plant-herbivore interactions. J Plant Growth Regul. 2007;26:201–209. [Google Scholar]
- 99.Harfouche AL, Shivaji R, Stocker R, Williams PW, Luthe DS. Ethylene signaling mediates a maize defense response to insect herbivory. Mol Plant-Microbe Interact. 2006;19:189–199. doi: 10.1094/MPMI-19-0189. [DOI] [PubMed] [Google Scholar]
- 100.Winz RA, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV.Insect-induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol. 2001;125:189–202. doi: 10.1104/pp.125.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Philippe RN, Ralph SG, Mansfield SD, Bohlmann J. Transcriptome profiles of hybrid poplar (Populus trichocarpa x deltoides) reveal rapid changes in undamaged, systemic sink leaves after simulated feeding by forest tent caterpillar (Malacosoma disstria). New Phytologist. 2010;188:787–802. doi: 10.1111/j.1469-8137.2010.03392.x. [DOI] [PubMed] [Google Scholar]
- 102.Kolomiets MV, Chen H, Gladon RJ, Braun EJ, Hannapel DJ. A leaf lipoxygenase of potato induced specifically by pathogen infection. Plant Physiol. 2000;124:1121–1130. doi: 10.1104/pp.124.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Veronesi C, Rickauer M, Fournier J, Pouenat ML, Esquerre-Tugaye MT. Lipoxygenase gene expression in the tobacco-Phytophthora parasitica nicotianae interaction. Plant Physiol. 1996;112:997–1004. doi: 10.1104/pp.112.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hwang IS, Hwang BK. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 2009;152:948–967. doi: 10.1104/pp.109.147827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Collins DW, Jukes TH. Rates of transition and transversion in coding sequences since the human-rodent divergence. Genomics. 1994;20:386–396. doi: 10.1006/geno.1994.1192. [DOI] [PubMed] [Google Scholar]
- 106.Thiel T, Michalek W, Varshney R, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet. 2004;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 107.Wei X, Wu Y, Reardon R, Sun TH, Lu M, Sun JH. Biology and damage traits of emerald ash borer (Agrilus planipennis Fairmaire) in China. Ins Sci. 2007;14:367–373. [Google Scholar]
- 108.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 109.Cogill P, Finn RD, Bateman A. Identifying protein domains with Pfam database. Curr Protoc Bioinformatics. 2008;2:2.5. doi: 10.1002/0471250953.bi0205s23. [DOI] [PubMed] [Google Scholar]
- 110.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 111.Conesa A, Goetz S, Garcia JM, Terol J, Talon M, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 112.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, et al. : KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;35:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thurston MI, Field D . 2005. Msatfinder: detection and characterization of microsatellites. Distributed by the authors at URL: http://www.genomics.ceh.ac.uk/msatfinder/.
- 117.Wallander E. Systematics of Fraxinus (Oleaceae) and evolution of dioecy. Plant Syst Evol. 2008;273:25–49. [Google Scholar]
- 118.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 119.Jian B, Liu B, Bi Y, Hou W, Wu C, et al. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol. 2008;9:59. doi: 10.1186/1471-2199-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- 121.Michel AP, Zhang W, Jung JK, Kang S, Mian MAR. Cross-species amplification and polymorphism of microsatellite loci in the soybean aphid, Aphis glycines. J. Econ. Entomol. 2009;102:1389–1392. doi: 10.1603/029.102.0368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of Fraxinus spp. sequences with the model plants Arabidopsis thaliana and Populus trichocarpa.
(XLSX)
Gene Ontology annotation results of Fraxinus spp. sequences.
(XLSX)
KEGG summary of Fraxinus spp. sequences.
(XLSX)
Pfam domain search of Fraxinus spp. sequences.
(XLSX)
Predicted SNPs in Fraxinus spp. sequences.
(XLSX)
Microsatellites (SSRs) loci in Fraxinus spp. sequences.
(XLSX)
List of primers for Fraxinus qPCR analysis
(DOC)
List of primers for SSRs identified from Fraxinus spp.
(DOC)