Abstract
Recent efforts have identified the p38α Ser/Thr kinase as a potential target for the treatment of inflammatory diseases as well as non-small cell lung carcinoma. Despite the significance of p38α, no direct activity probe compatible with cell lysate analysis exists. Instead, proxies for kinase activation, such as phosphospecific antibodies, which do not distinguish between p38 isoforms, are often used. Our laboratory has recently developed a sulfonamido-oxine (Sox) fluorophore that undergoes a significant increase in fluorescence in response to phosphorylation at a proximal residue, allowing for real-time activity measurements. Herein we report the rational design of a p38α-selective chemosensor using this approach. We have validated the selectivity of this sensor using specific inhibitors and immunodepletions and show that p38α activity can be monitored in crude lysates from a variety of cell lines, allowing for the potential use of this sensor in both clinical and basic science research applications.
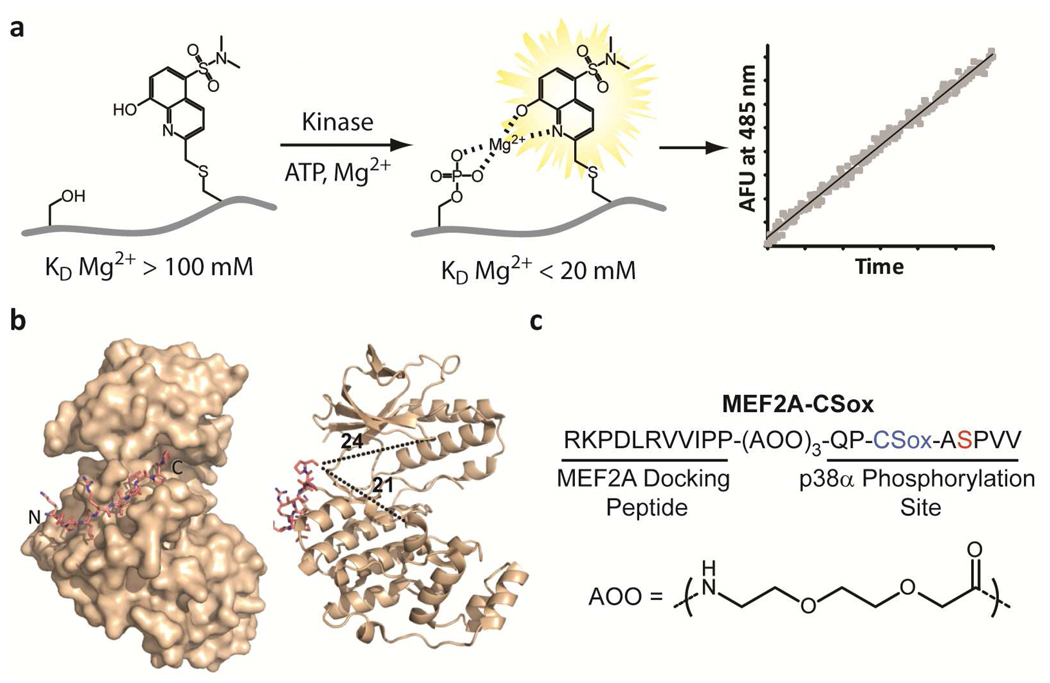
Methods for assessing kinase activities have relied on the transfer of a radioactive γ-phosphoryl moiety from ATP to substrate and have been very useful for in vitro studies of kinases. However, beyond being discontinuous, this assay is incompatible with unfractionated cell lysates since ATP is a common substrate for most kinases. This has prompted the development of protein-based fluorescence resonance energy transfer (FRET) sensors for probing kinase activity (1–3). While useful, these FRET-based sensors produce modest changes in fluorescence upon phosphorylation. Alternatively, the development of methods based on small organic fluorophores has provided sensitive probes for interrogating biological functions (4, 5). Recently our laboratory has introduced a direct kinase assay strategy based on chelation-enhanced fluorescence of a cysteine derivative of a sulfonamido-oxine fluorophore (6) which we term CSox (Figure 1, panel a). Placed (−)2 or (+)2 relative to the phosphorylation site in an optimized kinase substrate, the CSox amino acid provides a readily observable increase in fluorescence signal in response to phosphorylation due to chelation of Mg2+ between the newly installed phosphoryl group and CSox. These probes afford sensitive real-time fluorescence readouts of kinase activity in unfractionated cell lysates (7, 8), provided that selective substrate sequences for the kinase of interest can be identified (5, 9, 10).
Figure 1.
Rational design of a p38α chemosensor. a) A schematic of the chelation-enhanced fluorescence of the CSox amino acid upon phosphorylation. b) A crystal structure of the MEF2A docking peptide bound to p38α (left) and distances in Å from the C-terminus of the docking peptide to representative distal regions of the catalytic and substrate-binding domains of the kinase are shown (right) (18). c) The amino acid sequence of the MEF2A-CSox sensor with the position of CSox (blue) and the site of phosphorylation (red) indicated. The flexible 8-amino-3,6-dioxaoctanoic acid (AOO) linker is also shown.
Recently p38α, a member of the Mitogen-Activated Protein Kinase (MAPK) family, has been the target of a variety of drug development efforts (11, 12) since inhibitors of this kinase may provide treatments for inflammatory diseases (13). Additionally, increased activation of p38α in tumor tissue derived from patients with non-small cell lung carcinoma has been observed (14, 15). Consequently, with the goal of developing a direct p38α chemosensor which would be compatible with unfractionated cell lysates, we investigated strategies for generating selective substrates for MAPKs.
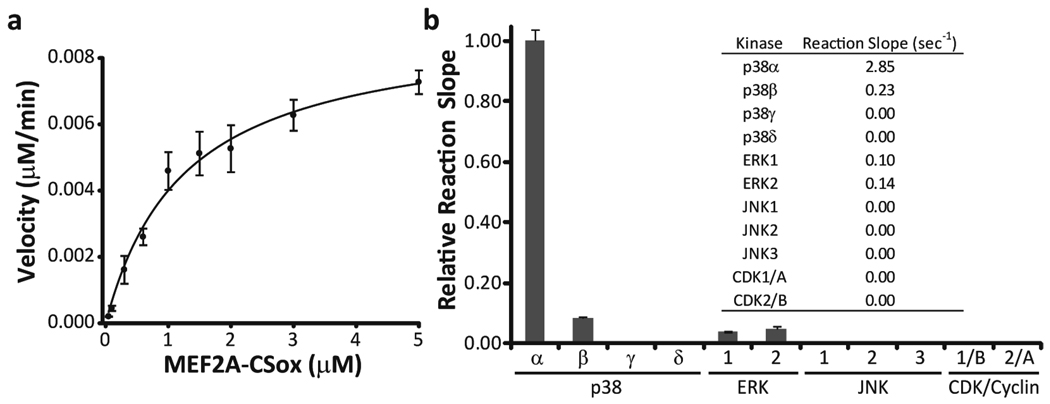
In the case of MAPKs the development of selective probes has proved more challenging due to the minimal local consensus phosphorylation sequence, S/T-P. This class of enzymes (including the ERK, JNK, and p38 family members) derives specificity through the use of extended protein or peptide docking domains that are distal to the phosphorylation site (16, 17). These docking domains serve to target a substrate to a particular kinase and can therefore be viewed as unique address elements. Due to the limited structural information concerning p38α substrates, we chose to employ a strategy in which a known docking peptide sequence (18, 19) (Figure 1, panel b) would be linked to a CSox-based phosphorylation site via a flexible linker (16) (Figure 1, panel c). Initial phosphorylation reactions indicated that this sensor, MEF2A-CSox, could act as a substrate for purified p38α (Supplementary Figure S1). Phosphorylation reactions containing differing amounts of MEF2A-CSox demonstrated a KM and Vmax for p38α of 1.3 µM and 1.1 µmol mg−1 min−1, respectively (Figure 2, panel a). We then assessed the specificity of MEF2A-CSox by exposing it to a panel of related kinases (Figure 2, panel b). MEF2A-CSox was selectively phosphorylated by p38α and showed minimal background activity in the presence of the closely related p38β isoform (8%) and the remaining kinase panel. Importantly, this difference in selectivity for p38α over p38β translated into a 17-fold enhancement in catalytic efficiency for p38α (Supplementary Figure S2). Encouraged by these in vitro studies, we investigated the ability of MEF2A-CSox to report p38α activation in unfractionated cell lysates.
Figure 2.
MEF2A-CSox is a substrate for recombinant p38α. a) A direct fit of a velocity versus MEF2A-CSox concentration plot using the Briggs-Haldane equation. b) Phosphorylation reactions were conducted with the indicated recombinant kinase (15 nM) using 1 µM substrate and demonstrate that MEF2A-CSox is selective for p38α among these kinases. The inset shows the average reaction slope for each kinase.
Several studies have demonstrated p38α activation in response to inflammatory cytokines or cellular stress (20). With this in mind, we treated HeLa cells with increasing amounts of the cytokine TNFα (Supplementary Figure S3). These initial experiments demonstrated that MEF2A-CSox was capable of reporting p38α activation despite appreciable signal due to phosphorylation by off-target kinases. A recent survey of kinase inhibitors (21) indicated that the broad spectrum inhibitor staurosporine is not effective against p38α, which was confirmed using recombinant enzyme (Supplementary Figure S4). Consequently we hypothesized that in this case, staurosporine may be used to reduce the off-target kinase activities allowing for discrimination of the p38α signal. Indeed, the addition of 1 µM staurosporine to assays using sorbitol-stimulated lysates demonstrated that a portion of off-target kinases could be suppressed by using this promiscuous inhibitor (Figure 3, panel a), therefore staurosporine was added to all subsequent lysate assays. A comparison between TNFα and sorbitol stimulation indicated an increase of 68% in the rate of MEF2A-CSox phosphorylation in cells stimulated by osmotic shock (Figure 3, panel b), consequently these lysates were used to optimize assay conditions. Optimal signal-to-noise for sorbitol-stimulated lysates was obtained using 10 µg of total protein (Supplementary Figure S5), which provided a clearly discernible enhancement in the rate of phosphorylation of MEF2A-CSox (Figure 3, panel c). Using small molecule inhibitors of p38α, specifically SB203580 which is ATP competitive (22–24) and BIRB796 a slow-binding allosteric inhibitor (25), the origin of the increase in the rate of MEF2A-CSox phosphorylation upon osmotic shock was investigated. The addition of 1 µM of each of these compounds, which completely abolishes activity of recombinant kinase (Supplementary Figure S6), reduced the rate of phosphorylation of MEF2A-CSox in sorbitol-stimulated HeLa lysates to levels observed for serum starved cells (Figure 3, panel d). Appropriate control measurements established that this effect was not due to the inhibitor solvent (DMSO) (Supplementary Figure S7).
Figure 3.
Reducing off-target phosphorylation of MEF2A-CSox in unfractionated cell lysates. a) Reactions were conducted using 1 µM substrate and 10 µg of the indicated HeLa lysate with or without the addition of staurosporine. The inset shows a western blot analysis of the indicated lysates, demonstrating p38 activation upon stimulation. b) The relative reaction slopes obtained using 1 µM substrate and 5 µg of the indicated lysates are shown. The inset shows a western blot analysis of each lysate. c) Initial phosphorylation reactions using 10 µg of total cellular protein from the indicated HeLa cell lysate demonstrated that MEF2A-CSox (1 µM) could report on the presence of active p38. Data is corrected for lag times, see Methods. d) Assays were performed using lysates from serum-starved or stimulated cells (10 µg) with MEF2A-CSox (1 µM). Inhibitors of p38α were added to the reactions containing lysates from stimulated cells where indicated. All reactions contained 1 µM staurosporine unless otherwise noted.
We sought to further verify the selectivity of MEF2A-CSox for p38α through a series of inhibition studies, immunodepletion experiments, and analyses across different cell lines. Accordingly, the phosphorylation of MEF2A-CSox was determined in the presence of varying concentrations of SB203580 and BIRB796 (Figure 4, panels a and b respectively). A concentration-dependent decrease in the rate of phosphorylation of MEF2A-CSox was observed in the presence of SB203580, yielding a Ki of 7.5 nM, which reflects the reported Ki of 21 nM (24). A similar dose-dependent response was observed for BIRB796 which was in good correlation with previously reported values (25). Importantly, these experiments indicate that the remaining background activity due to off-target kinases could be essentially eliminated through background subtraction of parallel reactions containing 1 µM SB203580 (see also Figure 3, panel d). Accordingly, to further verify the specificity of MEF2A-CSox, p38α immunodepletion studies were performed in which the activity remaining after the addition of SB203580 was used for background subtraction. These depletions clearly demonstrate that the increase in the rate of phosphorylation of MEF2A-CSox upon stimulation by osmotic shock is predominantly due to p38α (Figure 4, panel c). Moreover, similar depletions for the related kinase ERK5 (26) demonstrated no appreciable loss of signal (Supplementary Figure S8). In combination with the known selectivity profiles for the inhibitors used herein (21) (Supplementary Table S1), these results indicate that MEF2A-CSox is a p38α-selective activity probe and that off-target signal can be virtually eliminated using SB203580.
Figure 4.
MEF2A-CSox can selectively report on p38α activation in unfractionated cell lysates. Dose dependant responses to the addition of SB203580 (a) and BIRB796 (b) were observed in sorbitol-stimulated lysates (10 µg) using MEF2A-CSox (1 µM). The observed IC50 and structure of each compound are shown in the inset. c) Phosphorylation reactions were performed on lysates which had been depleted using a p38α-specific antibody. Values were background subtracted using the activity remaining in the input lysate after addition of 1 µM SB203580. The inset shows a western blot of the immunodepleted lysates. d) The indicated cell lines were stimulated with 300 mM sorbitol for 1 hr (S). Phosphorylation reactions were performed as above and compared to lysates from cells which had not been stimulated (C). The amount of p38α activity in each individual lysate was determined by subtracting the activity remaining after the addition of 1 µM SB203580. Western blots for both total and active p38 are shown above each sample. All reactions contained 1 µM staurosporine.
Finally, we investigated the ability of MEF2A-CSox to report the activation of p38α in a variety of cell lines isolated from different tissues and species. Indeed MEF2A-CSox was capable of reporting the activation of p38α in HeLa (human), Cos7 (simian), and NIH-3T3 (rodent) cells (Figure 4, panel d). The activity in each lysate correlated with western blot analyses indicating that MEF2A-CSox can be used in a variety of mammalian systems and tissues to directly interrogate p38α activity levels.
We have designed and validated the first isoform-selective p38 activity probe compatible with unfractionated cell lysates. This probe provides isoform-specific activity information that cannot be obtained through the use of currently available phosphospecific antibodies. Because the synthesis described herein produces 2.4 mg of MEF2A-CSox, sufficient material for 7,000 assays in 96-well plate format or 28,000 assays in 384-well format (7), this sensor could easily be utilized to rapidly screen compound libraries to identify p38α inhibitors. Furthermore, we envision that this sensor will be useful for detailing the changes in kinase signaling pathways during cellular transformations such as differentiation and cancer development (15).
Methods
General Reagents and Methods
Low metals grade chemicals were obtained from Sigma and Alfa Aesar. Lysates were normalized for total protein content using the Bio-Rad protein assay (500-0006) with BSA as a standard. Fluorescence emission was acquired at 485 nm using 360 nm excitation on either a HTS 7000 Bio Assay Reader (Perkin Elmer) or Spectramax Gemini XS (Molecular Devices, 455 nm cutoff) plate reader. All fluorescence assays were performed at 30 °C in 96-well plates (Corning, 3992).
Synthesis of MEF2A-CSox
MEF2A-CSox was synthesized using standard Fmoc-based solid phase peptide synthesis methods as described previously (6). The linker in MEF2A-CSox was installed by coupling three Fmoc-protected AOO linkers (Novabiochem, 851037) to the growing peptide chain on 100 mg of PAL-PEG-PS resin (0.19 mmol g−1 substitution, Applied Biosystems). The Sox fluorophore was incorporated via on-resin alkylation of a selectively deprotected cysteine residue (6). The resulting peptide was acetyl-capped at the N-terminus and included a C-terminal amide derived from the resin. Purification was carried out by standard reverse phase HPLC. Characterization was performed using ESI-MS and concentrations were determined based on the Sox chromophore using the absorbance of the peptide at 355 nm in 0.1 M NaOH containing 1 mM Na2EDTA (extinction coefficient = 8,427 M−1cm−1) (27).
MEF2A-CSox Reactions Containing Recombinant Enzyme
Reactions were carried out using 15 nM of the indicated enzyme (Invitrogen) and 1 µM MEF2A-CSox in a buffer containing 50 mM Tris-HCl (pH = 7.5 at 25 °C), 10 mM MgCl2, 1 mM EGTA, 2 mM DTT, 0.01% Triton X-100, and 1 mM ATP in a final volume of 120 µL. Well-to-well path length variation was corrected by normalizing to starting intensities.
Determination of Kinetic Parameters of MEF2A-CSox with p38α and β
Reactions were performed as above using 1 ng of recombinant human p38α or β (Invitrogen) with increasing concentrations of substrate. Initial reaction slopes were then converted to rates as described previously (27).
Recombinant Kinase Panel Assays
Reactions were conducted as described above with 1 µM MEF2A-CSox and 15 nM of the indicated kinase (Invitrogen).
Preparation of Cell Lysates
HeLa cells were propagated in 90% DMEM supplemented with 10% heat-inactivated FBS, 50 U mL−1 penicillin, and 50 µg mL−1 streptomycin. Cos7 and NIH-3T3 cells were propagated in 90% DMEM supplemented with 10% FBS, 50 U mL−1 penicillin, and 50 µg mL−1 streptomycin. Prior to stimulation, cells were starved overnight (14 hrs) by the addition of DMEM supplemented with 2 mM L-Gln, 50 U mL−1 penicillin, and 50 µg mL−1 streptomycin. Cells were stimulated by the addition of the indicated amount of TNFα (Cell Signaling) for 10 min or sorbitol to 300 mM for 1 hr. Cells were then washed with ice cold PBS and lysed on ice in 50 mM Tris (pH = 7.5 at 25 °C), 150 mM NaCl, 50 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 30 mM NaF, 1% Triton X-100, 2 mM EGTA, 100 µM Na3VO4, 1 mM DTT, protease inhibitor cocktail III (10 µL mL−1, Calbiochem, 539134), and phosphatase inhibitor cocktail 1 (10 µL mL−1, Sigma, P2825). Lysates were clarified by centrifugation and supernatants were flash frozen in liquid nitrogen and stored at −80 °C.
Immunodepletions
Immunodepletions were conducted as described previously (8). Briefly, sorbitol-stimulated HeLa lysates were aliquoted into separate samples (350 µg each) at 4 °C. Depletions were conducted using a rabbit anti-p38α (1 µg) antibody (Cell Signaling, 9218) along with a naïve rabbit IgG control (1 µg, GE Life Sciences). Antibody-bound complexes were precipitated by the addition of Protein A agarose conjugated beads (GE Life Sciences). Input (untreated) samples were used to determined the amount of activity lost due to handling while a separate sample was treated with Protein A beads alone to determine the amount of activity lost due to non-specific binding to the resin. Lysates were flash frozen and stored at −80 °C. ERK5 depletions were conducted in a similar manner using the appropriate antibody (Cell Signaling, 3372). The rate of phosphorylation by off-target kinases was background-subtracted using the activity remaining after the addition of 1 µM SB203580 to the input lysates.
MEF2A-CSox Lysate Assays
Assays were typically conducted using 1 µM MEF2A-CSox, 10 µg total protein from cell lysates, and 1 µM staurosporine, unless otherwise indicated. Reactions were prepared in bulk in a buffer consisting of 50 mM Tris-HCl (pH = 7.5 at 25 °C), 10 mM MgCl2, 1 mM EGTA, 2 mM DTT, and 0.01% Brij 35 P with the indicated concentration of p38α inhibitor and aliquoted into 96-well plates. After addition of lysate, reactions (final volume 120 µL) were incubated at 30 °C and fluorescence emission was monitored. Data were corrected for lag times (typically 5–10 min, after which fluorescence increases were linear with respect to time for at least 1 hr) as well as variations in well-to-well path lengths. For titrations with SB203580, the calculated IC50 was converted to a Ki using the Cheng-Prussof equation with the concentration of ATP in the assay and the reported KM of p38α for ATP (24). Slopes for assays containing the slow-binding inhibitor BIRB796 (25) were determined after a 10 min incubation of the entire solution at 30 °C. After this incubation fluorescence increases were linear with respect to time for at least 1 hr.
Slopes of phosphorylation reactions for assays containing lysates from different cell lines (Figure 4, panel d) were background corrected using the activity remaining in each individual lysate after the addition of 1 µM SB203580.
Western Blot Analysis
Lysates (20 µg total protein unless noted) were separated by SDS-PAGE and proteins were transferred to a nitrocellulose membrane. Blots were probed with primary antibodies for total p38 (Cell Signaling, 9212), phospho-p38 (Cell Signaling, 9215), p38α, p38β (Cell Signaling, 2339), total ERK5 (Cell Signaling, 3372), or phospho-ERK5 (Cell Signaling, 3371, 300 µg of total protein used for blot) which were detected using an HRP conjugated goat anti-rabbit secondary antibody (Pierce, 32460). Blots were visualized by enhanced chemiluminescence (Pierce, 34075).
Supplementary Material
Acknowledgement
C. Stains was supported by an NIH NRSA Fellowship (F32GM085909). This research was supported by a grant from the NIH (GM064346 – Cell Migration Consortium). We also acknowledge Professor K. Dalby for providing a sample of BIRB796 and the Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Harvey CD, Ehrhardt AG, Cellurale C, Zhong HN, Yasuda R, Davis RJ, Svoboda K. A genetically encoded fluorescent sensor of ERK activity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunkel MT, Toker A, Tsien RY, Newton AC. Calcium-dependent regulation of protein kinase D revealed by a genetically encoded kinase activity reporter. J. Biol. Chem. 2007;282:6733–6742. doi: 10.1074/jbc.M608086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato M, Kawai Y, Umezawa Y. Genetically encoded fluorescent indicators to visualize protein phosphorylation by extracellular signal-regulated kinase in single living cells. Anal. Chem. 2007;79:2570–2575. doi: 10.1021/ac062171d. [DOI] [PubMed] [Google Scholar]
- 4.Lee HM, Larson DR, Lawrence DS. Illuminating the Chemistry of Life: Design, Synthesis, and Applications of "Caged" and Related Photoresponsive Compounds. ACS Chem. Biol. 2009;4:409–427. doi: 10.1021/cb900036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothman DM, Shults MD, Imperiali B. Chemical approaches for investigating phosphorylation in signal transduction networks. Trends Cell Biol. 2005;15:502–510. doi: 10.1016/j.tcb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Lukovic E, Gonzalez-Vera JA, Imperiali B. Recognition-domain focused chemosensors: versatile and efficient reporters of protein kinase activity. J. Am. Chem. Soc. 2008;130:12821–12827. doi: 10.1021/ja8046188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukovic E, Vogel Taylor E, Imperiali B. Monitoring protein kinases in cellular media with highly selective chimeric reporters. Angew. Chem., Int. Ed. 2009;48:6828–6831. doi: 10.1002/anie.200902374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shults MD, Janes KA, Lauffenburger DA, Imperiali B. A multiplexed homogeneous fluorescence-based assay for protein kinase activity in cell lysates. Nat. Methods. 2005;2:277–283. doi: 10.1038/nmeth747. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-Vera JA, Lukovic E, Imperiali B. A rapid method for generation of selective Sox-based chemosensors of Ser/Thr kinases using combinatorial peptide libraries. Bioorg. Med. Chem. Lett. 2009;19:1258–1260. doi: 10.1016/j.bmcl.2008.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shults MD, Carrico-Moniz D, Imperiali B. Optimal Sox-based fluorescent chemosensor design for serine/threonine protein kinases. Anal. Biochem. 2006;352:198–207. doi: 10.1016/j.ab.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, Ding Y, Franklin GJ, Franzen KD, Gefter ML, Hale SP, Hansen NJV, Israel DI, Jiang JW, Kavarana MJ, Kelley MS, Kollmann CS, Li F, Lind K, Mataruse S, Medeiros PF, Messer JA, Myers P, O'Keefe H, Oliff MC, Rise CE, Satz AL, Skinner SR, Svendsen JL, Tang LJ, van Vloten K, Wagner RW, Yao G, Zhao BG, Morgan BA. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 12.Simard JR, Kluter S, Grutter C, Getlik M, Rabiller M, Rode HB, Rauh D. A new screening assay for allosteric inhibitors of cSrc. Nat. Chem. Biol. 2009;5:394–396. doi: 10.1038/nchembio.162. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S, Boehm J, Lee JC. p38 map kinases: Key signalling molecules as therapeutic targets for inflammatory diseases. Nat. Rev. Drug Discov. 2003;2:717–726. doi: 10.1038/nrd1177. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg AK, Basu S, Hu J, Yie TA, Tchou-Wong KM, Rom WN, Lee TC. Selective p38 activation in human non-small cell lung cancer. Am. J. Respir. Cell Mol. Biol. 2002;26:558–564. doi: 10.1165/ajrcmb.26.5.4689. [DOI] [PubMed] [Google Scholar]
- 15.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes N, Bailey DE, Vanvranken DL, Allbritton NL. Use of docking peptides to design modular substrates with high efficiency for mitogen-activated protein kinase extracellular signal-regulated kinase. ACS Chem. Biol. 2007;2:665–673. doi: 10.1021/cb700158q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldsmith EJ, Akella R, Min XS, Zhou TJ, Humphreys JM. Substrate and docking interactions in serine/threonine protein kinases. Chem. Rev. 2007;107:5065–5081. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CI, Xu BE, Akella R, Cobb MH, Goldsmith EJ. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 19.Barsyte-Lovejoy D, Galanis A, Sharrocks AD. Specificity determinants in MAPK signaling to transcription factors. J. Biol. Chem. 2002;277:9896–9903. doi: 10.1074/jbc.M108145200. [DOI] [PubMed] [Google Scholar]
- 20.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J. Biol. Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 21.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 22.Cuenda A, Rouse J, Doza YN, Meier R, Cohen P, Gallagher TF, Young PR, Lee JC. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 23.Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 24.Young PR, McLaughlin MM, Kumar S, Kassis S, Doyle ML, McNulty D, Gallagher TF, Fisher S, McDonnell PC, Carr SA, Huddleston MJ, Seibel G, Porter TG, Livi GP, Adams JL, Lee JC. Pyridinyl imidazole inhibitors of p38 mitogen-activated protein kinase bind in the ATP site. J. Biol. Chem. 1997;272:12116–12121. doi: 10.1074/jbc.272.18.12116. [DOI] [PubMed] [Google Scholar]
- 25.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 26.Barsyte-Lovejoy D, Galanis A, Clancy A, Sharrocks AD. ERK5 is targeted to myocyte enhancer factor 2A (MEF2A) through a MAPK docking motif. Biochem. J. 2004;381:693–699. doi: 10.1042/BJ20031940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shults MD, Imperiali B. Versatile fluorescence probes of protein kinase activity. J. Am. Chem. Soc. 2003;125:14248–14249. doi: 10.1021/ja0380502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.