Abstract
Glycoconjugation is a powerful tool to enhance the pharmacodymanics and/or pharmacokinetics of small molecule-based therapeutics, including natural products1. Yet, studies designed to systematically understand or exploit the attachment of carbohydrates in drug discovery remain limited by the availability of practical synthetic and/or biosynthetic tools2, 3. Here, we report the development of two prototype E. coli strains for the facile production of small molecule glucosides and glycosides. Through directed evolution, a model promiscuous glycosyltransferase (GT) (OleD-ASP)4, 5 was optimized for use as an in vivo glyco-catalyst to provide the OleD variant TDP16. A standard E. coli TDP16 overproduction strain, when subsequently grown in the presence of a diverse array of potential acceptors, led to the facile product of corresponding glucosides using endogenous host sugar donors (UDP/dTDP-Glc). Subsequent co-expression of the genes encoding for an engineered promiscuous anomeric kinase (GalK M173L/Y371H)6, engineered promiscuous nucleotidyltransferase (RmlA L89T)7, and TDP16 in E. coli, led to a prototype strain capable of generating novel glycosides via combining unnatural free sugars and aglycons fed to the strain under standard growth conditions. This work stands as the first proof of concept for in vivo glycorandomization wherein the demonstrated ability to mix and match non-natural sugars with a range of small molecule acceptors implicates vast combinatorial potential. In addition, prototype strains such as the ones described should open the door for simple large scale fermentation of novel complex glycosides not available via conventional biosynthetic methods.
Natural product glycosylation is accomplished by GTs, the donors for which are often exotic nucleotide sugars produced by rather lengthy (5-9 enzymatic transformations) biosynthetic pathways2, 8. Accordingly, the sugar biosynthetic pathways have been manipulated by metabolic engineering to produce novel natural product analogs. The first example of rational glycosyl-engineering involved the replacement of an endogenous daunorubicin sugar C-4′ reductase with one of inverting stereospecificity to enable a recombinant Streptomyces strain for the anticancer agent epirubicin9. Since this pioneering study roughly a decade ago, efforts have continued toward targeted metabolic glycosyl-engineering of select natural products and such efforts have more recently incorporated ‘sugar plasmids’ harboring entire gene sets encoding for the biosynthesis of specific novel sugar nucleotides10-13. While such studies have enabled the targeted production of non-natural glycosyl analogs of various natural products, the strategy is restricted by the inherent specificity of corresponding sugar nucleotide-forming enzymes and endogenous GTs and, when successful, the corresponding non-natural glycosides are often produced in low yield compared to the parent natural product.
As a response to these limitations, in vitro glycorandomization serves to produce diverse sets of sugar nucleotide donors via the combination of i) an engineered promiscuous anomeric kinase (GalK M173L/Y371H); ii) engineered promiscuous nucleotidyltransferase (RmlA L89T); and iii) an array of free reducing sugars readily accessible by chemical synthesis or commercial sources (Fig. 1a)6, 7, 14. The corresponding availability of these sugar nucleotide sets, in conjunction with the inherent promiscuity of several natural product GTs, enabled the generation of novel natural product-based glycoside arrays including those based upon aminocoumarins, enediynes, glycopeptides, macrolides and polyenes15-18. This approach has been further advanced via GT directed evolution to greatly expand donor and acceptor promiscuity and provide, for the first time, variant GTs capable of glycosylating natural products and small molecules for which natural GTs did not exist4, 19, 20. Yet, while in vitro glycorandomization has proven to be a useful tool for discovery scale synthesis of novel glycosides, the in vitro method requires expensive cofactors, purified proteins and often optimization of reaction conditions to prevent feed-back/forward inhibition by reactants in the coupled system.
Figure 1.
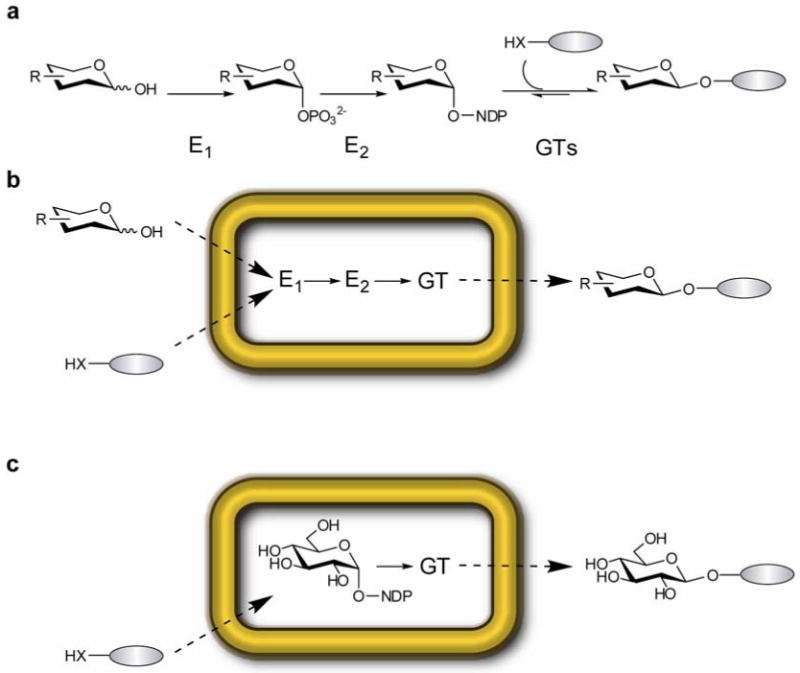
Comparison of methods for glycodiversification of natural products. (a) In vitro glycorandomization. Reducing sugars are converted to sugar-1-phosphates by E1, a flexible anomeric kinase. E2, A suitably flexible sugar-1-phosphate nucleotidyltransferase activates each sugar phosphate to the corresponding nucleotide sugar. Large panels of NDP-donors are used to probe the specificity of natural product GTs. Grey oval represents diverse natural product or natural product-like aglycons (X = O, S, or NH). (b) In vivo glycodiversification via a ‘non-natural glycoside host’ strain. Reducing sugars and aglycons are fed to a bacterial host engineered to express E1, E2, and a promiscuous GT. The endogenous biosynthetic machinery ensures recycling of necessary cofactors and aglycons decorated with non-natural sugars are collected from the culture media. (c) In vivo glucoside host. Aglycons are fed into a bacterial host engineered to express a GT which uses endogenous dTDP/UDPGlc as the glycosyl donor.
Previous studies using model plant GTs have demonstrated that simple aglycons can be taken up by E. coli, and the resulting glycosides secreted into the culture media21-24. Based upon this precedent, we envisioned feeding various aglycons and sugars to an E. coli strain overproducing the in vitro glycorandomization machinery in E.coli to provide for the in vivo production and utilization of novel sugar nucleotides en route to novel glycoside production. Unlike existing in vivo approaches that require discrete engineered strains for each different target glycoside to be produced, the approach described herein utilizes a single biocatalytic strain to generate an array of novel glycosides via simple alteration of the fermentation input (sugar and aglycon) (Fig. 1b). In a simpler version, small molecule glucosides could also be afforded via feeding suitable aglycons to a ‘glucoside’ host strain expressing OleD alone (Fig. 1c) wherein sufficient endogenous UDP-Glc (1) (Fig. 2) is provided by host. Here we report the rapid optimization of OleD to enhance its compatibility with the upstream GalK/RmlA TDP-sugar production pathway, and describe the first proof-of-principle demonstration of in vivo glycorandomization for the production of diverse natural product glycosides.
Figure 2.
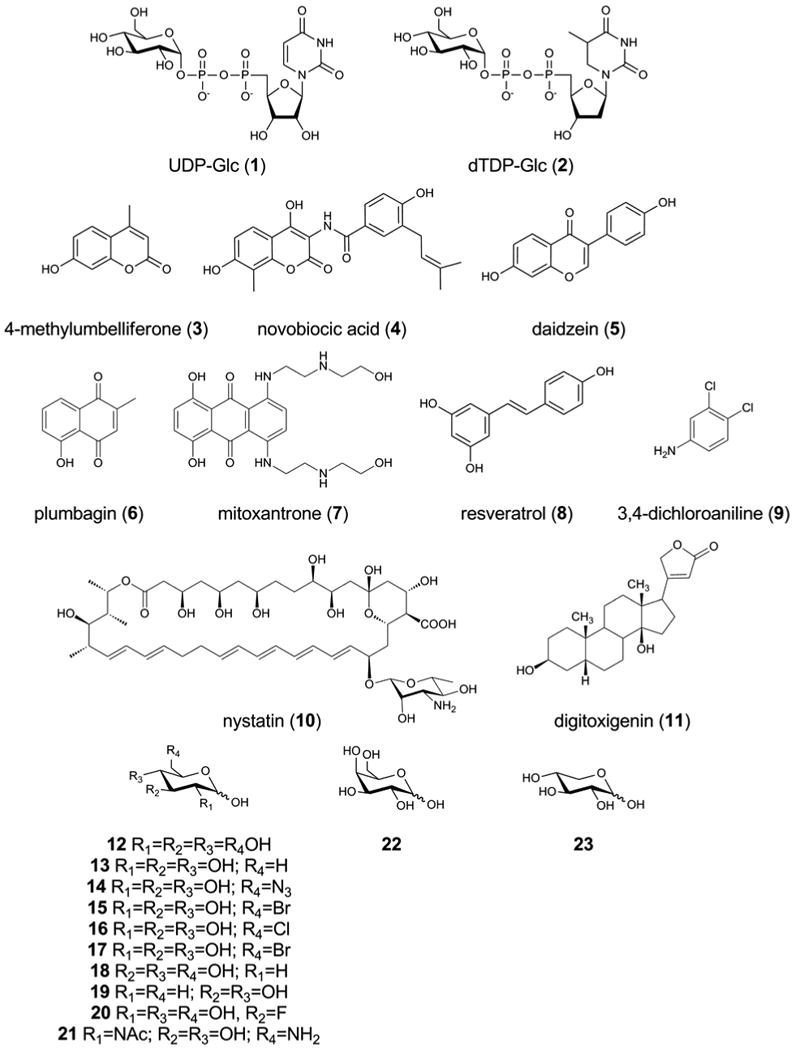
Structures of substrates used in this study.
While RmlA (i.e. E2, Fig. 1a) is more efficient with TTP than UTP when non-natural sugar-1-phosphates are used as substrates7, our previously described OleD variant ‘ASP’ displays only modest activity toward dTDP-Glc (2)4. To maximize the efficiency of OleD for glycosylation within the cytoplasm of E.coli, we therefore aimed to improve activity toward 2 by targeted saturation mutagenesis and screening using our recently described 4-methylumbelliferone (3, Fig. 2) fluorescence based assay19. The starting point for this mutagenesis was the OleD variant 3-1H12, itself identified from a saturation mutagenesis library19. Variant 3-1H12 differs from the well-characterized ASP variant by a single mutation (A242L) and displays several-fold improvement in activity toward UDP-donor 1 (see Supplementary Table 1 for description of OleD variants used in this study)19 and TDP-donor 2 (see Supplementary Table 2), compared to OleD ‘ASP’. We hypothesized that mutagenesis of active site residues that form the nucleotide binding site would result in the identification of variants with improved activity toward 2. Analysis of the WT OleD crystal structure revealed 8 residues within the N-terminal domain that were in contact or close to the nucleotide portion of 1 (Supplementary Fig. 1). Each of these positions was individually randomized by saturation mutagenesis, affording 8 libraries which were screened using a fluorescence based assay with 2 as donor, as described in the Supplementary Methods. The variant ‘TDP16’ was identified and DNA sequencing revealed the novel amino acid substitution Q268V. The OleD variants ‘TDP16’, ‘3-1H12’ and ‘ASP’ were compared by determining steady state kinetic parameters using either 1 or 2 as donor and the screening target 3 as acceptor (Supplementary Table 2), revealing TDP16 as a superior catalyst for conversion of 2. Subsequent substrate specificity analysis (Supplementary Table 3) revealed TDP16 to exhibit a similar donor specificity to that of ASP and a marked improvement in activity toward a representative aglycon panel, including 4-methylumbelliferone (3), daidzein (5), mitoxantrone (7), nystatin (10), and digitoxigenin (11) (Supplementary Table 4, see Fig. 2 for structures 3-7).
For the in vivo host construction, genes encoding GalK M173L/Y371H and RmlA L89T were cloned into pETDuet1 yielding the vector pDuet-GalK-Ep. The gene encoding TDP16 was cloned into the complimentary vector pCDFDuet1, giving pCDF-TDP16. Co-transformation of BL21(DE3) with pDuet-GalK-Ep and pCDF-TDP16 afforded the prototype ‘non-natural donor’ strain (Fig. 1b) which expressed soluble GalK, RmlA, and OleD TDP-16 in good yield (∼10 mg/ml culture, data not shown). Similarly, the corresponding universal ‘glucoside’ host (Fig. 1c) containing pCDF-TDP16 alone lead to soluble TDP16 production in similar yield.
A panel of known OleD aglycon substrates representing significant structural diversity and a dynamic range of proficiency with OleD5 was chosen to validate our ‘glucoside’ host (3-11, Fig. 2). Following protein expression, each aglycon 3-11 was added to a small volume of E.coli BL21(DE3) pCDFDuet-TDP16 that had been washed into phosphate buffered saline. Aliquots were removed at timely intervals and the culture supernatant analyzed for glucosides by HPLC (Fig. 3a and Supplementary Fig. 2). Putative glucoside products were compared to standards prepared by in vitro reactions using OleD ‘ASP’ and 1 as donor4, 5, and were also verified by LC-MS analysis (Supplementary Table 5). This analysis revealed that coumarin 3, aminocoumarin 4, flavonoid 5, quinone 6, polyphenol 8, amine 9, and polyene 10 were each converted to the expected glucoside(s) (Fig. 3a and Supplementary Fig. 2). Curiously, mitoxantrone 7 and digitoxigenin 11 were not converted (evidenced by the absence of product peak and no significant decrease in aglycon peak, as judged by HPLC), even though both are good in vitro substrates for TDP16 (Supplementary Table 2) and are likely poorly taken up by E.coli or modified within the cell. Expression of WT OleD in place of the variant TDP16 demonstrated that in most cases (except 11), the TDP16-based strain was a superior host and bioconversion using a strain which lacked OleD confirmed that glucoside formation in all cases was dependent on OleD (Supplementary Table 5). Product yields varied among these successful biotransformations (Fig. 3a) and, with the exception of 7 and 11, mirrored in vitro OleD aglycon specificity. Consistent with this, substitution of TDP16 in the glucoside host with 1C9, an OleD variant previously optimized for activity toward 4 20, led to a 7-fold higher conversion of 4 compared to the TDP16-based host (Fig. 3a).
Figure 3.
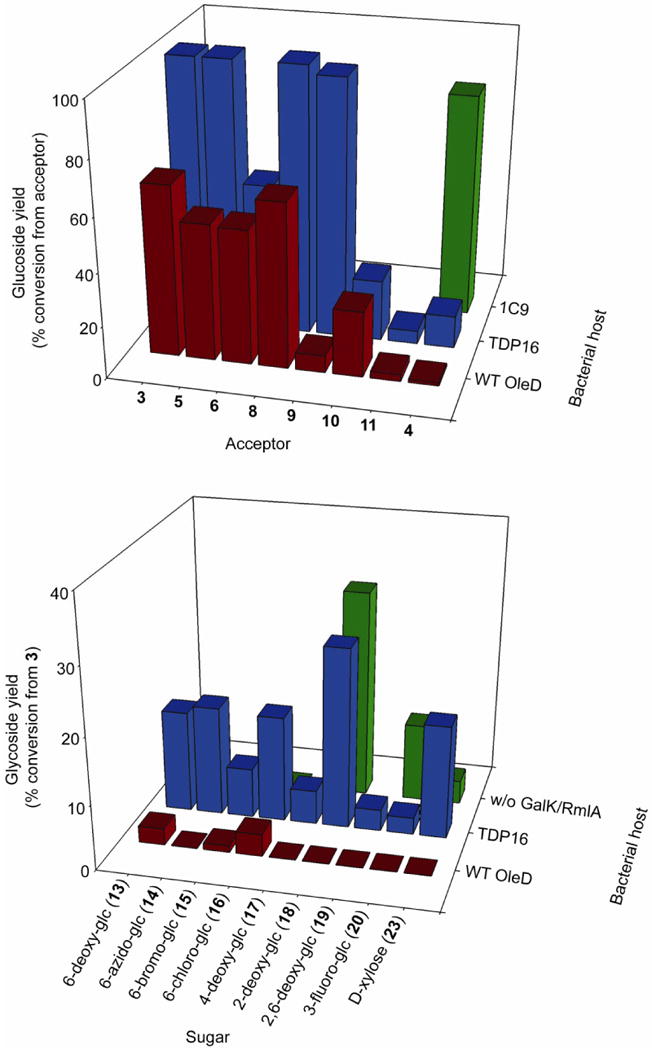
Activity of prototype glycoside producing strains. (a) Yields (% conversion from acceptor) of glucosides using the TDP16-, WT-, and 1C9-based glucoside host with a small panel of diverse acceptors. (b) Yields (% conversion from 3) of glycosides using the TDP16- and WT-based non-natural glycoside host using acceptor 3 and a panel of free sugars. ‘w/o GalK/RmlA’ refers to the TDP16-based host but which lacks the pDuet-GalK/RmlA vector. See Supplementary Materials for full description of the strains used, details of bioconversion conditions and detection. The standard deviation of the % conversions using data from three independent determinations was less than 20%.
Encouraged by the success of the in vivo glucoside host, efforts were next focused upon the prototype host for glycosylation with non-natural sugars (Fig. 1b). A panel of sugars was chosen to probe the efficiency and utility of our prototype ‘non-natural’-donor strain (12-23, Fig. 2). These sugars were chosen to represent diverse levels of proficiency with GalK, RmlA, and OleD. For example, 6-azido-glc (14) is a good substrate for the mutant GalK14, RmlA (as the 1-phosphate)25 and OleD (as the nucleotide)4, whereas d-galactose (22) is a relatively good substrate for the GalK14 and RmlA mutants26 (as the 1-phosphate) but a very poor substrate (as the UDP-sugar) for OleD (Supplementary Table 3). Cell suspensions of BL21(DE3) pDuet-GalK-Ep/pCDF-TDP16 were prepared as described in the Supplementary Methods, paying particular attention to wash the cells thoroughly in buffer in order to remove residual sugars from the culture medium. Acceptor 3 (100μM final concentration) and each sugar (at 4mM final concentration) were then added to the cell suspension, aliquots were removed at timely intervals and the culture supernatant analyzed for glucosides by HPLC (Fig. 3b and Supplementary Fig. 3). Putative glucoside products were compared to standards prepared by in vitro reactions using OleD ‘ASP’ and 3 as donor, and were also verified by LC-MS analysis (Supplementary Table 5). In addition to d-Glc (12), nine sugars were identified as substrates for the donor strain. Yields varied from 27% conversion for 18 to 2.5% for 3-fluoro-Glc 20. Notably, these results suggested that 3-fluoro-Glc (20) and 2,6-dideoxy-Glc (19) NDP-sugars were substrates for the OleD mutant, expanding on the previously established NDP-donor substrate promiscuity. Conversion with d-Gal (22) was not detected, likely reflecting the very poor activity of OleD toward UDP-Gal, and further suggesting that in vitro conversion with UDP-donor needs to be >2% for detectable in vivo bioconversion from the free sugar. Substitution of the OleD mutant TDP16 with the WT enzyme resulted in poorer conversions with every sugar tested and no discernable conversion with 14, 17, 18, 19, 20, and 23 (Supplementary Table 5). In the absence of added sugar, only the glucoside of acceptor 3 was detected, as expected (at only 26% conversion). In fact, 3-glc was detected when any of the sugars 12-22 was used, thus the preferred conversion of glucose to (U/T)DP-Glc by GalK/RmlA competes with conversion of the non-natural sugars. Nonetheless, 3-Glc is easily separated from the non-natural glycosides by HPLC (Supplementary Fig. 3), and total quantities of glycosides produced by the prototype strain are equivalent to 2-9 mg/L of cell suspension, yields which are comparable to other in vivo based systems. Additionally, a control strain which did not over-express the GalK/RmlA mutants only displayed conversion with 18, 20, and 23 (Supplementary Table 5), illustrating that these sugars are presumably processed by endogenous nucleotide-sugar biosynthesis machinery.
To assess the impact of host permeability upon conversion efficiency (e.g., in the case of validated in vitro substrates 7 and 11), detergent treatment and physical disruption failed to improve in vivo bioconversion (Supplementary Table 5). Subsequent deletion of the lpp gene encoding Brauns lipoprotein of BL21(DE3) - a mutation previously shown to produce marked improvement of permeability toward diverse small molecules27 - led to strains [BL21(DE3)/Δllp/pCDF-TDP16 and BL21(DE3)/Δllp/pDuet-GalK-Ep/pCDF-TDP16] capable of similar or slightly improved bioconversion compared to BL21(DE3)/pCDF-TDP16 and BL21(DE3)/pDuet-GalK-Ep/pCDF-TDP16, respectively (Supplementary Table 4). However, further analysis revealed the Δllp disruption mutants to rapidly lyse even under mild treatment such as washing and/or resuspension in PBS.
In summary, two novel prototype E.coli strains for the facile production of small molecule glucosides and glycosides were validated. These strains offer a number of advantages over prior microbial systems for small molecule glycoside production. First, E. coli is surprisingly permeable to a range of small molecule acceptors and sugars, and is readily amenable to further engineering for strain improvement. Second, OleD mutants can be created which are tailored toward specific aglycon acceptors and/or sugar donors, and can easily be substituted for OleD TDP16 within the prototype design described. Third, the in vivo glycoside system is amenable to standard large scale fermentation and, in most cases, the corresponding secretion of novel glycoside products greatly simplifies purification of the desired products. Furthermore, this in vivo approach circumvents the need for elaborate nucleotide sugar syntheses, cofactor regeneration, and/or enzyme purification required of existing in vitro strategies. Cumulatively, the ability to mix and match non-natural sugars with a range of small molecule acceptors offers vast combinatorial potential and also opens the door for similar strategies within important bioactive secondary metabolite-producing bacteria such as drug-producing actinomycetes.
Methods
For complete materials and methods, including construction of plasmids, mutant library preparation, screening, protein expression and purification, enzyme kinetics, and substrate specificity determinations, see the Supporting Information.
General
Bacterial strain E.coli BL21(DE3)pLysS was from Stratagene. NovaBlue was from Novagen. Plasmid pET28/OleD was a generous gift from Prof Hung-Wen Liu (University of Texas-Austin, Austin, USA) and pET28a was from Novagen. All other chemicals were reagent-grade purchased from Fluka, New England Biolabs, or Sigma, unless otherwise stated. Primers were ordered from Integrated DNA Technologies (Coralville, IA). Novobiocic acid (4) was prepared as previously described from novobiocin. UDP-Glc (1), TDP-glc (2) and acceptors 5-11 were from Sigma. Sugars 12, 18, 20, 21, 22, and 23 were from Sigma. 13-17 and 19 were synthesized as previously described. Analytical HPLC was performed on a Rainin Dynamax SD-2/410 system connected to a Rainin Dynamax UV-DII absorbance detector. To eliminate the need to purify acceptors and glucosides from the culture medium, the following optimal wavelengths were used: 254 nm for 3, 6, 8, 9, and 11; 325 nm for 4, 296 nm for 5, 590 nm for 7, and 300 nm for 10. At these wavelengths, the extinction coefficients of acceptor and glucoside were approximately equal. Mass spectra were obtained using electrospray ionization on an Agilent 1100 HPLC-MSD SL quadropole mass spectrometer connected to a UV/Vis diode array detector. For LC-MS analysis, quenched reaction mixtures were analyzed by analytical reverse-phase HPLC with a 250 mm × 4.6 mm Gemini 5μ C18 column (Phenomenex, Torrance, CA) using a gradient of 10-90% CH3CN in 0.1% formic acid/H2O in 20 min at 1ml/min, with detection at 254 nm unless otherwise stated.
In vivo bioconversions
For in vivo glucosylation of acceptors, a starter culture of BL21(DE3) pCDF-TDP16 or other control strain was used to inoculate a suitable volume of LB media containing 50 μg/ml streptomcyin and grown at 37 °C with shaking. Expression was induced by the addition of 0.1 mM IPTG when the OD600 was ∼0.6, and the cells were then incubated at 18 °C with shaking for 18 hrs. Cells were then washed four times with 10 × volume phosphate buffered saline (PBS) at 4 °C. Finally, cells were resuspended in a volume of PBS such that the OD600 was 7.0. Acceptor stock solutions (in DMSO) were added to suitable volume of cells to give 100 μM each of 3-9, 1 mM 10, and 0.2 mM 11 and the cell suspensions continued to incubate at 18 °C with rotation. Aliquots (100 μl) were removed at timely intervals. Cells were collected by centrifugation and the resulting supernatants analyzed directly by HPLC as described in the Supplementary Materials and Methods.
For in vivo glycosylation of 3 with non-natural sugars, a starter culture of BL21(DE3) pDuet-GalK-Ep pCDF-TDP16 or other control strain was used to inoculate a suitable volume of LB media containing 50 μg/ml ampicillin and 50 μg/ml streptomcyin and then grown at 37 °C with shaking. Expression was induced by the addition of 0.1 mM IPTG when the OD600 was ∼0.6, and the cells were then incubated at 18 °C with shaking for 18 hrs. Cells were then washed four times with 10 × volume phosphate buffered saline at 4 °C. Finally, cells were resuspended in a volume of PBS such that the OD600 was 7.0. Acceptor 3 (in DMSO) was added to suitable volumes of cell suspension, and 100 mM stock solutions of each sugar 12-23 added to a final concentration of 4 mM. Aliquots (100 μl) were removed at timely intervals. Cells were collected by centrifugation and the resulting supernatants analyzed directly by HPLC as described above.
Supplementary Material
Acknowledgments
The authors would like to dedicate this manuscript to the memory of the late Professor C. Richard Hutchinson, one of the pioneers of in vivo sugar engineering. We are grateful to the School of Pharmacy Analytical Instrumentation Center for analytical support. This work was supported in part by NIH AI52218 and NSF IIP-0740027. J.S.T is a UW HI Romnes Fellow and holds the Laura and Edward Kremers Chair in Natural Products.
Footnotes
Author Contributions: G.J.W., J.Y. and J.S.T. designed the research; G.J.W. and J.Y. performed the research; C.Z. provided reagents; G.J.W. and J.S.T. analyzed the data; G.J.W. and J.S.T. wrote the manuscript.
Competing Interests: The authors declare competing financial interests. JST is cofounder of Centrose, Madison, WI.
References
- 1.Williams GJ, Zhang C, Thorson JS. Natural product glycosyltransferases: Properties and applications. Adv Enzymol Relat Areas Mol Biol. 2008;76:55–119. doi: 10.1002/9780470392881.ch2. [DOI] [PubMed] [Google Scholar]
- 2.Thibodeaux CJ, Melancon CE, 3rd, Liu HW. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed Engl. 2008;47:9814–59. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boltje TJ, Buskas T, Boons GJ. Opportunities and challenges in synthetic oligosaccharide and glycoconjugate research. Nat Chem. 2009;1:611–622. doi: 10.1038/nchem.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams GJ, Zhang C, Thorson JS. Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nat Chem Biol. 2007;3:657–62. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- 5.Gantt RW, Goff RD, Williams GJ, Thorson JS. Probing the aglycon promiscuity of an engineered glycosyltransferase. Angew Chem Int Ed Engl. 2008;47:8889–92. doi: 10.1002/anie.200803508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmeister D, Yang J, Liu L, Thorson JS. Creation of the first anomeric D/L-sugar kinase by means of directed evolution. Proc Natl Acad Sci USA. 2003;100:13184–9. doi: 10.1073/pnas.2235011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton WA, Biggins JB, Jiang J, Thorson JS, Nikolov DB. Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering. Proc Natl Acad Sci USA. 2002;99:13397–402. doi: 10.1073/pnas.192468299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timmons SC, Thorson JS. Increasing carbohydrate diversity via amine oxidation: aminosugar, hydroxyaminosugar, nitrososugar, and nitrosugar biosynthesis in bacteria. Curr Opin Chem Biol. 2008 doi: 10.1016/j.cbpa.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madduri K, Kennedy J, Rivola G, Inventi-Solari A, Filippini S, Zanuso G, Colombo AL, Gewain KM, Occi JL, MacNeil DJ, Hutchinson CR. Production of the antitumor drug epirubicin (4′-epidoxorubicin) and its precursor by a genetically engineered strain of Streptomyces peucetius. Nat Biotechnol. 1998;16:69–74. doi: 10.1038/nbt0198-69. [DOI] [PubMed] [Google Scholar]
- 10.Blanco G, Patallo EP, Brana AF, Trefzer A, Bechthold A, Rohr J, Mendez C, Salas JA. Identification of a sugar flexible glycosyltransferase from Streptomyces olivaceus, the producer of the antitumor polyketide elloramycin. Chem Biol. 2001;8:253–63. doi: 10.1016/s1074-5521(01)00010-2. [DOI] [PubMed] [Google Scholar]
- 11.Fischer C, Rodriguez L, Patallo EP, Lipata F, Brana AF, Mendez C, Salas JA, Rohr J. Digitoxosyltetracenomycin C and glucosyltetracenomycin C, two novel elloramycin analogues obtained by exploring the sugar donor substrate specificity of glycosyltransferase ElmGT. J Nat Prod. 2002;65:1685–9. doi: 10.1021/np020112z. [DOI] [PubMed] [Google Scholar]
- 12.Lombo F, Gibson M, Greenwell L, Brana AF, Rohr J, Salas JA, Mendez C. Engineering biosynthetic pathways for deoxysugars: branched-chain sugar pathways and derivatives from the antitumor tetracenomycin. Chem Biol. 2004;11:1709–18. doi: 10.1016/j.chembiol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Perez M, Lombo F, Zhu L, Gibson M, Brana AF, Rohr J, Salas JA, Mendez C. Combining sugar biosynthesis genes for the generation of L- and D-amicetose and formation of two novel antitumor tetracenomycins. Chem Commun (Camb) 2005:1604–6. doi: 10.1039/b417815g. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Fu X, Liao J, Liu L, Thorson JS. Structure-based engineering of E. coli galactokinase as a first step toward in vivo glycorandomization. Chem Biol. 2005;12:657–64. doi: 10.1016/j.chembiol.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Albermann C, Fu X, Thorson JS. The in vitro characterization of the iterative avermectin glycosyltransferase AveBI reveals reaction reversibility and sugar nucleotide flexibility. J Am Chem Soc. 2006;128:16420–1. doi: 10.1021/ja065950k. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science. 2006;313:1291–4. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- 17.Albermann C, Soriano A, Jiang J, Vollmer H, Biggins JB, Barton WA, Lesniak J, Nikolov DB, Thorson JS. Substrate specificity of NovM: implications for novobiocin biosynthesis and glycorandomization. Org Lett. 2003;5:933–6. doi: 10.1021/ol0341086. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C, Fu Q, Albermann C, Li L, Thorson JS. The in vitro characterization of the erythronolide mycarosyltransferase EryBV and its utility in macrolide diversification. Chembiochem. 2007;8:385–390. doi: 10.1002/cbic.200600509. [DOI] [PubMed] [Google Scholar]
- 19.Williams GJ, Thorson JS. A high-throughput fluorescence-based glycosyltransferase screen and its application in directed evolution. Nat Protocols. 2008;3:357–362. doi: 10.1038/nprot.2007.538. [DOI] [PubMed] [Google Scholar]
- 20.Williams GJ, Goff RD, Zhang C, Thorson JS. Optimizing glycosyltransferase specificity via “hot spot” saturation mutagenesis presents a catalyst for novobiocin glycorandomization. Chem Biol. 2008;15:393–401. doi: 10.1016/j.chembiol.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol Bioeng. 2004;87:623–31. doi: 10.1002/bit.20154. [DOI] [PubMed] [Google Scholar]
- 22.Lim EK, Ashford DA, Bowles DJ. The synthesis of small-molecule rhamnosides through the rational design of a whole-cell biocatalysis system. Chembiochem. 2006;7:1181–5. doi: 10.1002/cbic.200600193. [DOI] [PubMed] [Google Scholar]
- 23.Caputi L, Lim EK, Bowles DJ. Discovery of new biocatalysts for the glycosylation of terpenoid scaffolds. Chemistry. 2008;14:6656–62. doi: 10.1002/chem.200800548. [DOI] [PubMed] [Google Scholar]
- 24.He XZ, Li WS, Blount JW, Dixon RA. Regioselective synthesis of plant (iso)flavone glycosides in Escherichia coli. Appl Microbiol Biotechnol. 2008;80:253–60. doi: 10.1007/s00253-008-1554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Antibiotic optimization via in vitro glycorandomization. Nat Biotechnol. 2003;21:1467–9. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]
- 26.Moretti R, Thorson JS. Enhancing the latent nucleotide triphosphate flexibility of the glucose-1-phosphate thymidylyltransferase RmlA. J Biol Chem. 2007;282:16942–7. doi: 10.1074/jbc.M701951200. [DOI] [PubMed] [Google Scholar]
- 27.Ni Y, Reye J, Chen RR. lpp deletion as a permeabilization method. Biotechnol Bioeng. 2007;97:1347–56. doi: 10.1002/bit.21375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


