Abstract
The green fluorescent protein (GFP) is a powerful genetic marking tool that has enabled virologists to monitor and track viral proteins during HIV infection. Expression-optimized Gag-GFP constructs have been used to study virus-like particle (VLP) assembly and localization in cell types that are easily transfected. The development of HIV-1 variants carrying GFP within the context of the viral genome has facilitated the study of infection and has been particularly useful in monitoring the transfer of virus between cells following virological synapse formation. HIV Gag-iGFP, a viral clone that contains GFP inserted between the matrix (MA) and capsid (CA) domains of Gag, is the first replication competent molecular clone that generates fluorescent infectious particles. Here, we discuss some methods that exploit HIV Gag-iGFP to quantify cell-to-cell transmission of virus by flow cytometry and to track the proteins during assembly and transmission using live cell imaging.
Introduction
HIV assembly, release, and entry are viral processes that depend upon complex cellular machineries that determine where and when each process occurs. Early efforts by virologists to study Gag localization used immunofluorescent staining of Gag in fixed cells and helped to map sequences that were important for plasma membrane binding and localization within the cell in the steady state [1]. These image-based studies supported a model of HIV membrane binding regulated by the MA domain of Gag during viral assembly. They defined clearly that certain sequences in MA are critical for the recruitment of Gag to the plasma membrane. A possible limitation of immunofluorescence studies, however, is the possible masking of some viral epitopes that may be hidden by the oligomerization of Gag during the assembly process [2].
GFP is a powerful tool for studying viral protein localization and has provided virologists with a complementary method to monitor the localization of viral proteins within human cells. Because expression of one viral protein, Gag, is sufficient to form virus-like particles (VLP), isolated Gag-GFP expressed from expression-optimized Gag constructs were used to map domains required for membrane association. These studies revealed a tight link between Gag oligomerization and binding to the plasma membrane of the cell in a manner that complemented immunofluorescence approaches [3–5]. Imaging of Gag-GFP allowed investigators to demonstrate the requirement for n-terminal myristoylation and basic sequences for recruitment of Gag to the plasma membrane [4] and determine residues within nucleocapsid (NC) critical to the assembly of Gag [6]. Color variants of Gag-GFP have been combined to perform fluorescence resonance energy transfer (FRET) imaging to demonstrate that Gag homo-oligomerization occurs at the plasma membrane [7].
Live cell imaging studies with Gag-GFP have begun to provide a more dynamic picture of how Gag moves within the cell before viruses are produced. Live confocal imaging of Gag-GFP constructs revealed that Gag fusion proteins can localize to the plasma membrane and endocytic compartments in virus-producing cells. This gave rise to a hypothesis that Gag may use endocytic compartments as an important assembly hub [8, 9]. Using fluorescence recovery after photobleaching (FRAP) approaches, Gomez et al showed that the mobility of Gag at the plasma membrane requires cholesterol [10]. Jouvenet et al used total internal reflection fluorescent microscopy (TIRFM) to follow the accumulation of Gag in individual virus particles as they formed at the plasma membrane in HeLa cells [11]. The authors demonstrated that there are two pools of VLPs at the membrane- one actively engaged in virus assembly and another which can be endocytosed from the surface of the cell.
While GFP enables one to track protein movements in cells with relative ease, it is critical to keep in mind that the activity of any protein may be altered in its biological activity by fusion with a large fluorescent moiety. Thus, functional validation of protein fusions is critical to ensure that the protein localization reflects that of the native, non-tagged protein. While Gag-GFP can be used to form VLPs, these fusions require the coexpression of helper virus with native Gag to generate infectious particles. This suggests that Gag-GFP is lacking in some biological feature that allows it to participate in infectious particle production. Part of this phenotype could be due to changes in the mRNA sequence itself, as the localization of Gag within the cell can be influenced by codon optimization or by the addition of RNA export signals [12–14]. Additionally, the large GFP moiety may also interfere more directly with assembly, thus causing the formation of irregular Gag-GFP VLP as seen in electron microscopy [15, 16]. For the above reasons, codon-optimized Gag-GFP fusions may not fully reflect the trafficking patterns of native HIV Gag.
To address these issues, fluorescent molecular clones of HIV that express the full complement of viral genes have been developed. These clones allow one to examine the behavior of the Gag proteins in a context that more closely resembles native infection. The Krausslich group produced a clone that carries GFP in the C-terminus of MA. While this virus was not infectious by itself, its infectivity could be rescued with helper virus [17]. If, however, the GFP moiety is inserted into the interdomain linker sequence between MA and CA, then the insertion of GFP only minimally perturbs the structure of the Gag precursor as well as the individual subunits of Gag. The result of this strategy, a viral clone called HIV Gag-iGFP, is a replication competent variant of pNL4.3 which is highly infectious in single round production from HEK 293 cells and able to replicate freely in the highly permissive T cell line, MT4. Time-lapsed confocal imaging of this virus has documented the changes in the subcellular distribution of Gag-iGFP within the context of a cell producing infectious viral particles [18]. Because of the stoichiometric loading of GFP into infectious particles, this construct is also well-suited to monitoring the efficiency of cell-to-cell HIV transmission in comparison to cell-free viral adsorption. Using a quantitative flow cytometry-based assay, we found that cell contact could transfer orders of magnitude more virus into a naïve cell population. The flow cytometry based-assay also revealed differential sensitivities to inhibitors of the actin cytoskeleton and some neutralizing antibodies [19]. We were also able to use the virus to study the dynamics of cell-to-cell transfer of HIV with high temporal resolution using spinning disk confocal microscopy. These studies revealed the rapid recruitment of micrometer-sized “buttons” of viral protein at the interface of the infected (donor) lymphocytes and the non-infected (target) lymphocytes, followed by transfer across these synapses into intracellular compartments of the target cells [20]. Live imaging of HIV Gag-iGFP revealed dynamic movements that occur when infectious HIV is produced within CD4+ T cells.
Because HIV Gag-iGFP is replication-competent in permissive cell lines, without the need of helper HIV Gag, it is more likely that the GFP signals that are tracked by flow cytometry or by live microscopy are indicative of functional pools of the recombinant Gag protein. Indeed, immunofluorescence studies of Gag-iGFP revealed that its localization is similar to that of the native Gag protein [18]. Here we provide detailed methods by which we use HIV Gag-iGFP and spectral variants of this construct to measure viral transfer from cell to cell and to image this process with live cell confocal microscopy.
2. Description of methods
2.1. Overview
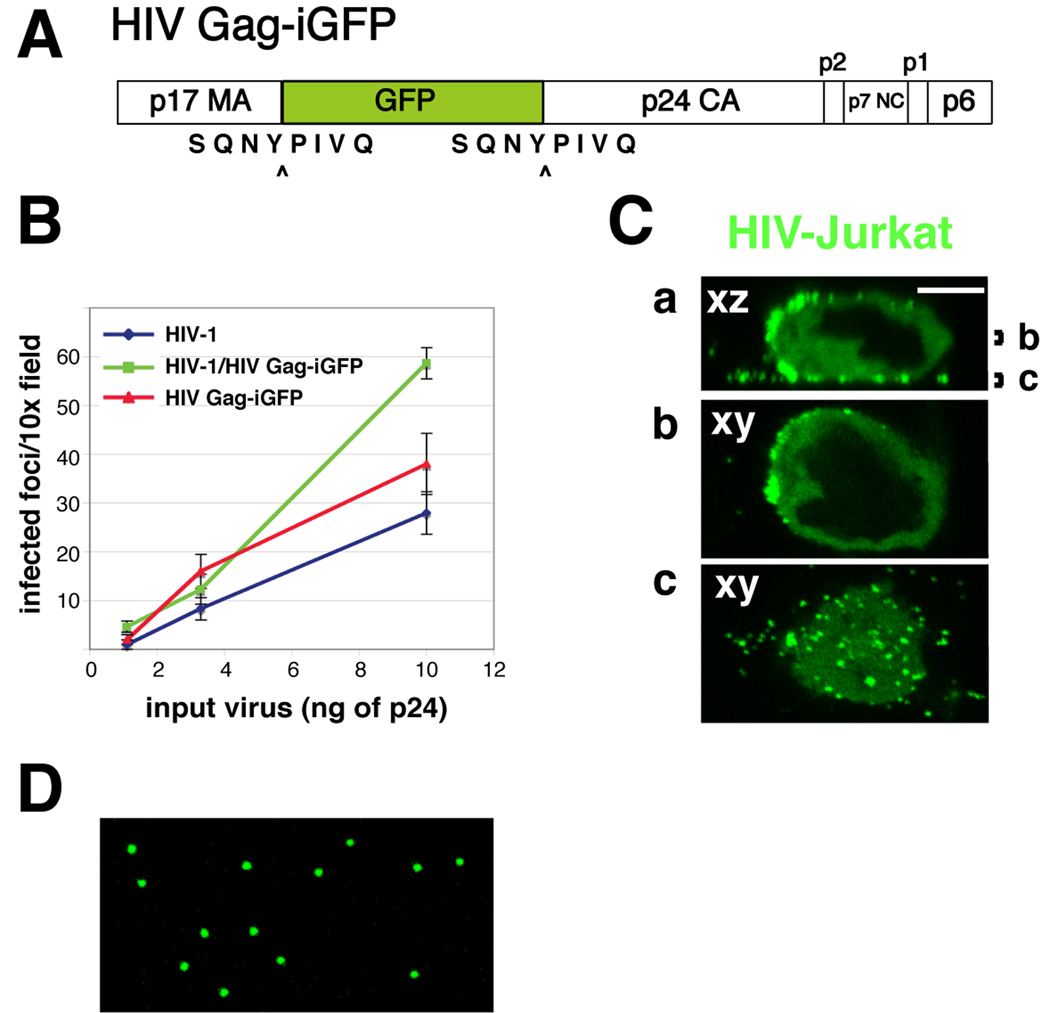
Cell to cell transmission of HIV was originally described using HIV-expressing Jurkat cells as donor cells and primary CD4+ T lymphocytes as acceptor cells [21–23]. In these studies, virus was detected in fixed cells using antibody staining. To track the transfer of virus from cell to cell with greater sensitivity, we exploited a recombinant, infectious reporter virus that carries the green fluorescent protein (GFP) inserted internally into the Gag protein between the MA and CA domains. In this clone, the protease cleavage sites are flanking the GFP leading to native-sized MA and CA after Gag processing occurs (Fig. 1A). This molecular clone of HIV retains infectivity without the need for helper virus (Fig. 1B) and in T cells the fluorescent Gag protein is localized primarily to the plasma membrane during the late stages of infection (Fig. 1C). Virus particles made from this clone are stoichiometrically loaded with GFP, providing improved signal of viral transfer (Fig. 1D). To monitor cell-to-cell transfer of the virus, flow cytometry (2.3.1) or live cell imaging (2.4.1) can be used to provide quantitative readouts of the amount and kinetics of viral transfer between cells. When used in conjunction with inert cell-tracking fluorescent dyes, recipient cells can readily be discriminated from input donor cells, and one can accurately measure or visualize the transfer of virus from cell to cell.
Fig. 1. HIV Gag-iGFP, an infectious, replication competent clone of HIV-1.
(A) Diagram of the Gag-internal, interdomain GFP insertion strategy into HIV Gag. GFP is placed between the MA and CA regions of Gag and flanked by viral protease cleavage sites. Upon maturation, the GFP is cleaved from Gag, but remains associated with the viral particle. (B) HelaT4 Magi cells [24] were infected with HIV Gag-iGFP, parent virus NL4-3, or a mix of the two viruses for 48 hr and then stained for β-galactosidase. (C) Jurkat T cells were transfected with HIV Gag-iGFP as described in the text. After 48 hrs, cells were plated on poly-l-lysine coated coverslips and fixed with 4% paraformaldehyde. Note the accumulation of Gag at the plasma membrane. (D) 293T cells expressing HIV Gag-iGFP following transfection using a calcium phosphate based method. After 48 hrs, supernatants were filtered and spotted onto glass coverslips. Viral particles were fixed and imaged on a confocal microscope using a 63× oil objective and 6× frame averaging.
The protocols below describe the methods used to express HIV Gag-iGFP in the Jurkat T cell line so that they may be used in cell-to-cell viral transfer studies with dye labeled primary CD4+ T cells.
2.2. Detailed Protocols
2.2.1 Preparation of HIV Gag-iGFP transfected Jurkat T cells
-
The human CD4+ T cell line Jurkat CE6.1 was obtained from American Type Culture Collection (ATCC, Manassas, VA) and is grown in Jurkat culture media (RPMI 1640, 10% Fetal Bovine Serum, 100 units/ml penicillin, and 100 µg/ml streptomycin). The cells are maintained in a humidified incubator at 37°C with 5% CO2 at a concentrations between 1 × 105 and 8 × 105 cells/ml. Maintaining cells within this density is often critical for achieving maximum transfection efficiency. Culture of the Jurkat cells at concentrations in excess of 8 × 105 cells/ml will result in a reduction in transfection efficiency.
-
1.1
Note: The transfection of HIV Gag-iGFP proviral DNA into T cells, as described below, results in the production of a replication-competent human immunodeficiency virus. As such, all procedures are to be undertaken only by trained laboratory personnel in certified biosafety level 2+ (BL2+) tissue culture hoods.
-
1.1
The HIV Gag-iGFP plasmid is relatively large (>15 KB) and thus requires optimal transfection conditions for efficient expression. Jurkats T cells are generally resistant to transfection by lipid-based transfection protocols. While we have had some success transfecting using a standard laboratory electroporator (BioRad Gene Pulser II), we observe the highest transfection efficiencies using nucleofection (Lonza, Walkersville, MD). We transfect Jurkat T cells with HIV Gag-iGFP using a modification of the method provided by the manufacturer. On the day of transfection, 5 × 106 cells are centrifuged for 10 min at 150 × g. The supernatant is aspirated and the cells are gently resuspended in sterile phosphate buffered saline (PBS). Cells are centrifuged again for 10 min at 150 × g before resuspending in 97 µl of prewarmed, supplemented nucleofector solution V. Three µl of HIV Gag-iGFP plasmid (1 µg/µl) are added to the cell suspension and gently mixed. For optimum transfection efficiency, it is essential to use endotoxin-free preparations of DNA (Qiagen, Valencia, CA). The cell suspension is transferred to a cuvette and nucleofected using program S-18. For the purposes of a control, one can transfect an aliquot of Jurkat cells with GFP alone. The cells are immediately transferred to 3 ml of prewarmed Jurkat culture media without antibiotics. We have found that we can “scale up” the above protocol by 40% without observing an adverse effect on transfection efficiency or cell viability. That is, 7 × 106 Jurkats can be resuspended in 136 µl of supplemented solution V with 4.2 µl of HIV Gag-iGFP plasmid (1 µg/µl) and transfected as described above.
On the day the Jurkat cells are transfected, an aliquot of frozen primary CD4+ T cells are thawed. We isolate these cells from peripheral blood mononuclear cells (PBMCs) using a CD4+ T cell isolation kit II (Miltenyi Biotec, Auburn, CA) and stored in liquid N2 in aliquots of 5×106 cells/500 µl of freezing media (90% fetal bovine serum/10% DMSO). Primary CD4+ T cells are rapidly thawed and cultured in Jurkat culture media supplemented with 10 units/ml of IL-2.
After allowing the Jurkat cells to recover overnight, cellular debris is removed by gradient centrifugation. In a 15 ml conical tube, 1.5 ml of Ficoll-Paque (GE Healthcare, Uppsala, Sweden) is added to a 15 ml tube and allowed to warm to room temperature. Transfected Jurkat cells are diluted to a 5 ml volume with RPMI and gently layered on top of the Ficoll. Cells are centrifuged at 400 × g for 20 min at room temperature (no brake). Cells are carefully removed from the media/Ficoll interface with a pipette and transferred to a new 15 ml conical tube. RMPI is added to a total volume of 15 ml and the cells are centrifuged at 300 × g for 10 min. Cells are resuspended in 3 ml of Jurkat culture media and returned to the tissue culture incubator. Recovery of the cells depends on a variety of factors, including the health of the cells at the time of transfection, the use of endotoxin-free DNA, and the use of proper technique during Ficoll purification. With optimum conditions, >90% will viable and able to be recovered. We rarely (< 0.1% of cells) see the formation of syncytia using Jurkat T cells.
To distinguish target cells from donor cells in cell-cell transfer experiments we prelabel the recipient cells with fluorescent dyes. Primary CD4+ T cells are labeled the evening before use. We have had excellent success labeling T cells with both CellTracker and CellTrace dyes (Invitrogen, Carlsbad, CA). While the dye concentration and incubation times must be optimized depending on the particular dye and cell type used, a representative protocol follows. Centrifuge 4 × 106 primary CD4+ T cells by spinning at 400 × g for 10 min. Wash the cells in 5 ml of PBS and resuspend in 2 ml of PBS. Add CellTracker Orange (cmtmr, Invitrogen, Carlsbad, CA) to a final concentration of 1.5 µM and incubate at 37°C for 20 min. Add 8 ml of complete Jurkat media and centrifuge at 400 × g for 10 min. Resuspend the cells in 3 ml of complete media supplemented with 10 units/ml of IL-2 and place in the tissue culture incubator overnight. We routinely use HIV Gag-iGFP transfected Jurkat cells 48 hours post-transfection. However, we have successfully used cells as early as 24 hours, and as late as 72 hours post-transfection. Below are detailed protocols for tracking virus using HIV Gag-iGFP by flow cytometry (2.3.1) and live cell imaging (2.4.1).
2.3.1 Monitoring cell-to-cell transfer of virus using HIV Gag-iGFP and flow cytometry
Approximately 48 hours post-transfection, HIV Gag-iGFP transfected Jurkat cells are counted and centrifuged at 300 × g. Resuspend cells in Jurkat cell media supplemented with 10 units/ml of IL-2 at a concentration of 2 × 106 cells/ml. For the purpose of a control reaction, a separate aliquot of non-transfected Jurkat cells are also counted, centrifuged, and resuspended at 2 × 106 cells/ml in Jurkat cell media. Supernatant from HIV Gag-iGFP-transfected Jurkats can be harvested and filtered through 0.45 µM filters. Supernatants from Jurkat T cells are usually low titer, yielding perhaps 20–40 ng/ml of virus as assessed by p24 ELISA. More concentrated preparations of HIV Gag-iGFP virus can be obtained by calcium phosphate-mediated transfection of 293T cells. Supernatants from 293T cells routinely have viral titers of 1 µg/ml.
Remove dye-labeled primary CD4+ T cells from the 37°C incubator, count them and centrifuge at 400 × g. Jurkat cell media supplemented with 10 units/ml of IL-2 at a concentration of 2 × 106 cells/ml.
Set up control and transfer reactions in sterile 5 ml round bottom FACS tubes (Becton Dickinson, Franklin Lakes, NJ). For a control reaction, combine 100 µl of GFP transfected Jurkat cells with 100 µl of dye-labeled primary CD4+ T cells. This condition allows the investigator to set flow cytometry gates during sample analysis. In a separate tube, combine 100 µl of HIV Gag-iGFP-transfected Jurkat cells with 100 µl of labeled primary CD4+ T cells.
Additional controls and experimental conditions are set up as needed. To set detectors for flow cytometry, individual tubes of unlabeled Jurkats, unlabeled primary CD4+ T cells, HIV Gag-iGFP Jurkat T cells, and labeled primary CD4+ T cells should be prepared. These tubes serve as references to identify individual cell populations and to facilitate spectral compensation. Additional control reactions in the presence of chemical inhibitors may be set up to show the specificity of the transfer, e.g. HIV Gag-iGFP-transfected Jurkat cells mixed with labeled primary CD4+ T cells in the presence of 2.5 µM cytochalasin D (Sigma Aldrich, St. Louis, MO) or 0.5 µg/ml of leu3a (CD4 blocking, BD Biosciences, San Jose, CA) antibody. These reagents have been shown to dramatically inhibit the cell-to-cell transfer of virus [19, 23].
Move all tubes to a 37°C tissue culture incubator and incubate for 3–4 hours. At the end of the incubation, add 3.5 ml of cold PBS to each tube and centrifuge at 4°C for 5 min at 400 × g. Aspirate the supernatant and resuspend the cells in 300 µl prewarmed 0.05% Trypsin-EDTA (Invitrogen, Carlsbad, CA) and incubate at 37°C for 10 min. After 10 min, rapidly add 300 µl of Jurkat culture media and 3.5 ml of cold PBS and centrifuge at 400 × g for 10 min. Aspirate the supernatant and resuspend the cell pellets in PBS with 4% paraformaldehyde. Incubate the cells in at room temperature in the dark for a minimum of 10 minutes. After fixation, add an additional 3.5 ml of PBS and centrifuge at 400 × g. Resuspend the pellets in 400 µl of PBS.
Each flow cytometry session should begin with the user running fluorescent and non-fluorescent single populations of both donor and acceptor T cells. This will allow the user to optimally set acquisition parameters and compensation. Ideally, this will enable the user to acquire information for both the donor and acceptor cells simultaneously (Fig. 2). Lasers should be set so that unstained cells land in the first log. This will allow for easy discrimination between the donor cells and the acceptor cells. Data is then acquired from the control and experimental samples analyzing 1 × 104 target cells per sample.
Following saving and export of FACS data, the user should open the data with a commercially available FACS analysis program such as CellQuest or FlowJo. A fluorescence gate set on the dual fluorescent population of cells may be set so that the control transfer condition (GFP transfected Jurkat cells with labeled primary CD4+ T cells) shows fewer than 0.5% dual positive cells. The same gate is applied to transfer conditions (Fig. 2). In a typical experiment, we successfully transfect 30% of Jurkat T cells with HIV Gag-iGFP. When these cells are mixed with primary CD4+ T cells at a 1:1 ratio for 3 hours, we typically see transfer of virus to approximately 15–20% of the primary CD4+ T cells.
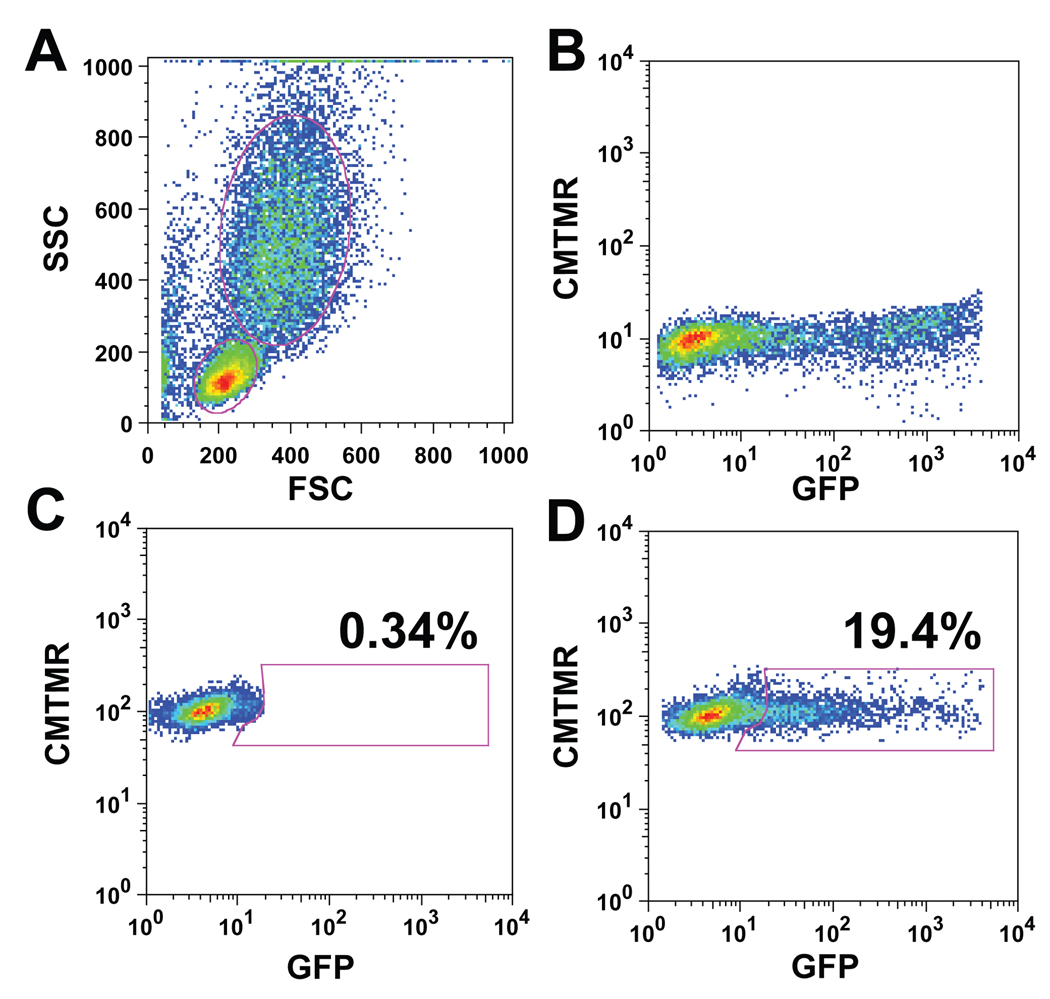
Fig. 2. Quantitation of cell-cell transfer of HIV Gag-iGFP using flow cytometry.
HIV Gag-iGFP transfected Jurkat T cells and cmtmr-labeled primary CD4+ T cells were co-cultured for three hours and processed for flow cytometry as described in the text. (A) The Jurkat T cells and the primary CD4+ T cells can be distinguished on the basis of their forward scatter (FSC) and side scatter (SSC); the Jurkat T cells are larger and thus are the population with the larger FSC/SSC profile. (B) Gating HIV Gag-iGFP transfected cells and plotting cmtmr versus GFP shows the donor cells to be cmtmr- population with approximately 30% GFP-positive cells. (C) Primary CD4+ target cells mixed with non-HIV-expressing, GFP-positive Jurkat T cells shows no GFP transfer to the target cells. Target cells may be gated based on FSC/SSC and by cmtmr label. A GFP-positive gate is selected based on this control so that the cmtmr(+)/GFP+ cells are fewer than 0.5%. (D) HIV Gag-iGFP transfected Jurkat T cells and cmtmr-labeled primary CD4+ T cells were co-cultured for three hours and processed for flow cytometry. Virus is typically transferred to 15–20% of the primary CD4+ T cells.
2.4.1 Monitoring cell-to-cell transfer of HIV Gag-iGFP using live cell imaging
The transfection of Jurkat T cells with HIV Gag-iGFP and labeling of primary CD4+ T cells is described in 2.2.1.
Approximately 48 hours post-transfection, HIV Gag-iGFP expressing Jurkats are counted, washed 1× with CO2-independent media (Invitrogen, Carlsbad, CA), and resuspended in live cell imaging media (CO2 independent media, 10% Fetal Bovine Serum, 100 U/ml penicillin, and 100 µg/ml streptomycin, 10 units/ml IL-2) at a concentration of 1–3 × 107 cells/ml.
In a similar fashion, wash and resuspend labeled primary CD4+ T cells at a concentration of 1–3 × 107 cells/ml in live cell imaging media.
Mix HIV Gag-iGFP transfected Jurkats with primary CD4+ T cells at a 1:2 or 1:3 ratio and load into a tissue culture treated, gas permeable, microchamber chamber (Ibidi, Verona, WI). For example, mix 20 µl of HIV Gag-iGFP transfected Jurkats with 40 µl of labeled primary CD4 T cells. Load 50 µl of this mixture into an Ibidi chamber and seal the chamber with tight plastic plugs. Secure the plugs by wrapping the plug/Ibidi chamber interface with parafilm. Wipe the entire surface of the Ibidi chamber with 70% ethanol. Move the Ibidi chamber to a sealable plastic container (e.g. Tupperware) for transport to the microscope.
2.5 Considerations for microscopy platforms: spinning disk confocal imaging
The above preparation is suitable for live cell imaging on any inverted fluorescence microscope platform outfitted with an appropriate slide holding device and thermostatic temperature regulation. We describe a simple, low-cost, but high-speed, high-resolution imaging platform, which employs a thermostatic heating device and a spinning disc confocal microscope. After loading an Ibidi imaging chamber with cells, we immediately mount the device onto an inverted optical microscope (iX71 Olympus, Center Valley, PA). The Ibidi chamber should be transferred to the microscope only by BL2+-trained laboratory personnel wearing appropriate safety garments. As noted above, appropriate approval for use of sealed imaging chamber must be approved by local biosafety committees. Although live-cell imaging commonly uses expensive incubation chambers to maintain a stable 37°C environment, this was achieved using a simple and economical thermostatic heater (ASI 400, Nevtek, Williamsville, VA) along with a T-type thermocouple tip mounted next to the sample for proper temperature feedback. We have found using CO2-independent media allows us to maintain high cell viability while avoiding the use of a CO2 chamber. To buffer the platform against temperature fluctuations in the room and to block out background room light, a large black shroud is draped over the whole microscope/heater setup creating an insulated air pocket around the system. This greatly reduces temperature variation and associated temperature-related sample drift, but requires pre-heating for 30 minutes prior to mounting the sample.
2.5.1 Data acquisition, transfer and image processing
Images are taken using a 60× 1.42 NA oil immersion objective (UPlanApo N, Olympus) as the high numerical aperture (NA) greatly improves resolution. To create 3D confocal images, scanning is performed by a spinning disk confocal unit (CSU-10, Yokagawa, Japan) that simultaneously uses >800 confocal spots to dramatically increase the scan rate. To compliment its speed, the CSU-10 is paired with a highly sensitive electron-multiplying charged-coupled device (EMCCD, iXon+ 897, Andor Technologies, Ireland) camera to allow fast, low light imaging which reduces both photobleaching and photoxicity. The fluorescence light source is a multi-wavelength ArKr ion gas laser (Innova 70C, Coherent, Santa Clara) where the desired wavelength (488 nm) and laser power measured right before the microscope objective (<1 mW) are selected by an acousto-optic tunable filter (AOTF), (Andor Technologies). To further reduce photobleaching and extend total imaging time, the AOTF rapidly (microseconds) shuts off the laser beam every time the camera is inactive (10–20 ms) while the newly acquired image is delivered from the CCD (15–30 ms exposure) to the computer. The laser light is coupled into the spinning disk system using a 405/488/568/647 quad band dichroic mirror (Semrock, Rochester, NY). Fluorescence emission is filtered using a 525/50 bandpass filter (Semrock) inside a filter wheel (Ludl Electronic Products, Hawthorne, NY) mounted between the EMCCD camera and the confocal spinning disk unit. All hardware and acquisition, including a z-stage (Mad City Labs, Madison, WI), are controlled by the Andor iQ v1.8 software. To maximize the speed of acquisition at around 1.2–1.9 second 3D image stacks, we crop the recording region of the EMCCD’s camera down to the cell-pair’s immediate area. Furthermore, at the expense of some information in z, a large z-step between 0.45–0.75 µm can be used to speed up acquisition. Imaging under these conditions can last up to 6 hours - using continuous 20–60 minute segments - with minimal photobleaching. Altogether, each data segment typically occupies 5–20 Gb of space as tens of thousands of images are taken. To facilitate the transport and analysis of the large data files, each acquisition set needs to be broken down and exported as 1 Gb TIFF image file segments using the Andor iQ v1.8 software. From here, many excellent software packages are available for image analysis, such as Metamorph, Volocity and ImageJ (we recommend its variation Fiji).
When two fluorescent markers are to be tracked, we simultaneously record both of them without slowing down acquisition by using a “split screen mode.” Both markers are excited by two different ArKr laser lines at once (often 488 and 568 nm). Inserted directly before the EMCCD camera is an image splitter (OptoSplit II, Cairn, Kent, UK) that separates the two different images. Each image is then independently filtered using fluorescence filters (Semrock) and projected side-by-side on the camera at once creating the "split-screen" effect. This would then require manual cropping and aligning of images later in image processing.
2.5.2 Considerations for image processing and data analysis
We typically perform image analysis with Volocity (PerkinElmer, Waltham, MA) on Macintosh computers (Apple, Inc). Spinning disk laser confocal microscopy images are first adjusted in their intensity to correct for photobleaching, then deconvolved with Volocity. Intensity measurements and tracking of Gag puncta is performed with the Volocity Quantification module. For image sets where the movement relative to a pre-formed synapse needs to be calculated, an automated tracking algorithm is employed which tracks the synaptic button throughout the entire sequence. Manual inspection of the regions of interest defined by the autotracking software must be performed to confirm that the software has correctly tracked the desired object. For objects where the contrast with the surrounding objects is too weak, manual tracking may be performed on a frame-by-frame basis. The Volocity software package allows the export of XYZ location, volume and integrated signal within the desired objects. The distance from the synapse and velocity of the tracked object are calculated by normalizing movements to the center of the synaptic button.
With this in vitro protocol, the user must use caution regarding the interpretation of results. For example, conditions used in this protocol are lacking in extracellular matrix (ECM) components that are likely to regulate cell-cell interactions in vivo. Receptors and signaling molecules that regulate the cell-cell transfer of HIV-1 may be influenced by cytokines and other factors that are not present in the in vitro environment. In addition, particularly with regard to the lymph nodes, it is likely that the cells are interacting at a much higher density in a 3D tissue environment. While we have demonstrated that cell-cell transfer of virus can occur between primary cells [20], the protocol here uses Jurkat cells as donor cells. Thus, while this protocol provides an excellent in vitro method for studying the cell-cell transfer of HIV-1, it is possible, that the results will differ from those found in vivo.
2.5.3 Creation of an Image Sequence in the Volocity software package
Open Volocity, create a new library, and under actions create a new image sequence.
Drag the TIFF stack into the image sequence window. We routinely acquire our images in a split screen mode and thus the number of channels at this point is one. Enter the number of z slices acquired and the number of time points taken. Arrange parameters in the correct order, e.g. z step, timepoints, channels.
Open the untitled image sequence and click on “image” tab. Use the rectangle tool and crop to selection. Save cropped images in an easily recognizable format.
Highlight the file containing the green cropped image and go to tools > split volumes. Be sure to “put all images in a single folder” and place in the appropriate order, e.g. z step, timepoints, channels. Repeat this process for the red cropped image.
Go to actions > create new image sequence. Double-click image sequence, drag both volumes in. Enter the parameters as above; e.g. z step, then timepoint, then channels for the newly created two channel image sequence.
Set relevant spatial (edit > properties) and temporal (sequence > set timepoints) properties
Use tools > change colors to put channels in green and red.
Use tools > contrast enhancement to adjust the leveling
Go to actions > create new registration. Adjust the overlay in all three planes.
Close registration file and open the image sequence. Go to tools > correct registration and apply the recently created registration.
Export the image sequence as a movie encoded in TIFF format if further corrections are needed. Otherwise, image sequences can be exported as Quicktime movies. To export a movie in extended focus, select file > export > view as Quicktime movie. Use the options tab to adjust the video compression and frame rate.
2.5.4 Manual tracking of virus particles
The diameter of a mature HIV-1 virus particle has been estimated to be approximately 120 nM. We have used structured illumination microscopy (SIM) to confirm that HIV Gag-iGFP particles are a similar size (McNerney et al, manuscript in preparation). However, the resolution limit of the spinning disk confocal microscope is limited by the wavelength of the diffracted light. Using the optics described here, this resolution limit is approximately 225 nM. Thus, we cannot determine whether some particles that appear as single particles, may in fact be aggregates of several particles. New technical advances, including the adaptation of stimulated emission depletion microscopy (STED) to GFP, have pushed resolution limits well under 80 nM. These new technologies will be able to be applied to HIV Gag-iGFP and thus increase our confidence that we are imaging single virions.
Using Volocity software, we have successfully used the automated tracking function to track objects as small as 20 pixels for extended periods of time. However, we have found that Volocity automated tracking programs have difficulty in tracking very small (2–8 pixels) viral particles when in proximity to intense GFP signals from infected cells. For this reason, when using Volocity software, we use manual tracking to monitor particle activity over time. A brief discussion of manual tracking using Volocity software is provided below.
Open an image sequence using Volocity software and click on the measurements tab. In this panel, select tools > track objects manually.
Use the zoom and inspection tool to find a particle and center it in all three planes.
Use the magic wand (tolerance set to 0) to select the center of a virus particle. Use edit > ROI > grow to adjust the size of the selection box so that it encompasses the particle. Use the inspection tool to verify that the particle has been selected in all three planes.
Once the particle has been selected, save the information by selecting measurements > make measurement item (current timepoint). Name this file, for example, particle track.
Click next to proceed to the next timepoint.
Select the particle in the next timepoint as described above and continue in this fashion for the desired amount of time.
Upon opening the “particle track” file in Volocity, the user has a large amount of data pertaining to the particle over time, including, fluorescence, area, and volocity.
Overview
The tagging of HIV proteins with GFP has allowed researchers to make novel observations regarding the localization and kinetics of individual viral processes. However, these studies may be compromised by the fact that the GFP-tagged viral proteins are not being expressed in the context of a fully infectious virus. HIV Gag-iGFP is replication competent, providing validation that the Gag-iGFP molecules that are being followed can contribute to the infectious cycle and are not revealing the movement of non-functional GFP fusions. Because HIV Gag-iGFP virus particles incorporate GFP at a stoichiometry equivalent to Gag, its signal is easily detectable by flow cytometry or using live cell imaging. As a viral tracking method, it is likely to compare favorably to alternative labeling strategies. Used in combination with the basic methodology outlined above, HIV Gag-iGFP will continue to reveal novel insights into the basic biology of the virus. We have found HIV Gag-iGFP to be especially useful for studies on the cell-cell transmission of the virus.
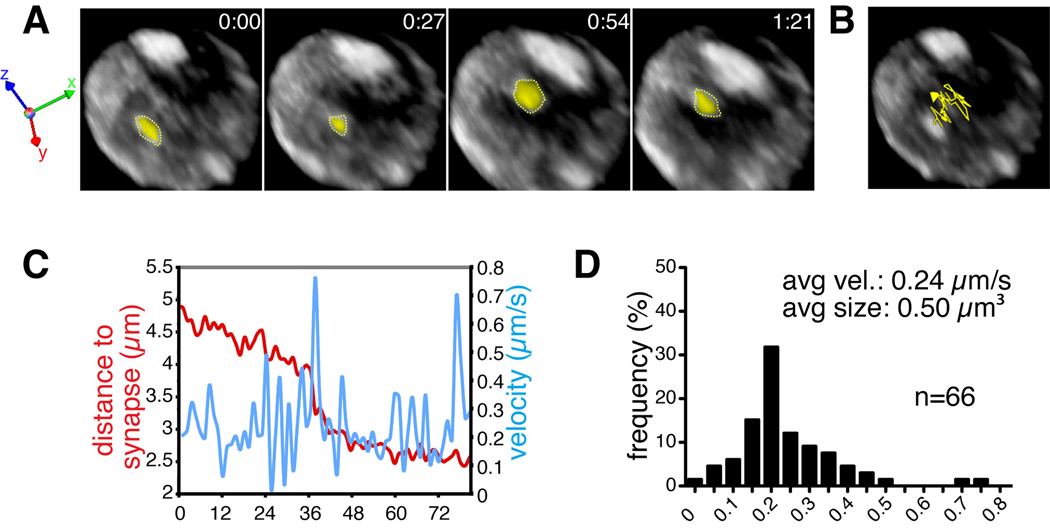
Fig. 3. Tracking puncta of HIV Gag-iGFP at the virological synapse.
HIV Gag-iGFP transfected Jurkat T cells and cmtmr-labeled primary CD4+ T cells were mixed and loaded into Ibidi chambers. Live cell imaging was performed with a spinning disk confocal microscope using methodology described in the text. (A) Volocity software was used to track the movement of Gag puncta moving towards a virological synapse. (B) A tracing of the movement of the Gag-iGFP puncta is shown over time. (C) Manual tracking was used to generate a measurement file that enabled one to calculate the position (with respect to the synapse) and velocity of the particle in microns/second. (D) Distribution of velocities of the object can be plotted to show the frequency with which the object moves at a given speed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhou W, Resh MD. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70(12):8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ono A, et al. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J Virol. 2005;79(22):14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ono A, Freed EO. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J Virol. 2004;78(3):1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hermida-Matsumoto L, Resh MD. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J Virol. 2000;74(18):8670–8679. doi: 10.1128/jvi.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono A, Demirov D, Freed EO. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J Virol. 2000;74(11):5142–5150. doi: 10.1128/jvi.74.11.5142-5150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandefur S, et al. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55(Gag) J Virol. 2000;74(16):7238–7249. doi: 10.1128/jvi.74.16.7238-7249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derdowski A, Ding L, Spearman P. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J Virol. 2004;78(3):1230–1242. doi: 10.1128/JVI.78.3.1230-1242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong X, et al. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120(5):663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Sherer NM, et al. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 2003;4(11):785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- 10.Gomez CY, Hope TJ. Mobility of human immunodeficiency virus type 1 Pr55Gag in living cells. J Virol. 2006;80(17):8796–8806. doi: 10.1128/JVI.02159-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454(7201):236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YJ, et al. Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J Virol. 2002;76(12):6332–6343. doi: 10.1128/JVI.76.12.6332-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson CM, et al. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 2004;23(13):2632–2640. doi: 10.1038/sj.emboj.7600270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin J, et al. HIV-1 matrix dependent membrane targeting is regulated by Gag mRNA trafficking. PLoS One. 2009;4(8):e6551. doi: 10.1371/journal.pone.0006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larson DR, et al. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci U S A. 2005;102(43):15453–15458. doi: 10.1073/pnas.0504812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pornillos O, et al. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003;162(3):425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller B, et al. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J Virol. 2004;78(19):10803–10813. doi: 10.1128/JVI.78.19.10803-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubner W, et al. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J Virol. 2007;81(22):12596–12607. doi: 10.1128/JVI.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen P, et al. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81(22):12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubner W, et al. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009;323(5922):1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco J, et al. High level of coreceptor-independent HIV transfer induced by contacts between primary CD4 T cells. J Biol Chem. 2004;279(49):51305–51314. doi: 10.1074/jbc.M408547200. [DOI] [PubMed] [Google Scholar]
- 22.Sourisseau M, et al. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J Virol. 2007;81(2):1000–1012. doi: 10.1128/JVI.01629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolly C, et al. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med. 2004;199(2):283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66(4):2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]