Abstract
The most common cause of cancer-related deaths in North America is lung cancer, 85% of which is non–small cell lung cancer (NSCLC). Gene therapy is a promising approach, but has been hindered by lack of methods for localizing and quantifying gene expression in vivo. Human somatostatin receptor subtype-2 (SSTR2)-based reporters can be used to follow gene expression in vivo using ligands with greater affinity for this subtype. NSCLCs can express SSTR subtypes, which may interfere with SSTR2-based reporters. We assessed whether a SSTR2-based reporter can serve as a reporter of gene transfer into NSCLCs. SSTR subtype expression was assessed in NSCLC cell lines A549, H460, and H1299 using RT-PCR. After infection with an adenovirus containing hemagglutinin-A-tagged-SSTR2 (Ad-HA-SSTR2) or control insert, expression was assessed by immunologic techniques and binding to clinically-approved 111In-octreotide. In vivo, after magnetic resonance (MR) imaging, intrathoracic H460 tumors were injected with Ad-HA-SSTR2 or control virus (n = 6 mice/group) under ultrasound guidance. Intravenous injection of 111In-octreotide 2 days later was followed by planar and single-photon emission computed tomography (SPECT) imaging. Biodistribution into tumors was assessed in vivo using anatomic MR and functional gamma-camera images and ex vivo using excised organs/tumors. In human lung tumor samples (n = 70), SSTR2 expression was assessed using immunohistochemistry. All three NSCLC cell lines expressed different SSTR subtypes, but none expressed SSTR2. Upon Ad-HA-SSTR2 infection, HA-SSTR2 expression was seen in all three cell lines using antibodies targeting the HA domain or 111In-octreotide targeting the receptor domain (p < 0.05). Intrathoracic tumors infected with Ad-HA-SSTR2 were clearly visible by gamma-camera imaging; expression was quantified by both in vivo and ex vivo biodistribution analysis and demonstrated greater uptake in tumors infected with Ad-HA-SSTR2 compared with control virus (p < 0.05). Immunohistochemistry found that 78% of NSCLCs are negative for and 13% have low levels of SSTR2 expression. It is concluded that SSTR2-based reporters can serve as reporters of gene transfer into NSCLCs.
Singh and colleagues describe the development of a new reporter system for the localization and quantification of gene transfer within the context of non–small cell lung cancer (NSCLC). Using this approach, the authors demonstrate successful application of this system for the imaging of gene transfer in three different NSCLC lines, in a mouse model of intrathoracic NSCLC and ex vivo in human NSCLC tumor samples.
Introduction
Lung cancer is the leading cause of cancer-related death in the United States for both men and women, with an estimated incidence and mortality of 219,440 cases and 159,390 deaths in 2009, respectively (American Cancer Society, 2009). Lung cancer patients suffer an overall 5-year survival of approximately 16% despite advances in chemotherapy, radiation therapy, and surgery. This low survival rate is commonly due to advanced stage upon diagnosis (Jett, 1993; Nesbitt et al., 1995). Chemotherapy and radiotherapy have had limited effect on improving survival even in patients with early-stage NSCLC, where some trials demonstrate that adjuvant chemotherapy prolongs overall survival by only approximately 5% at 5 years (Scagliotti, 2007). In patients with locally advanced NSCLC, frequent cross-sectional imaging does not alter survival after combined modality therapy (Benamore et al., 2007). New therapies, such as gene therapy (Vattemi and Claudio, 2004; Cross and Burmester, 2006), are promising for improving outcome, but they are hampered by an inability to noninvasively assess gene expression in patients. Reporter imaging systems that have the potential for clinical translation are needed for assessing expression at the target, for efficacy, and off target, which may cause toxicity.
For assessing expression, reporter systems may be used. These generally consist of a gene that encodes a protein that can be visualized directly or, more commonly, visualized after binding and/or entrapping a substrate. Poor tissue penetration of up to approximately 1 cm in light-based systems, such as green fluorescent protein (GFP) (Cote et al., 1997; de Martin et al., 1997; Meyer et al., 1998) and luciferase (Stables et al., 1999), limits their utility for percutaneous imaging of larger animals and humans. Nuclear medicine-based reporters do not suffer such a limitation. Human somatostatin receptor type 2 (hSSTR2)-based reporters (Rogers et al., 2000; Zinn et al., 2000; Kundra et al., 2002) have been proposed as reporter systems and have several advantages, including being of human origin, thus, limiting an immune response; change in expression can be quantified in vivo over time (Singh et al., 2009), and such reporters can be made signaling-deficient to inhibit untoward effects on transduced cells or on linked genes of interest (Han et al., 2007). We have previously demonstrated that hSSTR2 expression can be quantified using small animal cognates of clinical machines for functional nuclear imaging and anatomic imaging, such as single-photon emission computed tomography (SPECT) and magnetic resonance (MR) (Yang et al., 2005; Singh et al., 2009). Anatomic imaging of intrathoracic lung tumors is difficult due to poor tissue contrast using traditional methods, e.g., computed tomography (CT), as well as rapid breathing and cardiac motion, especially in mice, but can be approached using echo-planar imaging (EPI)-based MR (Bankson et al., 2008).
A potential limitation of using SSTR2-based reporter imaging in lung cancer is that there have been reports that lung cancers can express SSTRs (Fujita et al., 1994; Papotti et al., 2001; Herlin et al., 2009). This has the potential to confound hSSTR2-based imaging. Among the SSTR subtypes, clinically approved 111In-octreotide has high binding to SSTR2, intermediate binding to SSTR3 and 5, and poor binding to SSTR1 and 4 (Kluxen et al., 1992; O'Carroll et al., 1992; Yamada et al., 1992; Bell and Reisine, 1993; Panetta et al., 1994). We hypothesized that hSSTR2-based imaging could be performed in lung cancer using such a subtype-selective imaging agent. We further assessed the prevalence of subtype 2 in clinical lung cancer tumor samples. Thus, we assessed whether a SSTR2-based reporter can serve as a reporter of gene transfer into NSCLCs.
Materials and Methods
Cell lines and adenovirus infection
A549, H460, and H1299 human NSCLC cells were obtained from American Type Culture Collection (Manassas, VA) and maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine, and 1% penicillin–streptomycin mixture. Adherent monolayer cells were cultured at 37°C in a humidified atmosphere with 5% CO2 and 95% air. A549, H460, and H1299 (1 × 106 cells of each) were infected with or without 1010 pfu Ad-CMV-HA-hSSTR2 (Singh et al., 2009) diluted in 1 ml of serum-free RPMI medium in a six-well plate. Two milliliters of medium was added after 4 hrs and the cells incubated for 48 hrs. Ad-CMV-GFP served as a control. After 48 hrs, the cells were divided for immunofluorescence, ELISA, Western blotting, and receptor-binding studies.
Reverse transcriptase–polymerase chain reaction (RT-PCR)
Total RNA was isolated from A549, H460, and H1299 human NSCLC by extraction with TRIzol Reagent. One microgram of total RNA was reverse-transcribed, and PCR reactions were carried out using Superscript One-Step RT-PCR kit according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). Five SSTR primer sets (SSTR1–5) were used:
SSTR1 sense: AAATGCGTCCCAGAACGGGACCT
SSTR1 antisense: CAGGTTCTCAGGTTGAAGTCTT
SSTR2 sense: GATGATCACCATGGCTGTG
SSTR2 antisense: CAGGCATGATCCCTCTTC
SSTR3 sense: TCATCTGCCTCTGCTACCTG
SSTR3 antisense: GAGCCCAAAGAAGGCAGGCT
SSTR4 sense: CATGGTCGCTATCCAGTGCA
SSTR4 antisense: GTGAGACAGAAGACGCTGGTGAACAT
SSTR5 sense: GCTCTTGGTGTTCGCGGACGT
SSTR5 antisense: CAGGTTGACGATGTTGACGGTGAAG.
Ten microliters of each PCR product was subjected to electrophoresis on a 1% agarose gel and visualized under UV light.
Immunofluorescence and ELISA
Immunofluorescence, ELISA, and Western blotting were performed as described (Kundra et al., 2002; Yang et al., 2005). A mouse anti-HA primary antibody (Covance, Princeton, NJ) and fluorescein isothiocyanate (FITC)-labeled goat anti-mouse secondary antibody (Southern Biotech, Birmingham, AL) were used for immunofluorescence. A horseradish peroxidase (HRP) anti-HA antibody (clone 3F10; Roche, Indianapolis, IN) was used for ELISA, and positive cells exhibited a green-colored product after exposure to an HRP ELISA substrate (BioRad, Hercules, CA). Western blotting was performed using HRP anti-HA antibody and developed using a chemiluminescent HRP substrate (Pierce Biotech, Rockland, IL).
Binding assay
Receptor binding assays were performed in triplicate as described (Han et al., 2007). Assay reagents were added on 0.1% polyethylene amine (PEI)-treated and washed GF/B multi-screen plates (Millipore, Bedford, MA) in the following order: 160 μl of cell membrane (5 μg/well), 40 μl of 111In-octreotide (100 nM) (Mallinckrodt, Houston, TX) or 160 μl of membrane of SSTR2, 20 μl of cold ligand (1 μM unlabeled somatostatin), and 20 μl of 111In-octreotide (100 nM). After incubation for 40 min at room temperature and washing with Tris-HCl (50 mM), the plate filters were punched out and the associated radioactivity was quantified using a Beckman γ-counter.
Animal and orthotopic implantation of tumor cells
Male athymic nude mice (8 weeks old) were purchased from Charles River Laboratories (Wilmington, MA). All animals were cared for in accordance with guidelines set forth by the American Association for Accreditation of Laboratory Animal Care International and the U.S. Public Health Service Policy on Human Care and Use of Laboratory Animals. All experiments were approved and supervised by the MDACC Institutional Animal Care and Use Committee.
To produce tumors, H460 cells were harvested from subconfluent cultures by a brief exposure to 0.25% trypsin and 0.02% EDTA. Trypsinization was stopped by adding RPMI medium containing 10% FBS, washed once with PBS, then resuspended and counted in a Coulter counter (Beckman). H460 cells at 5 × 106 in 100 μl of medium were injected into the right side of the thoracic space.
Biodistribution and imaging
Twenty-four hours prior to injection, 12 male nude mice were irradiated with 3 Gy. Then mice were injected intrathoracically with 5 × 106 H460 cells in 100 μl of PBS. MR imaging was done at day 5 after inoculation to confirm tumor and at day 9 for measuring tumor size. Three days before mCAM (Siemens Medical Solutions, Brookfield, CT) and SPECT imaging (day 6 after tumor inoculation), the mice were randomized into two groups with six mice in each group. Under ultrasound guidance (Vevo 770, linear transducer, 40 mHz), the tumors in the first group were injected with 1010 pfu of Ad-CMV-HA-hSSTR2, and tumors in the control group were injected with 1010 pfu of Ad-CMV-GFP.
For MR imaging, a respiratory- and cardiac-gated, fat-suppressed EPI-based sequence was used (Bankson et al., 2008). Animals were anesthetized with 2% isoflurane. The animals were imaged supine with a 4.7 T small-animal MR scanner (Biospec USR47/40 NMR/MRI; Bruker Biospin Corp., Billerica, MA). A T2-weighted spin-echo EPI sequence (TE = 35 msec, TR >> T1; several respiratory cycles, bandwidth = 100 kHz, matrix size = 128 × 128; acquired in eight shots with 1.56-msec echo spacing in 25.27-msec EPI readout, field of view = 3 cm × 3 cm, slice thickness = 1.25 mm, and skip = 0.25 mm) with a frequency-selective fat-suppression pulse was synchronized with cardiac and respiratory cycles to acquire one shot from one slice at each of multiple triggers during the plateau of the respiratory cycle (with the lungs at full expiration) and at either the Q wave or R wave of the ECG signal, depending on their prominence. The spin-echo EPI sequence was optimized to maximize contrast between solid tumors, chest wall, and lung tissue while minimizing respiratory and cardiac motion artifacts. Using Image J program (version: Java 1.3.1-03, National Institutes of Health, Bethesda, MD), tumor volumes were obtained by drawing regions of interest (ROI) on each image containing tumor (Yang et al., 2005; Bankson et al., 2008; Singh et al., 2009) and converted to weight in grams, assuming a tissue density of 1 g/ml.
After MR imaging at day 9, anesthetized mice were injected intravenously with 300 μCi (13 MBq) of 111In-octreotide (Mallinckrodt, St. Louis, MO). Twenty-four hours later, planar and SPECT imaging was acquired as described (Yang et al., 2005). In brief, planar 10-min acquisitions were performed of anesthetized, prone mice using a gamma camera (mCAM) fitted with a medium-energy parallel-hole collimator. SPECT imaging (120 views, 7.5 sec per view, pixel size of 2.4 mm, 128 × 128 matrixes) of each animal was acquired using a 360o rotation of a fixed 1/15 rpm rotational device attached to the front of the medium-energy collimator (15-min acquisition) and reconstructed. For both the planar and SPECT images, ROIs were drawn over the periphery of the tumor and uptake was subtracted from the left lung, which did not have a tumor. The counts per pixel per minute obtained from the ROI were converted to microcuries by using a calibration constant derived from imaging of standard activity phantoms by the gamma-camera scanner. Uptake by each tumor was divided by the weight of the tumor derived from MR imaging to obtain the percentage of injected dose per gram (%I.D./g) in vivo biodistribution.
The mice were euthanized after imaging, and organs and tumors were excised and weighed. Samples of these tissues were then weighed, and associated radioactivity was determined using a Cobra gamma counter to determine %I.D./g ex vivo biodistribution.
Immunohistochemistry of mouse samples
Sections of the excised mouse tumors were fixed in 10% formalin. Paraffin-embedded sections were processed for immunohistochemistry using the Vector MOM immunodetection peroxidase kit (catalog no. PK2200; Vector Laboratories, Burlingame, CA). The sections were probed with a primary mouse anti-HA antibody (1:250) and stained with the DAB Peroxidase Substrate kit (catalog no. SK-4100; Vector Labs). They were counterstained with Mayer's hematoxylin (catolog no. H-3404; Vector Labs).
Immunohistochemistry of human samples
To determine the expression of SSTR2 in primary lung cancer tumors, we selected and evaluated 70 NSCLCs (48 adenocarcinomas, 22 squamous cell carcinomas). These samples were archived, formalin-fixed, paraffin-embedded tumor tissue from surgically resected lung cancer specimens from the Lung Cancer Specialized Program of Research Excellence Tissue Bank at The University of Texas M.D. Anderson Cancer Center. This study was approved by the M.D. Anderson Cancer Center institutional review board. Tumor tissues were histologically analyzed and classified using the 2004 World Health Organization classification system. Samples were placed in a TMA, using three 1-mm-diameter cores that included tissue from the center, intermediate, and peripheral areas of the tumor.
Five micron-thick formalin-fixed, paraffin-embedded tissue histology sections from TMAs were deparaffinized, hydrated, heated in a steamer for antigen retrieval for 30 min with 10 mM sodium citrate (pH 6.0), and washed in Tris buffer. Peroxide blocking was performed with 3% H2O2 in methanol at room temperature for 15 min, followed by 10% bovine serum albumin in Tris-buffered saline with Tween 20 for 30 min at room temperature. Next, samples were incubated in anti-somatostatin receptor 2A rabbit polyclonal antibody (SSTR2A) (catalog no. PA3-109; Affinity BioReagents, Inc., Golden, CO), at 1:2,000 dilution, for 1 hr at room temperature. After washing, incubation with the secondary antibody (EnVision + Dual Link labeled polymer; DAKO, Carpinteria, CA) was performed for 30 min, followed by application of diaminobenzidine chromogen for 5 min. The slides were then counterstained in hematoxylin and topped with a coverslip. As positive control, formalin-fixed and paraffin-embedded gastric mucosa tissue was used. As negative control, paraffin-embedded positive gastric mucosa tissue was subjected to SSTR2A omitting the primary antibody, which was replaced with PBS buffer. Immunostaining evaluation was performed by two pathologists (I.W. and L.S.) using a white light microscope. Positive immunostaining in control cells and lung cancer tumor cells was mainly cytoplasmic and membranous. The percentage of positive tumor cells was evaluated.
Statistical method
Groups for ELISA, RT-PCR, and in vivo or ex vivo biodistribution were compared by Student's t test. Linear regression was used to correlate tumor weight derived by MR versus that of excised tumors. The analyses were performed using Excel 2003 software (Microsoft Inc., Seattle, WA). For all the tests, p < 0.05 was considered statistically significant.
Results
Expression of hSSTR subtypes in lung cancer cell lines
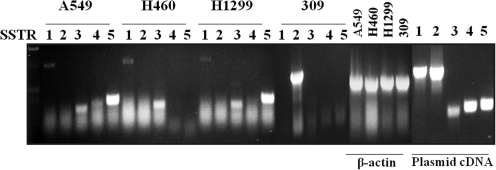
Expression of the SSTR subtypes (1–5) was tested in three human NSCLC cell lines (H460, H1299, and A549) using RT-PCR (Fig. 1). Both A549 and H1299 showed mRNA expression of SSTR1, 3, and 5, whereas H460 cells only expressed SSTR1 and 3. None expressed the subtype SSTR2. SSTR2 expression was seen in the positive control cell line 309 (Yang et al., 2005). RT-PCR of the “housekeeping gene” β-actin demonstrated the quality of the mRNA. Primer quality was confirmed by PCR of SSTR subtypes from subtype-specific plasmids.
FIG. 1.
H1299, H460, and A549 cells express combinations of SSTR subtypes other than subtype 2. RT-PCR of SSTR1–5 mRNA in NSCLC cell lines is presented. Plasmid cDNA and β-actin were used as positive controls and to demonstrate mRNA quality, respectively. As a second positive control for SSTR2, 309 cells were used.
In vitro expression of HA-hSSTR2 after adenoviral infection
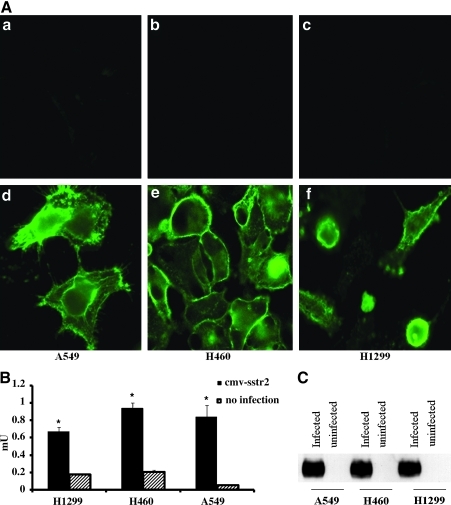
Expression of HA-hSSTR2 in H1299, H460, and A549 cells was assessed in vitro before and after Ad-CMV-HA-hSSTR2 infection using antibodies targeting the HA domain. Background is seen in uninfected cells, but expression on the cell membrane is clearly seen in all three cell types after Ad-CMV-HA-hSSTR2 infection (Fig. 2A). Quantitative ELISA confirmed expression in all three cell types after Ad-CMV-HA-hSSTR2 infection, but not in controls (p < 0.05, Fig. 2B). Western blot analysis confirmed that protein of the correct size was made in Ad-CMV-HA-hSSTR2 infected cells. As seen in Fig. 2C, a distinct band (60–72 kDa) is seen in cells expressing the HA-hSSTR2 gene product in all three cell lines infected with Ad-CMV-HA-hSSTR2, but not in uninfected cells.
FIG. 2.
Immunologic methods targeting the HA domain demonstrate that H1299, H460, and A549 cells express HA-SSTR2 after in vitro infection with Ad-CMV-HA-hSSTR2. In vitro HA-hSSTR2 expression by H1299, H460, and A549 cells with and without in vitro infection with Ad-CMV-HA-hSSTR2 using antibodies targeting the HA-domain was determined. (A) Immunofluorescent images of H1299, H460, and A549 cells without infection (a–c) or infected with Ad-CMV-HA-hSSTR2 (d–f). (B) ELISA of H1299, H460, and A549 cells with or without Ad-CMV-HA-hSSTR2 infection. Error bars represent SD of triplicate samples (*p < 0.05, uninfected cells vs. Ad-CMV-HA-hSSTR2 infected cells). (C) Western blots of H1299, H460, and A549 cells with or without Ad-CMV-HA-hSSTR2 infection. Cell protein extracted from cell lines was loaded at 20 μg per lane.
In vitro receptor binding of 111In-octreotide
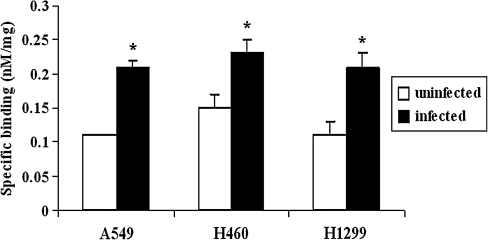
111In-Octreotide was used to target the receptor portion of the HA-SSTR2 fusion protein. Figure 3 shows the binding of 111In-octreotide at a saturating concentration of 100 nM to uninfected cells and cells infected with Ad-CMV-HA-hSSTR2. As seen, the radioligand binding increased significantly after Ad-CMV-HA-hSSTR2 infection.
FIG. 3.
Radioligand targeting of the receptor domain demonstrates that H1299, H460, and A549 cells express HA-SSTR2 after in vitro infection with Ad-CMV-HA-hSSTR2. Specific binding of 100 nM 111In-octreotide to cell membranes of NSCLC cell lines with or without infection with Ad-CMV-HA-hSSTR2 is shown. Error bars represent SD of triplicate samples (*p < 0.05, uninfected cells vs. Ad-CMV-HA-hSSTR2 infected cells).
In vivo imaging of 111In-octreotide and ex vivo biodistribution
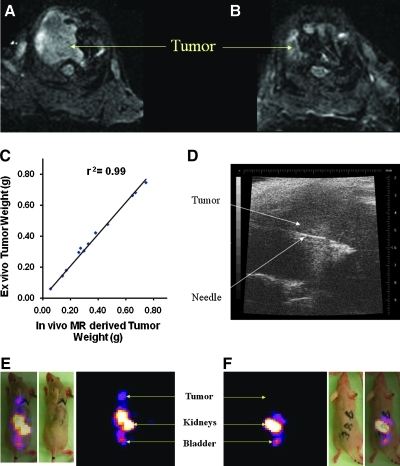
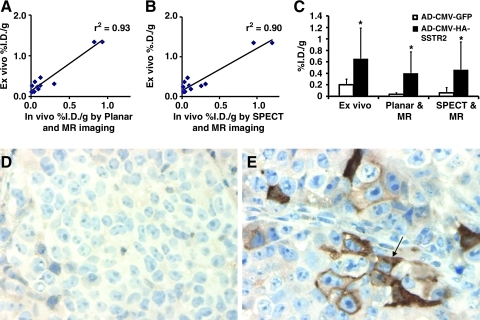
Representative EPI-based MR images (Fig. 4A and B) demonstrate tumor as increased signal in the superior aspect of the thorax. Tumor weights derived from volume assessment of the MR imaging correlated highly with that from excised tumors (r2 = 0.99, n = 12, coefficient of the x variable = 1.03; Fig. 4C), as we have reported previously (Bankson et al., 2008). We found that we could visualize the intrathoracic tumor by ultrasound; therefore, we used this modality to guide injection of adenovirus into the tumor (Fig. 4D). Both planar and SPECT imaging (Fig. 4E and F) demonstrated expression of HA-SSTR2 in tumors infected with Ad-CMV-HA-hSSTR2, but only background signal was seen in tumors infected with control virus. As mentioned above, the tumor volume assessment by MR enabled us to derive tumor weight. As with classic biodistribution experiments that excise and weigh tumors to control for underlying tumor size, in vivo biodistribution also was normalized to tumor weight. Ex vivo and in vivo biodistribution analysis normalized to tumor weight correlated (%I.D./g ex vivo vs. %I.D./g in vivo using planar and MR imaging, r2 = 0.93, p < 0.05, n = 12; Fig. 5A); %I.D./g ex vivo vs. %I.D./g in vivo using SPECT and MR imaging, r2 = 0.90, p < 0.05, n = 12; Fig. 5B). Ex vivo and in vivo biodistribution analysis demonstrated greater uptake in tumors injected with Ad-CMV-HA-hSSTR2 compared with tumors injected with control virus, Ad-CMV-GFP (p < 0.05; Fig. 5C). This was confirmed by immunohistochemistry targeting the HA-tag (Fig. 5D and E). Thus, by both ex vivo biodistribution analysis and in vivo biodistribution analysis using small animal cognates of clinically used imaging devices, expression of SSTR2-based reporters can be imaged and quantified after in vivo gene transfer into intrathoracic NSCLC tumors.
FIG. 4.
HA-SSTR2 expression can be imaged after in vivo infection of intrathoracic H460 tumors with Ad-CMV-HA-hSSTR2. (A, B) EPI-based MRI imaging demonstrated tumor as increased signal in the lung apices. The tumor in (A) was injected with Ad-CMV-HA-hSSTR2, and the tumor in (B) was injected with Ad-CMV-GFP. (C) In vivo MR-derived lung tumor weight correlated with ex vivo tumor weight (n = 12). (D) Representative image of ultrasound-guided intratumoral injection of adenoviruses. (E, F) Representative coronal, gamma-camera SPECT images of uptake of 111In-octreotide by lung tumors injected with Ad-CMV-HA-hSSTR2 (E) or Ad-CMV-GFP (F). Normal clearance through the kidneys and bladder is seen. There is uptake by tumor injected with Ad-CMV-HA-hSSTR2 (E), but only background signal is seen in the tumor injected with Ad-CMV-GFP(F). The same mouse injected with Ad-CMV-HA-hSSTR2 is shown by MR (A) and SPECT (E), and the same mouse injected with Ad-CMV-GFP is shown by MR (B) and SPECT (F).
FIG. 5.
Quantitative in vivo and ex vivo biodistribution analysis demonstrating HA-SSTR2 expression by intrathoracic H460 tumors after in vivo infection with Ad-CMV-HA-hSSTR2. (A, B) In vivo and ex vivo biodistribution into tumors correlate. Correlation of in vivo %I.D./g of 111In-octreotide uptake into tumors by planar and MR (A; n = 12) or SPECT and MR (B; n = 12) correlated with %I.D./g assessed from excised tumors. (C) In vivo and ex vivo biodistribution analysis demonstrates increased uptake in tumors infected with Ad-CMV-HA-hSSTR2 compared with tumors infected with Ad-CMV-GFP. *Tumor infected with Ad-CMV-HA-hSSTR2 vs. tumors infected with Ad-CMV-GFP: p < 0.05, n = 6; error bars represent SD. (D, E) Immunohistochemistry directed at the HA-tag demonstrates expression of HA-SSTR2 in tumors infected with Ad-CMV-HA-hSSTR2 (arrow, E), but background activity in tumors infected with Ad-CMV-GFP (D).
SSTR2 expression in human lung cancer samples
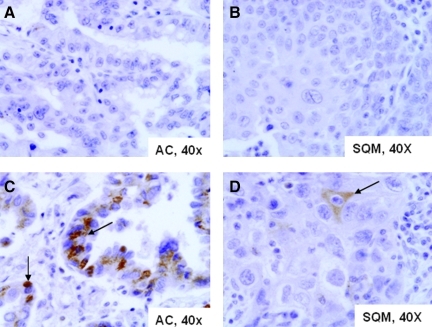
We evaluated whether SSTR2 is expressed in human lung cancer samples (Table 1, Fig. 6). As we noted in the cell lines, most NSCLCs do not express SSTR2. Only six (9%) NSCLCs expressed SSTR2A in more than 30% of the tumor cells, and all of them were adenocarcinomas (Fig. 6C). Among all NSCLC tumors, 79% lacked expression for this marker. Only one squamous cell carcinoma had focal and mild SSTR2A expression (Fig. 6D).
Table 1.
SSTR-2 Expression in Non–Small Cell Lung Cancer
| Diagnosis | Negative | 5–10% cells | 11–30% cells | 31–60% Cells | Total |
|---|---|---|---|---|---|
| Adenocarcinoma | 34 (70.8%) | 4 (8.3%) | 4 (8.3%) | 6 (12.5%) | 48 (100%) |
| Squamous cell carcinoma | 21 (95.5%) | 1 (4.5%) | 0 (0%) | 0 (0%) | 22 (100%) |
| Total NSCLC | 55 (78.5%) | 5 (7.1%) | 4 (5.7%) | 6 (8.6%) | 70 (100%) |
FIG. 6.
Immunohistochemical analysis of SSTR2 expression in human lung cancer. (A, B) Lung cancers that lack SSTR2 expression. (C, D) Lung cancers with SSTR2 expression (arrow): adenocarcinoma (AC) with high and diffuse SSTR2 expression (C), and squamous cell carcinoma (SQM), with focal and mild SSTR2 expression (D).
Discussion
Due to the poor prognosis of patients with lung cancer, novel therapies such as gene therapy are being explored. A limitation of gene therapy has been a lack of noninvasive, clinically applicable systems for assessing gene expression. Such tools are needed for multiple applications from vector development to assessing whether there is sufficient expression of a therapeutic gene product for efficacy. Reporters can be used for these purposes either alone or by linking to a therapeutic gene. SSTR2-based reporters have several advantages, including being of human origin, thus limiting an immune response; being amenable to longitudinal imaging (Singh et al., 2009); having clinically approved and new radioligands; and having a variant that is signaling-deficient to inhibit untoward effects on transduced cells or on linked genes of interest (Han et al., 2007). A potential disadvantage of using hSSTR2-based reporters in NSCLC is that there have been reports that NSCLC can express SSTRs (Fujita et al., 1994; Papotti et al., 2001; Herlin et al., 2009). This has the potential to confound hSSTR2-based imaging. We found that SSTR2-based reporters can be used for noninvasive imaging of in vivo gene transfer into intrathoracic NSCLCs.
The current work has several novelties, including showing SSTR subtype expression in three different NSCLC lines, using ultrasound guidance for gene transfer into an intrathoracic tumor, and demonstrating imaging and both in vivo and ex vivo quantification of SSTR2-based reporter gene transfer in an intrathoracic model of NSCLC. Moreover, our IHC dataset is the largest that assesses SSTR2 expression in human NSCLC tumor samples.
Data on SSTR expression in NSCLC is limited and has generally involved small numbers of samples. Papotti et al. (2001) found that 67% of NSCLCs were negative (6/12) or focally positive by immunohistochemistry for SSTR2 (2/12), whereas 33% were positive. Another report noted low (9/19) or moderate SSTR2 expression (7/19) in 84% of patients with NSCLC, and high levels in the remainder (Herlin et al., 2009). Using radiolabeled somatostatin or analogues, Reubi et al. (1990) found no SSTR expression in 12 cases by autoradiography. In cell lines, Fujita et al. (1994) evaluated SSTR1 expression, and Papotti et al. (2001) SSTR2 expression, by RT-PCR and noted low levels of SSTR1 and 2 expression in most of the six NSCLC lines tested. In comparison, we did not find SSTR2 expression in the three cell lines that we tested, but SSTR1 was noted in all three. We found that there can be expression of combinations of other SSTR subtypes in the NSCLC cell lines, depending on the cell line. One possible explanation for differences in observed SSTR2 expression in cell lines and histologic samples may be the degree of differentiation, because it has been reported that SSTR2 expression is greatest in well differentiated NSCLC (Herlin et al., 2009). Our IHC dataset including 70 patients is the largest evaluating SSTR2 expression in human NSCLC tumor samples of which we are aware. We found no SSTR2 expression in 78.5% of samples and low expression in 7.1%. Moderate to high expression was seen in 14.4%.
Clinically, the pattern of SSTR2 expression and 111In-octreotide background in patients is well known, and is mimicked in mice (Kundra et al., 2002). Tumors expressing SSTR2 can be seen even in areas of higher background, such as the liver (Lebtahi et al., 2002). By imaging, low levels of 111In-octreotide uptake have been reported in 10/10 patients with NSCLC by Lau et al. (2000); in 10/13 patients by Kirsch et al. (1994); and in 40/40 primary tumors, 5/15 lymph node metastases, and 1/7 distant metastases by Kwekkeboom et al. (1994). Using a 68Ga-labeled octreotide-based radiopharmaceutical, 68Ga-DOTATOC, Dimitrakopoulou-Strauss et al. (2006) reported moderate uptake in 7/9 primary tumors and no uptake in 8 metastases using PET. Thus, although there may be uptake of radiolabeled octreotide in NSCLC, it is generally at a low level in primary tumors, and often completely lost in metastases.
Herlin et al. (2009) noted low (9/19) or moderate (7/19) SSTR2 expression in 84% of patients with NSCLC. Upon imaging with the somatostatin analogue 99mTc-depreotide, uptake did not correlate with SSTR2 expression, likely because this agent has relatively equal affinity for subtypes 2, 3, and 5 (Virgolini et al., 1998). In this article, to partner with gene transfer of SSTR2-based reporters, we used 111In-octreotide because it preferentially binds SSTR2. In addition to subtype selectivity, our choice was also driven by the fact that neither 99mTc-depreotide nor other radiolabeled somatostatin analogues (Kwekkeboom et al., 1994) are commercially available currently in the United States. With 111In-octreotide, expression by intrathoracic NSCLC cells could be clearly visualized after in vivo gene transfer of an SSTR2-based reporter. Low uptake was seen in cells transfected with control virus, suggesting that expression of other SSTR subtypes, such as subtypes 1 and 3 by H460 cells, does not interfere with detection of the SSTR2-based reporter expression after gene transfer.
In vitro methods demonstrated expression by targeting either the HA domain or the receptor portion of the fusion protein after adenoviral infection in three different NSCLC lines. The HA domain allowed evaluation of expression regardless of any endogenous SSTRs that may bind to the radiopharmaceutical, using antibodies that are much less expensive than the radioligand. Expression of protein of the appropriate size and at the appropriate subcellular location was demonstrated. Next, the receptor portion of the fusion protein was targeted by 111In-octreotide, and again expression of the fusion protein after Ad-HA-SSTR2 infection was confirmed. Thus, SSTR2-based reporter expression could be clearly demonstrated in three different NSCLC cell lines despite expression of other SSTR subtypes. Supporting our findings, gene transfer of SSTR2 in an NSCLC cell line, A427, has been reported previously using a subcutaneous model (Zinn et al., 2000; Rogers et al., 2003, 2005; Parry et al., 2007), but SSTR subtype expression, an intrathoracic NSCLC model, and in vivo quantification were not demonstrated.
For in vivo studies, we used MR to identify the presence of tumors after intrathoracic injection in mice. In mouse models, CT does not provide sufficient soft-tissue contrast for distinguishing tumor from the chest wall or mediastinal structures. Use of MR has traditionally been limited for the evaluation of the thorax, and even more so in small animals due to rapid breathing and cardiac motion. An EPI-based, respiratory- and cardiac-gated sequence with fat saturation was used to generate T2-weighted images of the thorax that depicted the tumor (Bankson et al., 2008). This then allowed us to screen mice for tumors and measure tumor volume.
In a novel application of small-animal imaging, we used ultrasound to guide needle placement into intrathoracic tumors. Normally, one would consider the lung not to be in the realm of ultrasound imaging due to air that can disrupt propagation of sound waves. With the intrathoracic tumors, a window could be found for visualizing the tumors. A limitation of this technique is that small, deep tumors surrounded by lung will likely not be accessible due to surrounding air. In the current application, use of ultrasound ensured injection into the tumor.
After adenovirus injection, MR and gamma-camera imaging were performed. As we have demonstrated previously, MR image analysis correlated highly with tumor weight (Bankson et al., 2008), and we used these data to normalize in vivo %I.D. The in vivo %I.D./g correlated with ex vivo % I.D./g. Both methods demonstrated increased uptake after Ad-HA-SSTR2 infection compared with control virus. In vivo monitoring of gene transfer can be approached using reporter genes either alone or in conjunction with a gene of interest, e.g., for developing vectors (Hemminki et al., 2001), evaluating promoters, and inducing two separate genes (Sun et al., 2001), such as an imagable reporter and a therapeutic gene. The therapeutic gene of interest and reporter may be induced by separate promoters (Yaghoubi et al., 2001; Zinn et al., 2002) or may be linked, e.g., via an internal ribosome entry site (Tjuvajev et al., 1999; Liang et al., 2002) or a bidirectional promoter (Sun et al., 2001). Using such techniques, expression of the gene of interest and reporter correlate (Tjuvajev et al., 1999; Sun et al., 2001; Liang et al., 2002; Zinn et al., 2002; Yang et al., 2005). Furthermore, the vector itself may serve as a therapeutic; for example, SSTR2 imaging with 111In-octreotide has been used to demonstrate infection by a therapeutic vaccinia virus (McCart et al., 2004).
Because most lung cancers have no or little SSTR2 expression, they are good candidates for SSTR2-based reporter imaging. In the minority with SSTR2 expression, SSTR2-based reporter imaging should still be applicable if a prescan is performed to obtain baseline levels of radiopharmaceutical uptake. Then increased expression after gene transfer may be assessed, because different amounts of SSTR2-based reporter expression can be discerned (Kundra et al., 2002; Yang et al., 2005; Singh et al., 2009). Given that there is heterogeneity in radiopharmaceutical uptake among patients and among tumors/metastases within the same patient, when this technique is used in patients, it would be prudent to obtain a prescan with the radiopharmaceutical as a baseline before imaging gene transfer of SSTR2-based reporters.
To meet the promise of gene therapy, noninvasive imaging methods for assessing gene expression are needed that have the potential for clinical translation. In this work, we used clinically applicable tools such as adenovirus and 111In-octreotide to demonstrate the utility of an hSSTR2-based reporter in an intrathoracic model of NSCLC. The findings suggest that it should be possible to visualize and noninvasively quantify SSTR2-based reporter gene transfer in NSCLCs, enabling a host of gene therapy applications, such as developing vectors, designing dosing schedules, and assessing expression of a linked therapeutic gene. Importantly, because gene expression can vary among individuals and among tumors within an individual, the use of SSTR2-based reporters should enable the personalizing of therapy to individual patients.
Acknowledgments
This work was supported in part by the National Institutes of Health through RCA 123841, a Specialized Program of Research Excellence (SPORE) Grant CA 070907, and U.T.-M.D. Anderson's Cancer Center Support Grant P30-CA016672.
Author Disclosure Statement
There is a patent on the reporter imaging technology held by the institution.
References
- American Cancer Society. Cancer Facts & Figures 2009. American Cancer Society; Atlanta, GA: 2009. [Google Scholar]
- Bankson J.A. Ji L. Ravoori M. Han L. Kundra V. Echo-planar imaging for MRI evaluation of intrathoracic tumors in murine models of lung cancer. J. Magn. Reson. Imaging. 2008;27:57–62. doi: 10.1002/jmri.21221. [DOI] [PubMed] [Google Scholar]
- Bell G.I. Reisine T. Molecular biology of somatostatin receptors. Trends Neurosci. 1993;16:34–38. doi: 10.1016/0166-2236(93)90050-v. [DOI] [PubMed] [Google Scholar]
- Benamore R. Shepherd F.A. Leighl N. Pintilie M. Patel M. Feld R. Herman S. Does intensive follow-up alter outcome in patients with advanced lung cancer? J. Thorac. Oncol. 2007;2:273–281. doi: 10.1097/01.JTO.0000263708.08332.76. [DOI] [PubMed] [Google Scholar]
- Cote J. Bourget L. Garnier A. Kamen A. Study of adenovirus production in serum-free 293SF suspension culture by GFP-expression monitoring. Biotechnol. Prog. 1997;13:709–714. doi: 10.1021/bp970110i. [DOI] [PubMed] [Google Scholar]
- Cross D. Burmester J.K. Gene therapy for cancer treatment: past, present and future. Clin. Med. Res. 2006;4:218–227. doi: 10.3121/cmr.4.3.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martin R. Raidl M. Hofer E. Binder B.R. Adenovirus-mediated expression of green fluorescent protein. Gene Ther. 1997;4:493–495. doi: 10.1038/sj.gt.3300408. [DOI] [PubMed] [Google Scholar]
- Dimitrakopoulou-Strauss A. Georgoulias V. Eisenhut M. Herth F. Koukouraki S. Macke H.R. Haberkorn U. Strauss L.G. Quantitative assessment of SSTR2 expression in patients with non–small cell lung cancer using (68)Ga-DOTATOC PET and comparison with (18)F-FDG PET. Eur. J. Nucl. Med. Mol. Imaging. 2006;33:823–830. doi: 10.1007/s00259-005-0063-5. [DOI] [PubMed] [Google Scholar]
- Fujita T. Yamaji Y. Sato M. Murao K. Takahara J. Gene expression of somatostatin receptor subtypes, SSTR1 and SSTR2, in human lung cancer cell lines. Life Sci. 1994;55:1797–1806. doi: 10.1016/0024-3205(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Han L. Yang D. Kundra V. Signaling can be uncoupled from imaging of the somatostatin receptor type 2. Mol. Imaging. 2007;6:427–437. [PubMed] [Google Scholar]
- Hemminki A. Belousova N. Zinn K.R. Liu B. Wang M. Chaudhuri T.R. Rogers B.E. Buchsbaum D.J. Siegal G.P. Barnes M.N. Gomez-Navarro J. Curiel D.T. Alvarez R.D. An adenovirus with enhanced infectivity mediates molecular chemotherapy of ovarian cancer cells and allows imaging of gene expression. Mol. Ther. 2001;4:223–231. doi: 10.1006/mthe.2001.0446. [DOI] [PubMed] [Google Scholar]
- Herlin G. Kolbeck K.G. Menzel P.L. Svensson L. Aspelin P. Capitanio A. Axelsson R. Quantitative assessment of 99mTc-depreotide uptake in patients with non-small-cell lung cancer: immunohistochemical correlations. Acta Radiol. 2009;50:902–908. doi: 10.1080/02841850903127477. [DOI] [PubMed] [Google Scholar]
- Jett J.R. Current treatment of unresectable lung cancer. Mayo Clinic Proc. 1993;68:603–611. doi: 10.1016/s0025-6196(12)60376-0. [DOI] [PubMed] [Google Scholar]
- Kirsch C.M. Von Pawel J. Grau I. Tatsch K. Indium-111 pentetreotide in the diagnostic work-up of patients with bronchogenic carcinoma. Eur. J. Nucl. Med. 1994;21:1318–1325. doi: 10.1007/BF02426696. [DOI] [PubMed] [Google Scholar]
- Kluxen F.W. Bruns C. Lubbert H. Expression cloning of a rat brain somatostatin receptor cDNA. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4618–4622. doi: 10.1073/pnas.89.10.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundra V. Mannting F. Jones A.G. Kassis A.I. Noninvasive monitoring of somatostatin receptor type 2 chimeric gene transfer. J. Nucl. Med. 2002;43:406–412. [PubMed] [Google Scholar]
- Kwekkeboom D.J. Kho G.S. Lamberts S.W. Reubi J.C. Laissue J.A. Krenning E.P. The value of octreotide scintigraphy in patients with lung cancer. Eur. J. Nucl. Med. 1994;21:1106–1113. doi: 10.1007/BF00181066. [DOI] [PubMed] [Google Scholar]
- Lau S.K. Johnson D.S. Coel M.N. Imaging of non–small-cell lung cancer with indium-111 pentetreotide. Clin. Nucl. Med. 2000;25:24–28. doi: 10.1097/00003072-200001000-00006. [DOI] [PubMed] [Google Scholar]
- Lebtahi R. Le Cloirec J. Houzard C. Daou D. Sobhani I. Sassolas G. Mignon M. Bourguet P. Le Guludec D. Detection of neuroendocrine tumors: 99mTc-P829 scintigraphy compared with 111In-pentetreotide scintigraphy. J. Nucl. Med. 2002;43:889–895. [PubMed] [Google Scholar]
- Liang Q. Gotts J. Satyamurthy N. Barrio J. Phelps M.E. Gambhir S.S. Herschman H.R. Noninvasive, repetitive, quantitative measurement of gene expression from a bicistronic message by positron emission tomography, following gene transfer with adenovirus. Mol. Ther. 2002;6:73–82. doi: 10.1006/mthe.2002.0626. [DOI] [PubMed] [Google Scholar]
- McCart J.A. Mehta N. Scollard D. Reilly R.M. Carrasquillo J.A. Tang N. Deng H. Miller M. Xu H. Libutti S.K. Alexander H.R. Bartlett D.L. Oncolytic vaccinia virus expressing the human somatostatin receptor SSTR2: molecular imaging after systemic delivery using 111In-pentetreotide. Mol. Ther. 2004;10:553–561. doi: 10.1016/j.ymthe.2004.06.158. [DOI] [PubMed] [Google Scholar]
- Meyer K. Irminger J.C. Moss L.G. De Vargas L.M. Oberholzer J. Bosco D. Morel P. Halban P.A. Sorting human beta-cells consequent to targeted expression of green fluorescent protein. Diabetes. 1998;47:1974–1977. doi: 10.2337/diabetes.47.12.1974. [DOI] [PubMed] [Google Scholar]
- Nesbitt J.C. Putnam J.B., Jr. Walsh G.L. Roth J.A. Mountain C.F. Survival in early-stage non–small cell lung cancer. Ann. Thorac. Surg. 1995;60:466–472. doi: 10.1016/0003-4975(95)00169-l. [DOI] [PubMed] [Google Scholar]
- O'Carroll A.M. Lolait S.J. König M. Mahan L.C. Molecular cloning and expression of a pituitary somatostatin receptor with preferential affinity for somatostatin-28. Mol. Pharmacol. 1992;42:939–946. [PubMed] [Google Scholar]
- Panetta R. Greenwood M.T. Warszynska A. Demchyshyn L.L. Day R. Niznik H.B. Srikant C.B. Patel Y.C. Molecular cloning, functional characterization, and chromosomal localization of a human somatostatin receptor (somatostatin receptor type 5) with preferential affinity for somatostatin-28. Mol. Pharmacol. 1994;45:417–427. [PubMed] [Google Scholar]
- Papotti M. Croce S. Bellò M. Bongiovanni M. Allia E. Schindler M. Bussolati G. Expression of somatostatin receptor types 2, 3 and 5 in biopsies and surgical specimens of human lung tumours. Correlation with preoperative octreotide scintigraphy. Virchows Arch. 2001;439:787–797. doi: 10.1007/s004280100494. [DOI] [PubMed] [Google Scholar]
- Parry J.J. Eiblmaier M. Andrews R. Meyer L.A. Higashikubo R. Anderson C.J. Rogers B.E. Characterization of somatostatin receptor subtype 2 expression in stably transfected A-427 human cancer cells. Mol. Imaging. 2007;6:56–67. [PubMed] [Google Scholar]
- Reubi J.C. Waser B. Sheppard M. Macaulay V. Somatostatin receptors are present in small-cell but not in non–small-cell primary lung carcinomas: relationship to EGF-receptors. Int. J. Cancer. 1990;45:269–274. doi: 10.1002/ijc.2910450211. [DOI] [PubMed] [Google Scholar]
- Rogers B.E. Zinn K.R. Buchsbaum D.J. Gene transfer strategies for improving radiolabeled peptide imaging and therapy. Q. J. Nucl. Med. 2000;44:208–223. [PubMed] [Google Scholar]
- Rogers B.E. Chaudhuri T.R. Reynolds P.N. Della Manna D. Zinn K.R. Non-invasive gamma camera imaging of gene transfer using an adenoviral vector encoding an epitope-tagged receptor as a reporter. Gene Ther. 2003;10:105–114. doi: 10.1038/sj.gt.3301853. [DOI] [PubMed] [Google Scholar]
- Rogers B.E. Parry J.J. Andrews R. Cordopatis P. Nock B.A. Maina T. MicroPET imaging of gene transfer with a somatostatin receptor-based reporter gene and 94mTc-Demotate 1. J. Nucl. Med. 2005;46:1889–1897. [PubMed] [Google Scholar]
- Scagliotti G. Multimodality approach to early-stage non–small cell lung cancer. Lung Cancer. 2007;57(Suppl. 2):S6–S11. doi: 10.1016/S0169-5002(07)70421-X. [DOI] [PubMed] [Google Scholar]
- Singh S.P. Yang D. Ravoori M. Han L. Kundra V. In vivo functional and anatomic imaging for assessment of in vivo gene transfer. Radiology. 2009;252:763–771. doi: 10.1148/radiol.2531081825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stables J. Scott S. Brown S. Roelant C. Burns D. Lee M.G. Rees S. Development of a dual glow-signal firefly and Renilla luciferase assay reagent for the analysis of G-protein coupled receptor signalling. J. Receptor Signal Transduct. Res. 1999;19:395–410. doi: 10.3109/10799899909036660. [DOI] [PubMed] [Google Scholar]
- Sun X. Annala A.J. Yaghoubi S.S. Barrio J.R. Nguyen K.N. Toyokuni T. Satyamurthy N. Namavari M. Phelps M.E. Herschman H.R. Gambhir S.S. Quantitative imaging of gene induction in living animals. Gene Ther. 2001;8:1572–1579. doi: 10.1038/sj.gt.3301554. [DOI] [PubMed] [Google Scholar]
- Tjuvajev J.G. Joshi A. Callegari J. Lindsley L. Joshi R. Balatoni J. Finn R. Larson S.M. Sadelain M. Blasberg R.G. A general approach to the non-invasive imaging of transgenes using cis-linked herpes simplex virus thymidine kinase. Neoplasia. 1999;1:315–320. doi: 10.1038/sj.neo.7900053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattemi E. Claudio P.P. Gene therapy for lung cancer: practice and promise. Ann. Ital. Chir. 2004;75:279–289. [PubMed] [Google Scholar]
- Virgolini I. Leimer M. Handmaker H. Lastoria S. Bischof C. Muto P. Pangerl T. Gludovacz D. Peck-Radosavljevic M. Lister-James J. Hamilton G. Kaserer K. Valent P. Dean R. Somatostatin receptor subtype specificity and in vivo binding of a novel tumor tracer, 99mTc-P829. Cancer Res. 1998;58:1850–1859. [PubMed] [Google Scholar]
- Yaghoubi S.S. Wu L. Liang Q. Toyokuni T. Barrio J.R. Namavari M. Satyamurthy N. Phelps M.E. Herschman H.R. Gambhir S.S. Direct correlation between positron emission tomographic images of two reporter genes delivered by two distinct adenoviral vectors. Gene Ther. 2001;8:1072–1080. doi: 10.1038/sj.gt.3301490. [DOI] [PubMed] [Google Scholar]
- Yamada Y. Reisine T. Law S.F. Ihara Y. Kubota A. Kagimoto S. Seino M. Seino Y. Bell G.I. Seino S. Somatostatin receptors, an expanding gene family: cloning and functional characterization of human SSTR3, a protein coupled to adenylyl cyclase. Mol. Endocrinol. 1992;6:2136–2142. doi: 10.1210/mend.6.12.1337145. [DOI] [PubMed] [Google Scholar]
- Yang D. Han L. Kundra V. Exogenous gene expression in tumors: noninvasive quantification with functional and anatomic imaging in a mouse model. Radiology. 2005;235:950–958. doi: 10.1148/radiol.2353040108. [DOI] [PubMed] [Google Scholar]
- Zinn K.R. Buchsbaum D.J. Chaudhuri T.R. Mountz J.M. Grizzle W.E. Rogers B.E. Noninvasive monitoring of gene transfer using a reporter receptor imaged with a high-affinity peptide radiolabeled with 99mTc or 188Re. J. Nucl. Med. 2000;41:887–895. [PubMed] [Google Scholar]
- Zinn K.R. Chaudhuri T.R. Krasnykh V.N. Buchsbaum D.J. Belousova N. Grizzle W.E. Curiel D.T. Rogers B.E. Gamma camera dual imaging with a somatostatin receptor and thymidine kinase after gene transfer with a bicistronic adenovirus in mice. Radiology. 2002;223:417–425. doi: 10.1148/radiol.2232010501. [DOI] [PubMed] [Google Scholar]