Abstract
A20 negatively regulates inflammation by inhibiting the nuclear factor κB (NF-κB) transcription factor in the tumor necrosis factor receptor (TNFR) and Toll-like receptor (TLR) pathways. A20 contains deubiquitinase and E3 ligase domains and thus has been proposed to function as a ubiquitin-editing enzyme downstream of TNFR1 by inactivating ubiquitinated RIP1. However, it remains unclear how A20 terminates NF-κB signaling downstream of TLRs. We have shown that A20 inhibited the E3 ligase activities of TRAF6, TRAF2, and cIAP1 by antagonizing interactions with the E2 ubiquitin conjugating enzymes Ubc13 and UbcH5c. A20, together with the regulatory molecule TAX1BP1, interacted with Ubc13 and UbcH5c and triggered their ubiquitination and proteasome-dependent degradation. These findings suggest a mechanism of A20 action in the inhibition of inflammatory signaling pathways.
The zinc finger protein A20 (also known as TNFAIP3) has an essential role in limiting the strength and duration of NF-κB signaling (1). A20-deficient mice die prematurely from multiorgan inflammation and cachexia, and A20-deficient cells exhibit a defect in the termination of tumor necrosis factor–α (TNF-α) and lipopolysaccharide (LPS)–induced NF-κB signaling (2, 3). A20 requires several regulatory proteins, including Tax1 binding protein 1 (TAX1BP1), and the E3 ubiquitin ligases Itch and ring finger protein 11 (RNF11), to restrict NF-κB activation (4-6). A20 functions as a ubiquitin-editing enzyme with both deubiquitinating (DUB) and ubiquitin E3 ligase activity toward the adaptor protein and death-domain containing protein kinase, receptor-interacting protein 1 (RIP1) in the TNFR pathway (7). A20 first cleaves lysine 63 (K63)–linked polyubiquitin chains on RIP1 and then conjugates lysine 48 (K48)–linked polyubiquitin chains that target RIP1 for degradation by the proteasome (7). A20 also inhibits the polyubiquitination and activation of the E3 ubiquitin ligase TNF receptor–associated factor 6 (TRAF6) in the Toll-like receptor 4 and interleukin-1 receptor (TLR4/IL-1R) pathways (3); however, it is unclear whether A20 functions by a similar mechanism to inhibit TRAF6.
To investigate the mechanism of TRAF6 regulation by A20, we examined TRAF6 protein-protein interactions by coimmunoprecipitation in cells stimulated with IL-1. Wild-type mouse embryonic fibroblasts (MEFs) or MEFs that lack expression of A20 (A20−/−) or TAX1BP1 (Tax1bp1−/−) were treated with IL-1 for various times, and the interactions between TRAF6, A20, and TAX1BP1 were monitored by immunoprecipitations and protein immunoblotting (Fig. 1 and fig. S1). A20 and TAX1BP1 were recruited to TRAF6 with distinct kinetics. Whereas TAX1BP1 interacted with TRAF6 after 15 min of IL-1 treatment, A20 recruitment to TRAF6 was delayed until 45 min of stimulation (Fig. 1A). No binding was observed when immunoprecipitations were performed with a control rabbit immunoglobulin antibody (Fig. 1B). TAX1BP1 recruitment to TRAF6 was impaired in A20-deficient MEFs (Fig. 1A). Interaction of A20 with TRAF6 was also dependent on TAX1BP1 (fig. S1A). The A20 and TAX1BP1 regulatory proteins Itch and RNF11 were also recruited to TRAF6 at early time points, together with TAX1BP1, although the interactions with TRAF6 were transient (Fig. 1A). TRAF6 polyubiquitination and activation is dependent on the E2 enzyme Ubc13 (8, 9). TRAF6 interaction with Ubc13 was stimulus-dependent and was lost after 45 min of IL-1 stimulation, coinciding with the recruitment of A20 to TRAF6. The TRAF6-Ubc13 interaction was more persistent in A20−/− and Tax1bp1−/− MEFs treated with IL-1 (Fig. 1A and fig. S1A). Because Ubc13 may regulate NF-κB in stimulus- and cell type–specific ways (10), we also examined interactions of TRAF6 with the E2 enzyme UbcH5c (also known as Ube2D3), which functions with TRAF6 to synthesize unanchored polyubiquitin chains that activate IkBkinase (IKK) (11). TRAF6 and UbcH5c interacted transiently after IL-1 stimulation in control MEFs (Fig. 1A). However, binding of TRAF6 to UbcH5c was prolonged in A20-deficient MEFs (Fig. 1A). The persistent interactions between TRAF6 and the E2 enzymes Ubc13 and UbcH5c coincided with enhanced degradation of IκBα and activation of NF-κB in A20−/− and Tax1bp1−/− MEFs (Fig. 1A) (3, 4).
Fig. 1.
Disruption of interactions between E2 and E3 enzymes in the TNFR and TLR4/IL-1R pathways by A20 and TAX1BP1. (A) Kinetics of TRAF6, Ubc13, UbcH5c, Itch, RNF11, A20, and TAX1BP1 interactions in control and A20-deficient MEFs. A20+/+ and A20−/− MEFs were stimulated with IL-1 for the indicated times. Proteins from lysates were immunoprecipitated with TRAF6 antibody and detected by immunoblotting with antibodies to A20, Ubc13, UbcH5c, Itch, RNF11, TAX1BP1, or TRAF6. Lysates were subjected to immunoblotting with antibodies to IκBα, A20, TAX1BP1, Ubc13, UbcH5c, RNF11, Itch, and β-actin. (B) Specificity of TRAF6, Ubc13, A20, UbcH5c, and TAX1BP1 interactions. A20+/+ MEFs were stimulated with IL-1 for the indicated times. Proteins from lysates were immunoprecipitated with TRAF6 or control rabbit antibody [Cont. IgG (immunoglobulin G)] and detected by immunoblotting with antibodies to A20, Ubc13, UbcH5c, TAX1BP1, or TRAF6. Lysates were subjected to immunoblotting with antibodies to IκBα and β-actin. (C) Kinetics of TRAF2, Ubc13, Itch, RNF11, A20, and TAX1BP1 interactions in control and A20-deficient MEFs. A20+/+ and A20−/− MEFs were stimulated with TNF-α, and proteins from lysates were immunoprecipitated with TRAF2 antibody followed by immunoblotting with antibodies to A20, Ubc13, Itch, RNF11, TAX1BP1, and TRAF2. Lysates were subjected to immunoblotting with antibodies to IκBα, TAX1BP1, A20, Ubc13, RNF11, Itch, and β-actin. (D and E) Interaction of TAX1BP1 with Ubc13. A20+/+ and A20−/− MEFs were stimulated with TNF-α (D) or LPS (E) for 30 and 60 min, and proteins from lysates were immunoprecipitated with antibody to TAX1BP1, followed by immunoblotting with antibodies to Ubc13, UbcH5c, or TAX1BP1. Lysates were subjected to immunoblotting with antibodies to IκBα, A20, TAX1BP1, Ubc13, and β-actin. The results shown are representative of three independent experiments. IP, immunoprecipitation; IB, immunoblot.
Ubc13 also functions as an E2 enzyme for other E3 ligases, including TRAF2 and cIAP1 in the TNFR pathway (12, 13). Therefore, we investigated whether A20 targeted TRAF2-Ubc13 and cIAP1-Ubc13 complexes for inactivation in the TNFR1 pathway. TRAF2 interacted transiently with Ubc13 in a TNF-α–dependent manner, and the interaction was disrupted upon recruitment of A20 to TRAF2 (Fig. 1C). However, in A20−/− and Tax1bp1−/− MEFs, the TRAF2-Ubc13 interaction was persistent after stimulation with TNF-α (Fig. 1C and fig. S1B). Similar results were obtained with cIAP1 and Ubc13 (fig. S1C). We also confirmed that A20 interacted with Ubc13 in a stimulus-dependent manner in both primary bone marrow–derived macrophages (BMDMs) and dendritic cells (BMDCs) (fig. S1, D and E). A20 and TAX1BP1 were dependent on each other to interact with Ubc13 in cells treated with TNF-α or LPS (Fig. 1, D and E, and fig. S1, F and G). Collectively, these results indicate that A20 and TAX1BP1 function together to disrupt interactions among the E3 ligases TRAF6, TRAF2, and c-IAP1 and the E2 enzymes Ubc13 and UbcH5c.
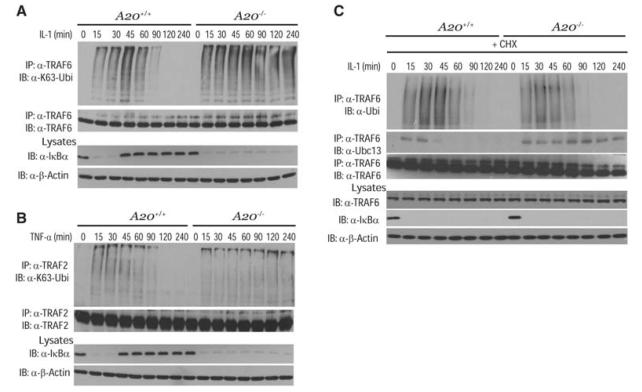
TRAF6 is activated in cells treated with LPS or IL-1 and is auto-ubiquitinated in a Ubc13-dependent manner (8, 14, 15). As expected, TRAF6 ubiquitination was transient in control MEFs, in sharp contrast to the persistent TRAF6 ubiquitination observed in A20−/− and Tax1bp1−/− MEFs upon IL-1 stimulation (Fig. 2A and fig. S2A). Interleukin-1 receptor-associated kinase 1 (IRAK1) functions upstream of TRAF6 in the IL-1R pathway and is regulated by K63-linked polyubiquitination (16). However, A20 or TAX1BP1 deficiency had no effect on the ubiquitination or degradation of IRAK1 (fig. S2B). TRAF2 and RIP1 ubiquitination were also prolonged in A20−/− and Tax1bp1−/− MEFs upon stimulation with TNF-α (Fig. 2B and fig. S2D). Finally, the stability of ubiquitinated TRAF6 was similar in control and A20−/− or Tax1bp1−/− MEFs treated with IL-1 and the protein synthesis inhibitor cycloheximide (CHX) (Fig. 2C and fig. S2F).
Fig. 2.
Negative regulation of TRAF2 and TRAF6 polyubiquitination by A20. (A) Kinetics of TRAF6 ubiquitination. A20+/+ and A20−/− MEFs were stimulated with IL-1 for the indicated times. Proteins from lysates were immunoprecipitated with antibody to TRAF6, eluted with 1% SDS, diluted in lysis buffer, reimmunoprecipitated with TRAF6 antibody followed by immunoblotting with K63-specific antibody to Ubi or antibody to TRAF6. Lysates were subjected to immunoblotting with antibodies to IκBα or β-actin. (B) Kinetics of TRAF2 ubiquitination. A20+/+ and A20−/− MEFs were stimulated with TNF-α for the indicated times. Proteins from lysates were immunoprecipitated with antibody to TRAF2, eluted with 1% SDS, diluted in lysis buffer, and reimmunoprecipitated with TRAF2 antibody, followed by immunoblotting with antibodies to Ubi or TRAF2. Lysates were subjected to immunoblotting with antibodies to IκBα and β-actin. (C) Stability of ubiquitinated TRAF6. A20+/+ and A20−/− MEFs were pretreated with cycloheximide, followed by IL-1 stimulation for the indicated times. Proteins from lysates were immunoprecipitated with antibody to TRAF6 as described (A), followed by immunoblotting with antibodies to Ubi, Ubc13, or TRAF6. Lysates were subjected to immunoblotting with antibodies to TRAF6, IκBα, and β-actin. The results shown are representative of three independent experiments.
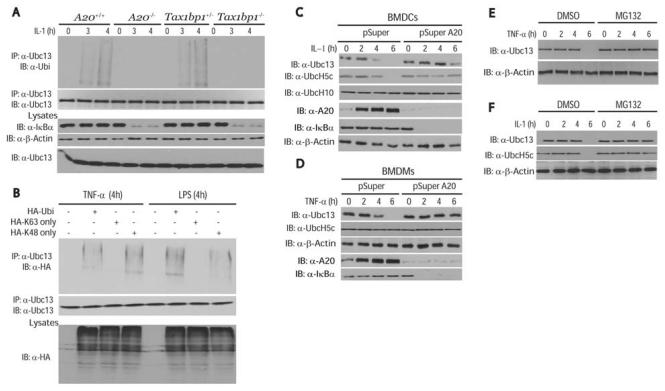
Because A20 promotes K48-linked ubiquitination and proteasomal degradation of RIP1 in the TNFR pathway (7), but does not promote degradation of TRAF6 in cells stimulated with IL-1 (Fig. 2C), we next investigated whether A20 targeted Ubc13 for degradation. Treatment of cells with IL-1 or TNF-α triggered the ubiquitination of endogenous Ubc13 in control MEFs but not in A20−/− or Tax1bp1−/− MEFs (Fig. 3A and fig. S3). To determine the type of Ubc13 polyubiquitin chains, we transfected MEFs with a hemagglutinin (HA)–tagged wild-type ubiquitin or ubiquitin mutants with substitution of arginine for all lysine residues except the lysine at position 48 (K48-only mutant) or the lysine at position 63 (K63-only mutant). Ubc13 polyubiquitination was observed in cells treated with either TNF-α or LPS and expressing either wild-type HA-Ubi or HA-Ubi K48-only (Fig. 3B), which suggests that Ubc13 ubiquitination was predominantly K48-linked, leading to proteasome-mediated degradation. Indeed, Ubc13 degradation was observed in primary BMDCs or BMDMs after treatment with IL-1 (Fig. 3C) or TNF-α (Fig. 3D). Stimulus-dependent degradation of UbcH5c also occurred, but only in response to IL-1 and not to TNF-α (Fig. 3, C and D, and fig. S4, A and C). Knockdown of A20 with small interfering RNA (siRNA) prevented the degradation of both Ubc13 and UbcH5c (Fig. 3, C and D). However, the E3 ubiquitin ligases RNF11 and Itch were both dispensable for Ubc13 degradation, despite the requirement of Itch for the attenuation of TRAF6 ubiquitination (fig. S5). Ubc13 was also degraded in response to IL-1 and TNF-α in MEFs in an A20- and TAX1BP1-dependent manner (fig. S4). Finally, TNF-α and IL-1–induced Ubc13 and UbcH5c degradation was blocked by the proteasome inhibitor MG-132 (Fig. 3, E and F).
Fig. 3.
The ubiquitination and degradation of Ubc13 is promoted by A20 and TAX1BP1. (A) Polyubiquitination of Ubc13 in cells stimulated with IL-1. Control, A20−/−, or Tax1bp1−/− MEFs were treated with IL-1 for the indicated times. Proteins from lysates were immunoprecipitated with antibody to Ubc13, eluted with 1% SDS, diluted in lysis buffer, and reimmunoprecipitated with Ubc13 antibody, followed by immunoblotting with antibodies to Ubi or Ubc13. Lysates were subjected to immunoblotting with antibodies to IκBα, Ubc13, and β-actin. (B) K48-linked polyubiquitination of Ubc13. Control MEFs were transiently transfected with plasmids encoding HA-Ubi, HA-Ubi-K63 only, or HA-Ubi-K48 only. After 36 hours, cells were stimulated for 4 hours with either TNF-α or LPS. Ubiquitination assays were performed as described in (A). Lysates were subjected to immunoblotting with antibody to HA. (C and D) Requirement of A20 for the degradation of Ubc13 and UbcH5c. BMDCs (C) or BMDMs (D) were transiently transfected with pSuper or pSuper A20 siRNA plasmids. After 36 hours, cells were stimulated with either IL-1 or TNF-α for the indicated times. Lysates were subjected to immunoblotting with antibodies to Ubc13, UbcH5c, UbcH10, A20, IκBα,or β-actin. (E and F) Proteasome-dependent degradation of Ubc13. Control MEFs were treated with TNF-α (E) or IL-1 (F) for the indicated times in the presence of MG132 or vehicle [dimethyl sulfoxide (DMSO)]. Lysates were subjected to immunoblotting with antibodies to Ubc13, UbcH5c, and β-actin. The results shown are representative of three independent experiments.
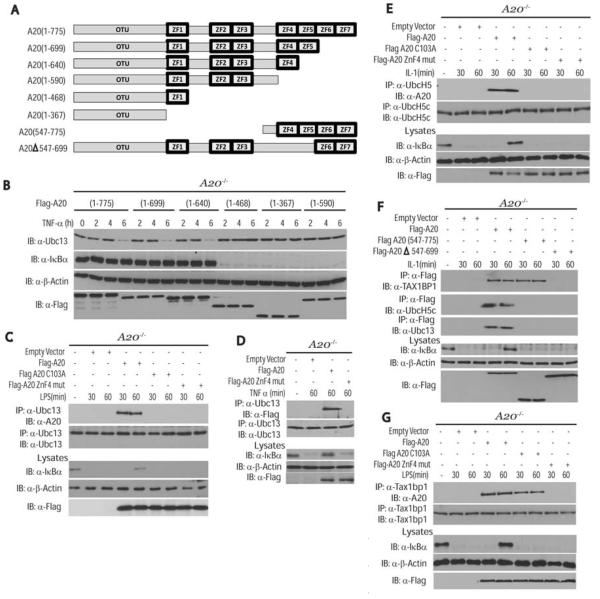
A20 inhibits RIP1 activation in the TNFR pathway through its deubiquitinase and E3 ligase activities; however, whether these activities are required for the down-regulation of Ubc13 and UbcH5c is unknown. A20 contains an N-terminal ovarian tumor (OTU) DUB domain and seven C-terminal zinc finger domains (Fig. 4A). A20-deficient MEFs were reconstituted with various Flag-tagged A20 deletion mutants to determine the domain(s) important for the degradation of Ubc13 and UbcH5c. Deletion of zinc fingers 5 through 7 had no effect on the degradation of Ubc13 or UbcH5c (Fig. 4B and fig. S6). However, deletion of zinc finger 4 (ZnF4) abrogated A20-mediated degradation of Ubc13 and UbcH5c (Fig. 4B and fig. S6), correlating with published reports on the importance of ZnF4 in A20-mediated inhibition of NF-κB (17). The degradation of Ubc13 and UbcH5c by A20 appeared to be specific because UbcH10 (also known as Ube2c) was not degraded in cells treated with IL-1 (fig. S6). Reconstitution studies in A20−/− MEFs revealed that wild-type A20, but not A20 C103A, disrupted the binding of TRAF6 and Ubc13 (fig. S7A). Overexpression of the deubiquitinating enzyme cylindromatosis (CYLD) exerted no effect on TRAF6 and Ubc13 binding, which suggests that CYLD uses a distinct mechanism to inhibit TRAF6 ubiquitination (fig. S7A). Similarly, A20, but not A20 C103A or CYLD, efficiently inhibited TRAF2-Ubc13 binding in TNF-α–stimulated A20−/− MEFs (fig. S7B). Although wild-type A20 interacted with endogenous Ubc13 and UbcH5c in cells stimulated with LPS or IL-1, both A20 C103A and A20 ZnF4 mutants were defective for binding to Ubc13 and UbcH5c (Fig. 4, C to E) and thus were unable to inhibit TRAF6 ubiquitination or promote Ubc13 degradation (fig. S8, A to C). A20 C103 and ZnF4 were also required for TAX1BP1 to interact with Ubc13 (fig. S8D). Furthermore, A20 deletion mutants lacking either the OTU domain or zinc fingers 4 and 5 were not associated with Ubc13 or UbcH5c (Fig. 4F). However, for TAX1BP1 binding, the OTU domain was dispensable and only ZnF4 was critical (Fig. 4, F and G) (17). Collectively, these results indicate that A20 ZnF4 is important for binding to TAX1BP1, whereas the OTU domain and ZnF4 are important for binding to Ubc13 and UbcH5c.
Fig. 4.
Requirement of A20 zinc finger 4 for TAX1BP1 binding and degradation of Ubc13 and UbcH5c. (A) Schematic of A20 deletion mutants. (B) Requirement of A20 ZnF4 for the degradation of Ubc13. A20−/− MEFs were transiently transfected with the indicated mutants. After 36 hours, cells were treated with TNF-α for the indicated times. Lysates were subjected to immunoblotting with antibodies to Ubc13, IκBα, β-actin, or Flag. (C) A20 DUB and ZnF4 mutants do not interact with Ubc13. A20−/− MEFs were transiently transfected with Flag-A20, Flag-A20 (C103A), and Flag-A20 ZnF4 mutant (C624A and C627A). After 36 hours, cells were stimulated with LPS for the indicated times, and proteins from lysates were immunoprecipitated with Ubc13 antibody and detected by immunoblotting with antibodies to A20 or Ubc13. Lysates were subjected to immunoblotting with antibodies to Flag, IκBα, and β-actin. (D) A20 ZnF4 mutant does not interact with Ubc13. A20−/− MEFs were transiently transfected with Flag-A20 and Flag-A20 ZnF4 mutant (C624A and C627A). After 36 hours, cells were stimulated with TNF-α for 60 min, and proteins from lysates were immunoprecipitated with Ubc13 antibody and detected by immunoblotting with antibodies to Flag or Ubc13. Lysates were subjected to immunoblotting with antibodies to Flag, IκBα,and β-actin. (E) A20 DUB and ZnF4 mutants do not interact with UbcH5c. A20−/− MEFs were transiently transfected with plasmids as described in (C). After 36 hours, cells were stimulated with IL-1 for the indicated times, and proteins from lysates were immunoprecipitated with UbcH5c antibody and detected by immunoblotting with antibodies to A20 or UbcH5c. Lysates were subjected to immunoblotting with antibodies to Flag, IκBα,and β-actin. (F) Differential requirement for the A20 OTU domain and C-terminal zinc fingers in binding to TAX1BP1, Ubc13, and UbcH5c. A20−/− MEFs were transiently transfected with empty vector, Flag-A20, Flag-A20 (547-775) or Flag-A20Δ547-699. After 36 hours, cells were treated with IL-1 for either 30 or 60 min. Proteins from lysates were immunoprecipitated with antibody to Flag, followed by immunoblotting with antibodies to TAX1BP1, UbcH5c, or Ubc13. Lysates were subjected to immunoblotting with antibodies to IκBα, Flag, and β-actin. (G) A20 ZnF4 mutant does not interact with TAX1BP1. A20−/− MEFs were transiently transfected with plasmids as described in (C). After 36 hours, cells were stimulated with LPS for the indicated times, and proteins from lysates were immunoprecipitated with TAX1BP1 antibody and detected by immunoblotting with antibodies to A20 and TAX1BP1. Lysates were subjected to immunoblotting with antibodies to Flag, IκBα,and β-actin. The results shown are representative of three independent experiments.
The human T cell leukemia virus type I (HTLV-I) Tax oncoprotein promotes a persistent NF-κB response, in part, by interacting with TAX1BP1 and disrupting the interactions between A20, TAX1BP1, and Itch (5). Because Tax interacts with Ubc13 (18), we hypothesized that Tax would prevent A20 from binding to Ubc13, thereby protecting Ubc13 from degradation. Indeed, Tax impaired the TNF-α–dependent interaction of A20 with Ubc13 (fig. S9A), as well as TNF-α–mediated degradation of Ubc13 (fig. S9B). Tax also prevented A20 from binding to TRAF6 in response to IL-1 stimulation (fig. S9C). The Tax point mutant Tax M22, but not Tax M47, is defective for NF-κB activation (19). Tax M22, but not Tax M47, failed to bind to Ubc13 (fig. S9D). Thus, Tax preserves E2:E3 enzyme complexes essential for NF-κB activation and prevents Ubc13 degradation, which is essential for Tax polyubiquitination (18).
The importance of A20 in limiting inflammation is underscored by the numerous human auto-immune diseases associated with polymorphisms in the A20 genomic region (20-22). A20 also functions as a tumor suppressor gene for B cell lymphomas (23, 24). Our findings indicate that A20 disrupts key E2 and E3 ubiquitin enzyme complexes in both TNFR and TLR pathways. A20 binding to E2 or E3 enzymes may sterically interfere with their interactions, or alternatively A20 may directly modify E2 or E3 enzymes that antagonize their interactions.
Supplementary Material
Acknowledgments
We thank S. C. Sun, L. Matesic, D. Abbott, C. Vincenz, P. Storz, R. Beyaert, and the Belgian Coordinated Collections of Microorganisms/Laboratory of Molecular Biology of Ghent University (BCCM™/LMBP) for reagents and H. Ishikawa for assistance in the preparation of BMDMs and BMDCs. The project described was supported by NIH grants RO1GM083143 and RO1CA135362 awarded to E.W.H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute/National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/327/5969/1135/DC1
Materials and Methods
Figs. S1 to S9
References
References and Notes
- 1.Coornaert B, Carpentier I, Beyaert R. J. Biol. Chem. 2009;284:8217. doi: 10.1074/jbc.R800032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee EG, et al. Science. 2000;289:2350. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boone DL, et al. Nat. Immunol. 2004;5:1052. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 4.Shembade N, Harhaj NS, Liebl DJ, Harhaj EW. EMBO J. 2007;26:3910. doi: 10.1038/sj.emboj.7601823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shembade N, et al. Nat. Immunol. 2008;9:254. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- 6.Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. EMBO J. 2009;28:513. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wertz IE, et al. Nature. 2004;430:694. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 8.Lamothe B, et al. J. Biol. Chem. 2007;282:4102. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukushima T, et al. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6371. [Google Scholar]
- 10.Yamamoto M, et al. Nat. Immunol. 2006;7:962. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 11.Xia ZP, et al. Nature. 2009;461:114. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi CS, Kehrl JH. J. Biol. Chem. 2003;278:15429. doi: 10.1074/jbc.M211796200. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand MJ, et al. Mol. Cell. 2008;30:689. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Wooff J, Pastushok L, Hanna M, Fu Y, Xiao W. FEBS Lett. 2004;566:229. doi: 10.1016/j.febslet.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 15.Deng L, et al. Cell. 2000;103:351. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 16.Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Mol. Cell. Biol. 2008;28:3538. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klinkenberg M, Van Huffel S, Heyninck K, Beyaert R. FEBS Lett. 2001;498:93. doi: 10.1016/s0014-5793(01)02504-2. [DOI] [PubMed] [Google Scholar]
- 18.Shembade N, Harhaj NS, Yamamoto M, Akira S, Harhaj EW. J. Virol. 2007;81:13735. doi: 10.1128/JVI.01790-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith MR, Greene WC. Genes Dev. 1990;4:1875. doi: 10.1101/gad.4.11.1875. [DOI] [PubMed] [Google Scholar]
- 20.Dieguez-Gonzalez R, et al. Arthritis Res. Ther. 2009;11:R42. doi: 10.1186/ar2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musone SL, et al. Nat. Genet. 2008;40:1062. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fung EY, et al. Genes Immun. 2009;10:188. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 23.Compagno M, et al. Nature. 2009;459:717. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato M, et al. Nature. 2009;459:712. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.