Abstract
Conventional 3D culture is typically performed in multi-well plates (e.g. 12 wells). The volumes and dimensions necessitate relatively large numbers of cells and fluid exchange steps are not easily automated limiting throughput. 3D microchannel culture can overcome these challenges simplifying 3D culture processes. However, the adaptation of immunocytochemical endpoint measurements and the validation of microchannel 3D culture with conventional 3D culture are needed before widespread adoption can occur. Here we use a breast carcinoma growth model governed by complex and reciprocal interactions between epithelial carcinoma cells and mesenchymal fibroblasts to validate the 3D microculture system. Specifically, we report the use of a 3D microchannel co-culture assay platform to interrogate paracrine signalling pathways in breast cancer.
Using a previously validated 3D co-culture of human mammary fibroblasts and T47D breast carcinoma cells, we demonstrate the use of arrayed microchannels to analyze paracrine signalling pathways and screen for inhibitors. Results in both conventional format (multiwell plate) and microchannels were comparable. This technology represents a significant advancement for high-throughput screening in individual patients and for drug discovery by enabling the use of 3D co-culture models via smaller sample requirements and compatibility with existing HTS infrastructure (e.g. automated liquid handlers, scanners).
Keywords: Breast cancer, fibroblasts, paracrine signaling, 3D co-culture, high throughput screening, assay development
Introduction
It is becoming increasingly evident that a proper response to many signalling molecules requires the cells to be situated in a physiologically relevant topographic context1, 2 and more technical approaches are being used to explore the cell-matrix interactions3-5. Particularly in epithelial cell mitogenesis and morphogenesis experiments, 3D microenvironment assays, where cells are embedded in basement membrane-type material or collagen matrices have become the state-of-the-art. Unfortunately, these assays are typically performed in open well plates (e.g. 12 well) and are cumbersome and not amenable to high-throughput screening (HTS) adaptation.
The development of microfluidic cell culture devices may provide a technological solution to this problem. However, while a number of groups have demonstrated the potential of microfluidic-based 3D culture6-8, the complexity and requirement for tubes and specialized instrumentation has limited their use. Recently, arrayed, tubeless microfluidic devices (i.e. no physical tube connections are required as fluid exchange is accomplished via pipetting and surface tension driven pumping) have been shown capable of testing multiple experimental conditions in a reproducible fashion9, 10. The channels can be loaded with cells suspended in extracellular matrix (ECM) gels of choice and gel contraction facilitates rapid fluid exchanges11. Standard, automated fluid handling equipment can be used for treating and staining the cells12. The arrayed arrangement of the cell-containing channels permits the automated measurement of experimental endpoints13. However, it is important to validate this new culture platform against conventional methods prior to use to ensure no biases or artifacts are introduced via the differences (e.g. geometry, materials)14.
Increasingly, interactions between multiple cell types (particularly epithelial and stromal) are implicated in a wide variety of diseases. This is particularly true for cancer which for decades has been viewed largely as a disease caused by renegade cells that spread through the body in an autonomous fashion. Only relatively recently has it become clear that cancer should instead be viewed as an “organ system”, characterized by active reciprocal communications between the carcinoma cells and normal cells in the adjacent microenvironment, collectively referred to as stroma15. Fibroblasts appear to play a dominant role in carcinoma-stroma interactions16. Carcinoma cell-derived factors induce a number of changes in the stromal fibroblasts, leading to altered protein expression and morphology. Conversely, the activated fibroblasts of carcinoma stroma stimulate carcinoma cell proliferation and invasion via the secretion of extracellular matrix components and paracrine growth factors. It appears plausible that the therapeutic disruption of these paracrine pathways would retard breast carcinoma growth, which could be a promising treatment strategy particularly in difficult-to-treat cancer subtypes.
Some of the factors participating in breast carcinoma-stroma interactions have been identified. For example, transforming growth factor beta (TGFβ) and platelet-derived growth factor (PDGF) are known to activate fibroblasts17, 18. Our lab and other investigators have identified the matrix metalloprotease MT1-MMP and stromal cell-derived factor 1 (SDF1) as critical, fibroblast-derived factors that stimulate breast carcinoma cell proliferation19-21, however, it is likely that many other factors play a role. The identification of these factors has become a priority in breast cancer research.
The goal of the present study was to validate microfluidics-based 3D co-culture by applying the technology to a well-characterized biological model. Our group recently observed that human mammary fibroblasts stimulate T47D breast carcinoma cell growth in 3D collagen gels and that this effect depends on the activity of SDF1 and matrix metalloproteases (MMPs)20, 21. We show that the microfluidic and conventional 3D co-culture approaches provide equivalent results and that immunocytochemical endpoint measurements can be adapted to the innovative platform. Thus, microfluidic 3D co-culture is a feasible, innovative technology to interrogate paracrine interactions in cancer. In a HTS setting, it may be used to identify potential drug candidates.
Materials and Methods
Cells and Culture Devices
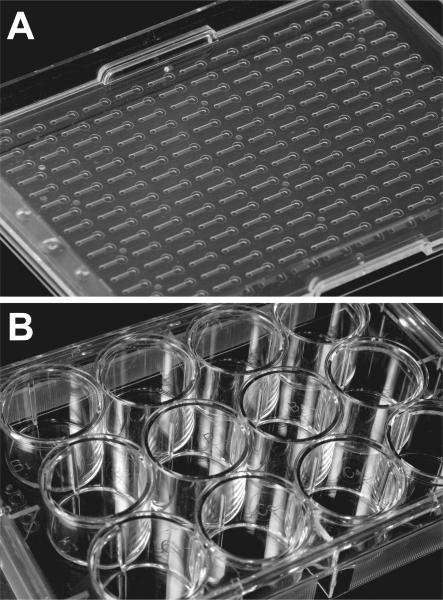
Polydimethylsiloxane (PDMS) microchannel plates were fabricated as described previously22. Each plate contained 48 microchannels (channel dimensions, 1.0 × 9.0 × 0.25 mm, W × L × H). The inlet/outlet ports are typically 1 mm/ 2 mm in diameter respectively and the thickness of the top PDMS layer is 200-500 um. In addition, arrays of 192 polystyrene microchannels (Bellbrook Labs, Madison, WI) were used for confirmatory experiments as indicated. Conventional cultures were grown in 12-well plates (Corning, Costar 3512). Figure 1 shows the polystyrene channels (Fig. 1A) used along side conventional 12-well plates (Fig. 1B). The human breast carcinoma cell line T47D was obtained from Dr. M. Gould (University of Wisconsin, Madison). The normal human mammary fibroblasts (RMF/EG) immortalized with human telomerase and labeled with GFP were generously provided by Dr. C. Kuperwasser23. In the following RMF/EG fibroblast will be referred to as human mammary fibroblasts (HMF). Both cell types were maintained in DMEM (Mediatech, 10-013-CV) supplemented with 10% fetal bovine serum (Gemini Bio-Products, 900-108), 2 mM L-glutamine (Fisher Scientific, BP379-100), and penicillin/streptomycin (Mediatech, 30-002-CI) in 37°C incubators in a humidified atmosphere containing 5% CO2.
Fig. 1. Comparison of microfluidic and conventional culture devices.
Microfluidic co-culture device with 96 arrayed single channels (photo courtesy of Steve Hayes, Bellbrook Labs) (A). Conventional 12-well culture vessel (B).
Conventional 3D Co-Culture
Conventional three-dimensional collagen gel co-cultures were prepared using a method described previously20, 21. In brief, T47D cells and HMF were mixed at a ratio of 2:1 in collagen type I at a final collagen concentration of 1.3 mg/ml. MMP activity was inhibited by adding the broad-spectrum inhibitor GM6001 (500 nM; Chemicon); paracrine signalling of SDF1 via its receptor CXCR4 was blocked by adding the inhibitor AMD3100 (5 ng/ml; Chemicon) to the collagen I gels as well as to the culture medium. The final total volume (gel plus media overlay) was 2 ml/well.
Microchannel 3D Co-Culture
Microchannel 3D collagen gel co-cultures were prepared consistent with the conventional cultures. 2 μl of the cell suspension in collagen (600 T47 cells per μl; 300 HMF per μl) were added to each microchannel via passive pumping10, and a minimum of eight channels were used for each experimental condition. Both 3D culture types were maintained at 37°C in a humidified atmosphere containing 5% CO2 for 5 days. The culture medium was changed every other day, and for the inhibition experiments, GM6001 and AMD3100 were added fresh to the culture medium each time at the concentrations described above.
Cell Analysis
For the quantification of cell growth and proliferation, collagen gels in conventional 3D co-cultures were fixed and stained as described previously20, 21. The primary antibodies were used at a 1:100 dilution (mouse monoclonal anti-human pancytokeratin, LabVision, MS-356-P0; rabbit polyclonal anti-human vimentin, Lab Vision, RB-9063-P0) and 1:200 dilution (rabbit monoclonal anti-human Ki67, Lab Vision, RM-9106-S0). The secondary antibodies (Alexa Fluor 594 goat anti-mouse, Molecular Probes, A 11032; Alexa Fluor 488 goat anti-rabbit, Molecular Probes, A11008) were added at a 1:100 dilution. For counterstaining of nuclei, Hoechst 33342 was used at 2.5 μg/ml (Invitrogen, H3570). Collagen gels in the PDMS microchannel devices were fixed and stained in situ by pipetting 20 μl of each solution onto the outport and 4 μl onto the input ports of each microchannel. Gels were fixed in 2% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature, and then permeabilized with 0.5% TritonX-100 for 15 min at 4°C. After 3 washes with 100 mM glycine in PBS, gels were blocked with 10% goat serum in immunofluorescence buffer (PBS with 0.1% bovine serum albumin, 0.2% Triton X-100, and 0.05% Tween-20) for 1 hour at room temperature. 3D-cultures were incubated with primary antibodies in block buffer at 4°C overnight. After 3 washes with PBS, gels were incubated with the secondary antibodies for 2 hours at room temperature. Primary and secondary antibodies were used at dilutions as in the conventional cultures. For the negative control, the primary antibodies were omitted. Gels in the microchannels were washed twice in PBS for 5 min, and nuclei were stained with Hoechst 33342 for 15 min at room temperature. After washing with PBS, all visible liquid droplets were aspirated and microchannel plates were dried overnight until all liquid in the channels had evaporated. Mounting media consisting of 90% glycerol in 100 mM Tris was forced into the channels and plates were stored protected from light until immunostaining was analyzed.
Fluorescence images were acquired with an Olympus inverted microscope equipped with a SPOT imaging system (Diagnostic Instruments, Inc.). For quantification of cell growth, the total area of tumor cell clusters was measured using customized macros generated in NIH ImageJ version 1.37 (http://rsbweb.nih.gov/ij/). To confirm the validity of the area measurements, 30 carcinoma cell clusters in both conventional and microfluidic cultures were randomly selected and nuclei were counted manually and correlated with area size. To assess the proliferative activity of carcinoma cells, the Ki67 proliferation index was determined by counting 500 nuclei manually for each microfluidic and conventional culture condition.
Results
In our previous work we developed a 3D co-culture system that combines human breast carcinoma cells with human mammary fibroblasts in a collagen matrix20, 21. In this organotypic model, we observed a significant growth stimulation of T47D carcinoma cells caused by the presence of fibroblasts compared to T47D monoculture. Furthermore, we demonstrated that the growth stimulation was reversed by blocking the paracrine SDF1-CXCR4 signalling pathway20 or by inhibiting matrix metalloproteases (MMPs)21. Importantly, the inhibition of either pathway did not affect T47D cell growth in monoculture. The aim of this study was to validate a recently developed microfluidic-based cell culture platform10, 12 by transferring this established biological model system from conventional 3D co-culture.
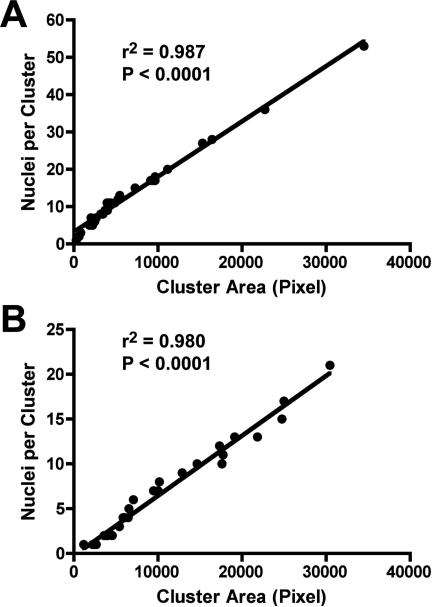
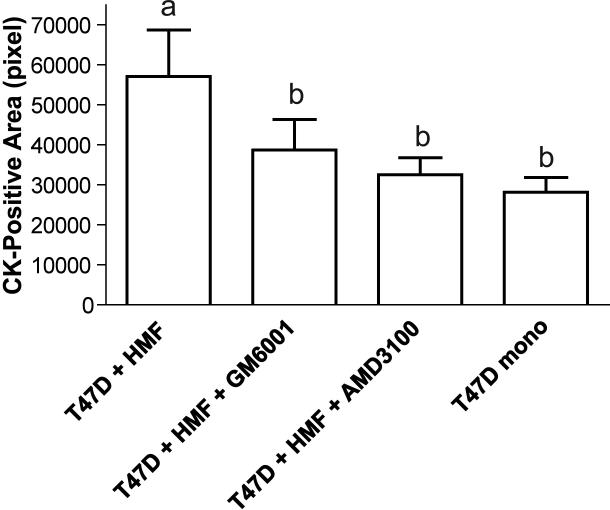
An initial quantitative assessment of cell growth was performed by measuring the total area occupied by carcinoma cell clusters. Tumor cells were identified and distinguished from fibroblasts by immunocytochemical labelling for pancytokeratin (CK). The immunofluorescently stained gels were then analyzed by image analysis. Carcinoma cell clusters composed of one cell to more than a hundred cells were included in the analysis. To determine whether the CK-positive area is an acceptable surrogate measure of cell number, carcinoma nuclei were counted manually in randomly selected clusters and the number of nuclei was correlated with the absolute CK-positive area. Linear regression analysis demonstrated a significant correlation between both parameters, thus validating the use of cytokeratin immunolabeling combined with image analysis (Fig. 2).
Fig. 2. Relationship between carcinoma cell cluster size and cell number.
T47D breast carcinoma cells were grown in 3D collagen gels in monoculture and in co-culture with HMF for 5 days. T47D cells were immunofluorescently labelled with antibodies to pancytokeratin and the nuclear dye Hoechst 33342. Thirty tumor cell clusters were randomly selected and nuclei were counted manually. The cell cluster area was determined using Image J software. Both in conventional co-culture (A) and in microfluidics co-culture (B), cluster size correlated significantly with the nucleus count and thus cell number.
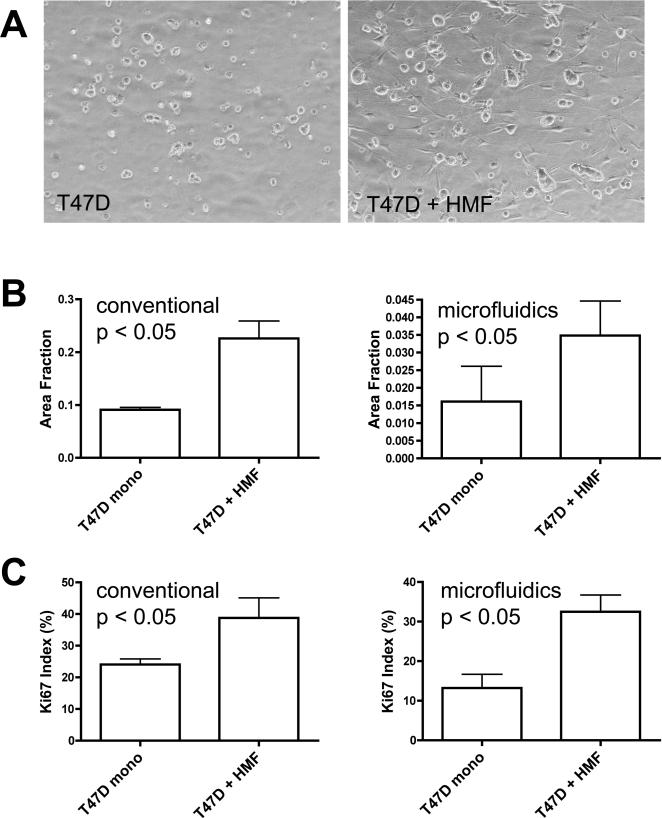
The co-culture experiments in both conventional 12-well plates and PDMS microchannel devices were set up simultaneously using carcinoma and stroma cells with identical passage numbers. After 5 days of culture, T47D cells formed irregular clusters in both culture systems and the carcinoma cell clusters were larger in co-cultures than in mono-cultures (Fig. 3 A). The quantitative assessment of cell growth revealed a 1.5 to 2.5-fold induction of carcinoma cell growth in the co-cultures (Fig. 3 B). This growth stimulation caused by the presence of fibroblasts was statistically significant in both culture systems. The increase in cell number could result from the stimulation of mitogenesis or a decrease in cell death. To distinguish between these possibilities, we determined carcinoma cell labelling for the proliferation marker Ki67. The Ki67 proliferative index was significantly increased in the presence of fibroblasts both in conventional and microfluidic co-cultures compared to the respective T47D mono-culture controls (Fig. 3 C).
Fig. 3. HMF stimulate T47D breast carcinoma cell growth.
Phase contrast images of 3D collagen gel cultures after 5 culture days in PDMS microfluidics channels (A). Original magnification 100x. T47D cell clusters in co-culture with HMF (right panel) are bigger than in monoculture (left panel). Quantification of total carcinoma cell cluster area as surrogate of cell number (B) and Ki67-proliferation index (C) in conventional and microfluidic 3D collagen-cultures as indicated (Error bars indicate standard deviation).
Prior work from our lab defined a reciprocal growth-promoting signalling pathway between normal mammary fibroblasts and breast carcinoma cells. Using conventional 3D-co-cultures we demonstrated that blocking of SDF1 signalling by adding a neutralizing anti-SDF1 antibody or a small molecule inhibitor of the SDF1 receptor CXCR4 abolished the growth advantage caused by the presence of fibroblasts20. Similarly, MMP enzyme activity is required for the paracrine growth stimulation of T47D breast carcinoma cells, as the MMP family member MT1-MMP is responsible for the proteolytic release of the paracrine mediator syndecan-1 from the fibroblast surface21, 24.
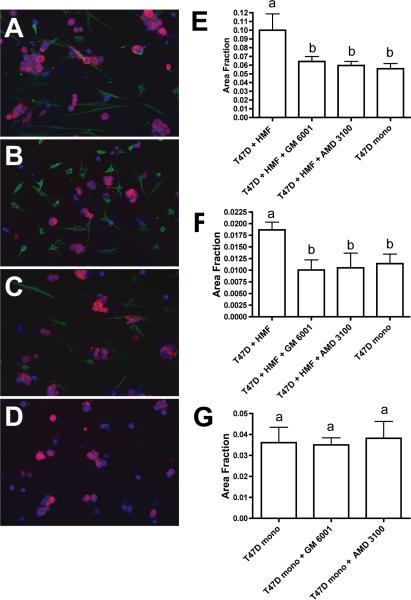
In our present study we inhibited paracrine SDF1 signalling by adding the small molecule CXCR4 receptor inhibitor AMD3100 to collagen gels and culture media. After a co-culture period of five days, a statistically significant suppression of the growth promoting effect of HMFs on T47D cells was observed (Fig. 4 A-F)). AMD3100 reduced T47D carcinoma cell growth in co-cultures to the level of T47D mono-cultures in both conventional and microfluidic systems. Similarly, the broad-spectrum MMP inhibitor GM6001 abolished HMF-induced T47D growth. To ascertain that the growth-blocking effect of the pharmacological inhibitors was the result of disrupted paracrine signalling pathways rather than due to a direct effect on the carcinoma cells, we tested AMD3100 and GM6001 in T47D mono-cultures. After 5 days of culture, neither compound suppressed the growth of T47D cells in monoculture, demonstrating that the inhibitors disrupted fibroblast-derived signals rather than autonomous T47D cell growth (Fig. 4 G).
Fig. 4. Growth induction of T47D breast carcinoma cells by HMF is reversed by blocking SDF-1 signalling or MMP activity.
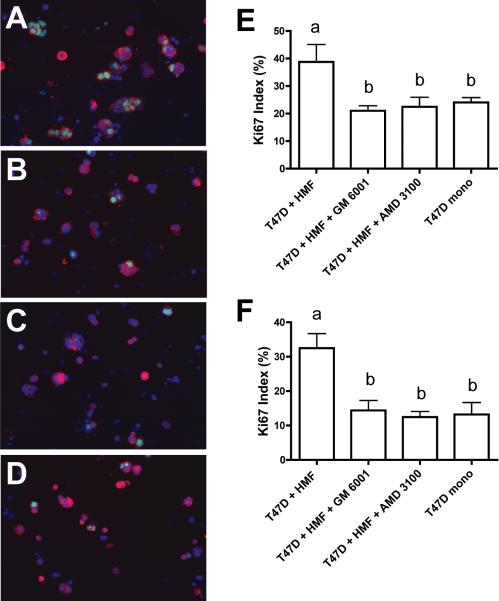
A-D. Immunofluorescence labelling of 3D collagen cultures in PDMS microchannel devices. T47D cells were labelled with an antibody to pancytokeratin (red) and HMF were labelled with an antibody to vimentin (green). Nuclei were counterstained with Hoechst 33342 (blue). A. T47D cell clusters in co-cultures with HMF. Inhibition of MMP activity by GM6001 (B) or blocking SDF-1 signalling by addition of AMD3100 (C) reduces growth of T47D cell clusters to levels seen in T47D monoculture (D). Quantification of T47D cell growth in conventional (E) and microfluidics (F) 3D collagen cultures. G. The addition of GM6001 or AMD3100 does not influence T47D cell growth in monoculture. Error bars indicate standard deviation; letters above the columns indicate the results of statistical comparisons by ANOVA. Columns sharing the same letter are not significantly different; columns labelled with different letters are significantly different (at least P< 0.05).
The proliferative activity of the carcinoma cells was quantified by measuring Ki67 labelling. In both culture systems a statistically significant reduction of carcinoma cell proliferation in co-cultures was observed during suppression of the SDF1 signalling pathway or MMP activity. T47D cell Ki67 labelling in co-culture was reduced by approximately 50% and indistinguishable from the proliferation seen in monoculture. (Fig. 5). This result confirmed that AMD3100 and GM6001 abolished fibroblast-induced carcinoma cell mitogenesis.
Fig. 5. Proliferation of T47D breast carcinoma cells in co-cultures is suppressed by inhibition of SDF-1 signalling or MMP activity.
A-D. Immunofluorescence labelling of 3D collagen cultures in PDMS microchannel devices after 5 culture days. Proliferative activity was assessed by labelling with an antibody to Ki67 (green). Nuclei were counterstained with Hoechst 33342 (blue). T47D cells, which were identified with an antibody to pancytokeratin (red), show increased proliferation in co-culture with HMF (A) compared to T47D cells in monoculture (D). Proliferative activity in co-cultures is suppressed by inhibition of MMP activity with GM6001 (B) or inhibition of SDF-1 signalling by the addition of AMD3100 (C). Quantitative analysis of Ki67 labelling in conventional (E) and microfluidics (F) cultures. Error bars indicate standard deviation; letters above the columns indicate the results of statistical comparisons by ANOVA. Columns sharing the same letter are not significantly different; columns labelled with different letters are significantly different (at least P< 0.05).
PDMS devices afford the advantage of flexible microchannel design and simple manufacturing in the user lab. On the other hand, PDMS is a suboptimal material for cell culture vessels because its inherent high adsorption may lead to the sequestration of secreted factors25-27. Therefore, we repeated the co-culture and inhibition experiments with simple, single-channel polystyrene devices. The results were identical to those obtained with the PDMS devices, demonstrating the feasibility of deciphering paracrine signalling pathways with inhibitor screens of co-cultures in polystyrene microchannel devices (Fig. 6).
Fig. 6. Single-channel polystyrene devices can be used for co-culture experiments and inhibitor screens; results are similar to those obtained with dual-channel PDMS devices.
T47D cells and HMF were co-cultured in single-channel polystyrene devices under conditions described for figure 3. Error bars indicate standard deviation; letters above the columns indicate the results of statistical comparisons by ANOVA. Columns sharing the same letter are not significantly different; columns labelled with different letters are significantly different (at least P< 0.001).
Discussion
This proof-of-principle study demonstrates the feasibility of using microfluidics devices for in-gel 3D co-culture. The inhibition experiments show that the device can be applied to interrogate complex, heterotypic cellular interactions, as they occur between breast carcinoma cells and stromal fibroblasts. This is important because the microfluidics co-culture platform affords a number of advantages over conventional culture devices: 1) The culture volume is significantly smaller than in conventional vessels (2 μl vs. 50-100 μl), which reduces reagent consumption and thus cost. 2) Microfluidics co-cultures can be set up using much smaller cell numbers (102 to 103) making it feasible to use primary cells from patient samples and test multiple experimental conditions. 3) The effect of convection is greatly reduced in the microchannels and thus, the distribution of soluble molecules is governed almost exclusively by diffusion28. Therefore, microchannel co-culture is the platform of choice to model paracrine interactions with high fidelity29. 4) Importantly, the microchannel devices used in this study can be easily adapted to HTS12. The arrangement of the channels in an array format and fluid handling via passive pumping make it possible to set up the assays in regular screening labs and use standard fluid handling equipment (multichannel pipettors or pipette robots) for media changes, reagent delivery and staining.
Recently, the potential influences of materials on cell behavior in microchannel culture has begun to be explored25-27. Specifically, issues of adsorption and monomer leaching from PDMS devices can influence or bias experimental outcomes. The results here suggest that for this specific assay there are no significant biases or artifacts from PDMS. Importantly, the method described here is not in any way dependent on PDMS microchannels. Thus, should one want to apply the method to other biological model systems where bias from PDMS is likely (e.g. estrogen responsiveness), the microchannels can be fabricated out of other materials such as polystyrene. In fact, we have demonstrated similar results in polystyrene based microchannel arrays (see Fig. 5). The observation of similar results in both PDMS and polystyrene suggests that for this particular assay the role of matrix in regulating cell behavior is dominant as compared to any material biases.
A few groups have also started to explore any inherent differences between microchannel culture and macro culture13, 30 as well as issues of evaporation-mediated osmolarity shifts31. In some studies differences were observed. Our results here suggest that the potential sources of those differences (e.g. transport via diffusion vs. convection, materials differences) maybe mitigated in 3D culture. For example, in 3D culture the presence of the matrix limits convective flow in both micro and macro formats. Further, the cell-matrix and molecule-matrix interactions are also similar in both formats likely limiting the effects of material differences. While the focus of this work was on soluble factor signalling, it will be important for future studies to explore the potential influence of the microchannel structure (e.g. boundary conditions) on the matrix mechanical properties.
The application of small volume microfluidic 3D co-culture in an HTS format opens the door to a number of exciting applications. Drug discovery screens can be performed to identify inhibitors of paracrine, growth stimulatory signalling pathways. This approach may reveal compounds that would be dismissed as failures in 2D monoculture carcinoma growth inhibition assays. If cell lines (as used in this study) are replaced with primary cells (carcinoma cells and/or stromal fibroblasts) from actual clinical patient samples, information about patient-to-patient variability of paracrine signalling pathways and the susceptibility or resistance to individual drugs could be obtained. Hereby the concept of personalized medicine could be taken to the level of heterotypic intercellular interactions. Recent reports which demonstrate a modulatory effect of stromal cells or stromal cell-derived matrix on the response of tumor cells to chemotherapeutic agents underscore the importance of the tumor microenvironment in drug discovery and drug sensitivity testing32-34.
In summary, we show for the first time the feasibility of using an arrayed microfluidics assay platform for the interrogation of paracrine interactions in breast carcinomas and demonstrate results equivalent to state-of-the-art conventional 3D co-culture.
Insight Box.
We present a significant advancement in 3D culture (and co-culture) capabilities by reducing the number of cells needed per endpoint and by moving 3D culture into a platform that is easily automated. With the increasing realization that proper response to many signalling molecules requires the cells to be situated in a physiologically relevant topographic context, the ability to perform 3D cultures in higher throughput and with smaller numbers of cells is a critical enabling step. This technology has great potential for high-throughput screening in individual patients and for drug discovery by enabling the use of 3D co-culture models via smaller sample requirements and compatibility with existing HTS infrastructure (e.g. automated liquid handlers, scanners). The technology integrates micro channel 3D culture with tube-less microfluidics which is then biological validated using a breast carcinoma growth model governed by complex and reciprocal interactions between epithelial carcinoma cells and mesenchymal fibroblasts.
Acknowledgements
The work was funded in part by the Wisconsin Partnership program and the National Institutes of Health (2R01 CA107012). Maret Bauer was supported by a scholarship from the Dr. Mildred Scheel Stiftung. D. J. Beebe has an ownership interest in Bellbrook Labs LLC, which has licensed technology reported in this publication.
References
- 1.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. Nature cell biology. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. The Journal of cell biology. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaBarge MA, Nelson CM, Villadsen R, Fridriksdottir A, Ruth JR, Stampfer MR, Petersen OW, Bissell MJ. Integr Biol (Camb) 2009;1:70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jongpaiboonkit L, King WJ, Lyons GE, Paguirigan AL, Warrick JW, Beebe DJ, Murphy WL. Biomaterials. 2008;29:3346–3356. doi: 10.1016/j.biomaterials.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khetani SR, Bhatia SN. Curr Opin Biotechnol. 2006;17:524–531. doi: 10.1016/j.copbio.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Lab Chip. 2009;9:269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 7.Vickerman V, Blundo J, Chung S, Kamm R. Lab Chip. 2008;8:1468–1477. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong AP, Perez-Castillejos R, Christopher Love J, Whitesides GM. Biomaterials. 2008;29:1853–1861. doi: 10.1016/j.biomaterials.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puccinelli JP, Su X, Beebe DJ. Lab Automation. 2009;15:25–32. [Google Scholar]
- 10.Walker GM, Beebe DJ. Lab Chip. 2002;2:131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 11.Sung KE, Su G, Pehlke C, Trier SM, Eliceiri KW, Keely PJ, Friedl A, Beebe DJ. Biomaterials. 2009 doi: 10.1016/j.biomaterials.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyvantsson I, Warrick JW, Hayes S, Skoien A, Beebe DJ. Lab Chip. 2008;8:717–724. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 13.Yu H, Alexander CM, Beebe DJ. Lab Chip. 2007;7:388–391. doi: 10.1039/b612358a. [DOI] [PubMed] [Google Scholar]
- 14.Paguirigan AL, Beebe DJ. Bioessays. 2008;30:811–821. doi: 10.1002/bies.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bissell MJ, Radisky D. Nature reviews. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalluri R, Zeisberg M. Nature reviews. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 17.Ronnov-Jessen L, Petersen OW. Laboratory investigation; a journal of technical methods and pathology. 1993;68:696–707. [PubMed] [Google Scholar]
- 18.Shao ZM, Nguyen M, Barsky SH. Oncogene. 2000;19:4337–4345. doi: 10.1038/sj.onc.1203785. [DOI] [PubMed] [Google Scholar]
- 19.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 20.Su G, Blaine SA, Qiao D, Friedl A. The Journal of biological chemistry. 2007;282:14906–14915. doi: 10.1074/jbc.M611739200. [DOI] [PubMed] [Google Scholar]
- 21.Su G, Blaine SA, Qiao D, Friedl A. Cancer Res. 2008;68:9558–9565. doi: 10.1158/0008-5472.CAN-08-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jo BH, Van Lerberghe LM, Motsegood KM, Beebe DJ. Journal of Microelectromechanical Systems. 2000;9:76–81. [Google Scholar]
- 23.Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endo K, Takino T, Miyamori H, Kinsen H, Yoshizaki T, Furukawa M, Sato H. The Journal of biological chemistry. 2003;278:40764–40770. doi: 10.1074/jbc.M306736200. [DOI] [PubMed] [Google Scholar]
- 25.Paguirigan A, Beebe DJ. Integr. Biol. 2009;1:182–195. doi: 10.1039/b814565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ. Lab Chip. 2009;9:2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toepke MW, Beebe DJ. Lab Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 28.Atencia J, Beebe DJ. Nature. 2005;437:648–655. doi: 10.1038/nature04163. [DOI] [PubMed] [Google Scholar]
- 29.Domenech M, Yu H, Warrick J, Badders NM, Meyvantsson I, Alexander CM, Beebe DJ. Integr Biol (Camb) 2009;1:267–274. doi: 10.1039/b823059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stangegaard M, Petronis S, Jorgensen AM, Christensen CB, Dufva M. Lab Chip. 2006;6:1045–1051. doi: 10.1039/b603379b. [DOI] [PubMed] [Google Scholar]
- 31.Heo YS, Cabrera LM, Song JW, Futai N, Tung YC, Smith GD, Takayama S. Anal Chem. 2007;79:1126–1134. doi: 10.1021/ac061990v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serebriiskii I, Castello-Cros R, Lamb A, Golemis EA, Cukierman E. Matrix Biol. 2008;27:573–585. doi: 10.1016/j.matbio.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent T, Mechti N. Leuk Lymphoma. 2005;46:803–811. doi: 10.1080/10428190500051448. [DOI] [PubMed] [Google Scholar]
- 34.Grigorieva I, Thomas X, Epstein J. Exp Hematol. 1998;26:597–603. [PubMed] [Google Scholar]