Abstract
Aldehyde dehydrogenases (ALDHs) represent a superfamily of NAD(P)+-dependent enzymes which catalyze the oxidation of a wide variety of endogenous and exogenous aldehydes to their corresponding acids. Some ALDHs have been identified as corneal crystallins and thereby contribute to the protective and refractive properties of the cornea. ALDH3A1 is highly expressed in the cornea of most mammals with the exception of rabbit, who abundantly expresses ALDH1A1 in the cornea instead of ALDH3A1. In this study, we examined the gene expression of other ALDHs and found high messenger levels of ALDH1B1, ALDH2 and ALDH7A1 in mouse cornea and lens. Substantial evidence supports a protective role for ALDH3A1 and ALDH1A1 against ultraviolet radiation (UVR)-induced oxidative damage to ocular tissues. The mechanism by which this protection occurs includes UVR filtering, detoxification of reactive aldehydes generated by UVR exposure and antioxidant activity. We recently have identified ALDH3A1 as a nuclear protein in corneal epithelium. Herein, we show that ALDH3A1 is also found in the nucleus of rabbit keratocytes. The nuclear presence of ALDH3A1 may be involved in cell cycle regulation.
INTRODUCTION
The human eye is exposed daily to solar ultraviolet radiation (UVR). UVR is subdivided into three wavebands: UVA (315–400 nm), UVB (280–315 nm) and UVC (100–280 nm). The cornea absorbs all UVC and most UVB, whereas UVA is primarily absorbed by the lens. No UVC or UVB (and very little UVA (<1%)) reach the retina [1]. According to “Draper’s law”, only the fraction of energy that is absorbed by a tissue can change or damage the tissue. High level UVR exposure may lead to thermal damage in the eye; chronic low level UVR exposure, particularly that associated with the shorter and higher energy wavelengths of UVC and UVB, damages the ocular tissues by photochemical reactions that induce oxidative stress through the generation of reactive oxygen species (ROS) [1]. ROS have the capacity to attack important macromolecules including proteins, DNA and lipids and thereby cause protein modification, DNA damage and lipid peroxidation, the sequelae of which is cellular damage or death [2]. It is known that UVR-induced formation of ROS is involved in various types of eye pathologies including cataract formation, corneal and retinal degeneration [3].
The cornea, the outermost layer of the eye, serves two fundamental functions: (i) it is the initial barrier that protects the inner ocular tissues against environmental insults and (ii) it forms the “refraction” unit (together with lens) to permit light entry and focusing on the retina [4]. The cornea consists of a stratified squamous epithelium, a thick stroma containing collagen fibers, proteoglycans, glycosaminoglycans and keratocytes, and a posterior single layer of endothelium. Due to its ability to absorb UVC and UVB, the cornea plays a pivotal role in protecting internal ocular tissues (lens and retina) from UVR-induced damage and in so doing makes itself more vulnerable to damage. Seemingly to withstand this challenge, the cornea is equipped with robust antioxidant systems. Enzymatic antioxidants present in the cornea include catalase (CAT), glucose-6-phosphate dehydrogenase (G6PDH), glutathione peroxidase (GPX), glutathione reductase (GR) and superoxide dismutase (SOD). Non-enzymatic antioxidants in the cornea include ascorbate, glutathione (GSH), α-tocopherol and NAD(P)H [5].
Several studies have shown that a few water-soluble enzymes are abundantly expressed in the cornea in a taxon-specific manner. These enzymes are given the name “corneal crystallins” to emphasize their similarity to the lens crystallins [6] in that: (i) they are in most cases diverse, cytoplasmic proteins with metabolic functions, (ii) they display taxon-specificity, and (iii) they accumulate to high levels in transparent tissues, while they are present at lower levels in other tissues. These crystallins are believed to contribute to the transparent and refractory properties of the cornea and lens [7]. In addition to such physicochemical roles, corneal and lens crystallins may, through their metabolic properties, serve to protect against UVR-induced damage through detoxification, chaperone activity and generation of the antioxidant NAD(P)H [5]. Such “enzyme” crystallins include the aldehyde dehydrogenase isozymes (ALDHs).
ALDHs represent a superfamily of NAD(P)+-dependent enzymes that catalyze the oxidation of a wide variety of endogenous and exogenous aldehydes to their corresponding acids. Some aldehydes play vital physiological roles, including vision, embryonic development and neurotransmission. Others, such as the major products of lipid peroxidation viz. 4-hydroxy-2-nonenal (4-HNE) and malondialdehyde (MDA), are cytotoxic and carcinogenic [5]. 4-HNE and MDA possess terminal carbonyl groups, making them strong electrophiles and highly-reactive. These aldehydes are relatively long-lived, allowing them to react with cellular components more distant from their site of generation [8]. The ALDH superfamily (and other enzymes involved in aldehyde metabolism) plays critical roles in protecting cells from these toxic metabolites by regulating their levels. The pathophysiological significance of this protective role is exemplified by human diseases associated with mutations and polymorphisms of ALDH genes [9].
Some members in the ALDH superfamily have been identified as crystallins in the cornea and lens of both vertebrates and invertebrates (Table 1). In vertebrates, it was first reported that bovine corneal protein 54 (BCP54) comprised 20–40% of total soluble protein of the bovine cornea; later studies identified BCP54 as ALDH3A1 [10]. It is now well established that ALDH3A1 is a major soluble protein in the cornea of most mammalian species including human; by contrast, rabbits express ALDH1A1 in the cornea [11;12]. Other vertebrates, including chicken, frog, and fish, also express ALDH1A1 and, in some cases, ALDH2 rather than ALDH3A1 in the cornea [13]. Another member of ALDH class 1 proteins, ALDH1A8 (η-crystallin), is a lens crystallin in the elephant shrew [14]. In invertebrates, ALDH1A9 (Ώ –crystallin) and ALDH1C1/2 (Ώ –crystallins) are lens crystallins in scallops and cephalopods, respectively. Interestingly, ALDH enzyme activities is extremely low in lens extracts from these invertebrate species [15]. Although expressed in a taxon-specific fashion, members of ALDH class 1 and ALDH3A1 are well conserved in mammals, showing ~90% homology in amino acid sequence among human, rabbit, cow, sheep, mouse and rat [16]. The essential residues for NAD(P)+-binding (Lys-192, Gly-245, Gly-250, Glu-399, and Phe-401; numbering based on human ALDH1A1) and for catalytic activity (Cys-302 and Glu-268) are strictly conserved in these species [16].
Table 1.
ALDH isozymes identified as corneal and lens crystallins *
| ALDHs | Species | |
|---|---|---|
| Corneal cystallins | ALDH3A1/BCP54 | Most mammals |
| ALDH1A1 | Rabbit, human, pig, chicken, fish | |
| ALDH2 | Rabbit, fish | |
| Lens crystallins | ALDH1A1 | Mammals |
| ALDH1A8/η-crystallin | Elephant shrew | |
| ALDH1A9/Ώ-crystallin | scallops | |
| ALDH1C1/2/Ώ-crystallin | cephalopods | |
See text for references.
In this paper, we review the current knowledge on corneal ALDH3A1 and ALDH1A1, report the mRNA expression profile of nine ALDH isozymes in mouse cornea and lens and present experimental data showing the novel nuclear localization of ALDH3A1 in rabbit corneal keratocytes.
MATERIALS AND METHODS
Animals
New Zealand White rabbits (2–4 Kg) were purchased from Western Oregon Rabbit Company (Philomath, Oregon) and male C57BL/6J mice of approximately 8-wk old were purchased from Jackson Laboratory (Bar Harbor, Maine). All procedures involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Colorado Denver and the University of California, Irvine.
ALDH3A1 stably transfected cell lines
The development and characterization of the rabbit corneal keratocytes (TRK43) stably transfected with human ALDH3A1 has been previously described [17]. Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 100 U/ml penicillin– 100 μg/ml streptomycin solution and maintained at 37°C in a humidified 5% CO2 incubator.
RNA isolation, reverse transcription and quantitative real-time PCR (Q-PCR)
Mice were euthanized by CO2 inhalation followed by cervical dislocation. Corneas and lenses were removed directly from enucleated eyes and pooled from 3–5 animals. Total RNA was isolated from respective ocular tissues using RNeasy Mini kit (Qiagen, Valencia, CA) according to manufacturer’s protocol. cDNA was synthesized using Superscript III RT kit (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions, using 5μg total RNA in a 20μl reaction volume. Q-PCR reactions were carried out using 30ng cDNA by the Taqman gene expression assay (ABI, Foster City, CA) according to manufacturer’s protocol. Gene-specific Q-PCR primers were purchased from ABI (Foster City, CA) and are summarized in Table 2. Relative mRNA levels were reported as fold of control (=1), which showed the least amount after normalization to β-actin (ACTB).
Table 2.
PCR primers used in Q-PCR analysis
| mRNA | Primer ID | Product size (bp) | Gene number |
|---|---|---|---|
| ALDH1A1 | Mm00657317_m1 | 116 | NM_013467.3 |
| ALDH1A2 | Mm00501306_m1 | 68 | NM_009022.3 |
| ALDH1A3 | Mm00474049_m1 | 86 | NM_053080.3 |
| ALDH1B1 | Mm00728303_s1 | 68 | NM_028270.4 |
| ALDH2 | Mm00477463_m1 | 59 | NM_009656.3 |
| ALDH3A1 | Mm00839312_m1 | 87 | NM_001112725.1; NM_007436.2 |
| ALDH3A2 | Mm00839320_m1 | 71 | NM_007437.4 |
| ALDH4A1 | Mm00615268_m1 | 105 | NM_175438.3 |
| ALDH7A1 | Mm00519645_m1 | 85 | NM_138600.4; NM_001127338.1 |
Generation of Chicken anti- rabbit ALDH1A1 antibody
Immunization of chickens using recombinant rabbit ALDH1A1 [16] and collection of eggs were performed by Calbiochem (San Diego, CA, USA). Egg yolks were carefully separated from the white by washing with deionised water and collected without the yolk skin in a graduated cylinder. Two volumes of 100mM phosphate buffer (pH 7.2) containing 0.02% (w/v) sodium azide (Sigma Aldrich) were added and mixed thoroughly. 3.5% (w/v) polyethylene glycol (PEG 6000, Sigma Aldrich) was introduced and stirred until the PEG was completely dissolved. After incubation at 4°C overnight, the sample was centrifuged at 4500g for 20min at room temperature. The supernatant was filtered through n.4 Whatman paper and solid PEG was added to a final concentration of 12% (w/v). Following incubation overnight at 4°C, the mixture was centrifuged at 12,000g for 20min. The pellet containing IgY was resuspended in KPO4 (1/6 of the initial volume of yolk) and dialyzed against the same buffer at 4°C with 10K molecular-weight Dialysis Cassettes (Pierce). The purity and specificity of anti-rabbit ALDH1A1 IgY was tested using whole cell lysates and recombinant ALDH1A1 by Western immunoblotting as previously described [18]. The pre-immune IgY which was prepared in the same manner as for anti-ALDH1A1 IgY was used as the serum control. The purified anti-rabbit IgY recognized a single band running approximately at a MW 55kDa (data not shown).
Immunohistochemistry (IHC)
IHC staining was carried out on 5μm thick paraffin embedded rabbit eye sections mounted on microscope slides (Fisher). Sections deparaffinized in xylol and rehydrated in graded ethanol solutions were pretreated in 10mM sodium citrate in a microwave at 95°C for 30min. Slices were then treated with 3% hydrogen peroxide (Sigma Aldrich) to block peroxidase activity and, after washing in PBS- Brij 0.2% (Sigma Aldrich), incubated in blocking buffer (Zymed) at room temperature in a humidified chamber for 1h (to minimize non-specific binding). Samples were incubated with anti-ALDH1A1 antibody at a dilution of 1:50 for 1.5h. Antibody binding was detected using horseradish peroxidase-conjugated anti-chicken antibody (1:100 dilution) for 45min. Sections were rinsed three times in PBS- Brij and incubated with avidin–biotin complex reagents (Dako-cytomation) for 1h at room temperature. Sections were then rinsed three times in PBS- Brij and diaminobenzidine (DAB) was used as a chromophore. Sections were then counterstained with Mayer’s haematoxylin, dehydrated with ethanol/xylol solutions and mounted with Permount (Fisher). Control samples performed on adjacent sections were incubated with PBS instead of anti-ALDH1A1 antibody. Images were obtained using a Nikon upright microscope linked to a Nikon digital camera.
Immunofluorescent staining (IHF) and confocal microscopy
IHF staining was carried out on 8 μm thick frozen sections of rabbit cornea and on cultured ALDH3A1-transfected rabbit keratocytes. Cells and tissues were initially fixed with 2% paraformaldehyde in PBS then rinsed and permeabilized with freezing acetone. Cells and tissues were rehydrated and incubated with 1% BSA in PBS for 30 min. Cells were then stained with rabbit anti-human ALDH3A1 [18] while tissue sections were incubated with chicken anti-rabbit ALDH1A1 (both diluted 1:100 in PBS). Samples were then washed and stained with FITC-conjugated goat anti-rabbit IgG (cells) or FITC-conjugated goat anti-chicken IgY (tissue). Cells and tissue sections were co-stained with 4′,6-diamidino-2-phenylindole (DAPI) to identify the nucleus. Tissue sections were evaluated using a Nikon Eclipse E600 fluorescence microscope. Cells were evaluated using a Zeiss 510 Meta LSM using a 40x, NA 1.4 objective.
RESULTS AND DISCUSSION
Expression of ALDH3A1/1A1 in the cornea
ALDH3A1 is a homodimer and highly expressed in mammalian corneal epithelial cells. It has been found to represent 5–50% (depending on the species) of the water-soluble proteins in mammalian corneal epithelial cells and it is considered the first enzyme to be categorized as a corneal crystalline [19]. A number of studies have shown constitutive expression of ALDH3A1 in the cornea of human, cow, pig, mouse, rat, baboon, opossum and kangaroo [19]. In addition to corneal epithelial cells, we have shown by immunolabeling that ALDH3A1 is also strongly expressed in stromal keratocytes of human cornea, but not in endothelial cells [18]. A similar expression pattern in pig and mouse corneas has also been observed [20]. No detectable ALDH3A1 protein or messenger was found in the rabbit cornea [13;21]. Interestingly, the SWR/J mouse strain exists as a ‘natural gene knockout’ model for ALDH3A1 by showing undetectable catalytic activity of corneal ALDH3A1 when compared to other inbred mouse strains that do possess constitutively high ALDH3A1 levels [22]. The molecular basis of this defect has been shown to be 13 nucleotide changes in the Aldh3a1c allele carried by SWR/J mice, four of which cause amino acid substitutions that may affect the catalytic activity and the stability of the protein [22]. Corneal expression of ALDH3A1 demonstrates a taxon-specific pattern, as shown by the absence of ALDH3A1 enzymatic activity from birds, such as the chicken and turkey [13]. Similarly, neither ALDH3A1 protein nor enzymatic activity were detected in corneal extracts from frog or a variety of fish species, including trout, zebrafish, chain pickerel and redhorse [13;23]. These results support the hypothesis that expression of ALDH3A1 in the cornea is specific to mammals.
ALDH1A1 is a homotetramer with 55 kDa subunits and well-noted for its constitutive expression in the liver. In humans, ALDH1A1 represents 3% and 2% of the soluble proteins in the cornea and lens epithelium, respectively [12]. Mouse ALDH1A1 is found mainly in the lens and to a lesser extent (< 5%) in the cornea epithelium. Rabbits primarily express ALDH1A1 in the cornea, with negligible amounts of ALDH3A1. In this species, ALDH1A1 constitutes 3, 16, and 11% of the total soluble proteins of the epithelium, stromal keratocytes, and endothelium, respectively [20]. It has been speculated that the absence of ALDH3A1 from the corneal epithelium of some species is counterbalanced by the expression of ALDH1A1 in order to provide the necessary transparent, refractive and protective properties of the cornea. ALDH1A1 protein has also been found in the adult retina of the mouse and rabbit [24;25]. The functional role of ALDH1A1 in the retina remains to be elucidated. It has a critical role in retinoic acid signaling [26], however, this isozyme appears dispensable for retinal development in mice in that Aldh1a1(−/−) knockout mice exhibit normal morphology of the retina [24].
Expression of other ALDH isozymes in the cornea
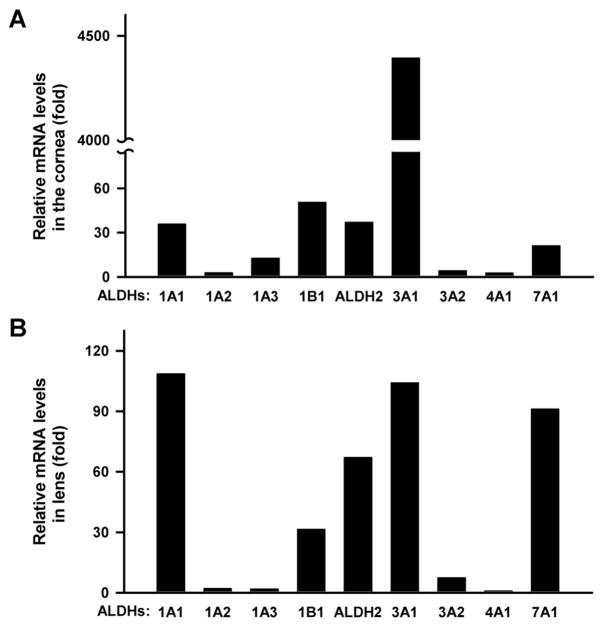
To this date, the majority of ALDH investigations in mammalian eyes have focussed on ALDH1A1 and ALDH3A1; other isozymes have been neglected. Nevertheless, in species where ALDH3A1 is absent from the cornea, the ALDH class 2 member (ALDH2) has been found at levels in the cornea comparable to those identified for ALDH1A1 [13]. Such species include chicken, turkey, trout and zebrafish. Human corneal epithelium and keratocytes have been shown to express ALDH2 mRNA [27]. However, no attempts have been made to determine the functional role of ALDH2 (or, for that matter, other ALDHs aside from ALDH1A1 or ALDH3A1) in the cornea in these species. To start to explore the possible roles of other ALDHs in the cornea and lens, we examined the mRNA expression of nine mouse ALDHs, namely ALDH1A1, ALDH1A2, ALDH1A3, ALDH1B1, ALDH2, ALDH3A1, ALDH3A2, ALDH4A1 and ALDH7A1 (Fig. 1). The mRNA level of each individual gene in the cornea and lens was normalized to that of β-actin (ACTB) in tissues. In addition, the mRNA level of keratin 12 (K12), a cornea-specific gene, was measured to control for potential corneal contamination of lens RNA; as expected, K12 mRNA was negligible in lens, but 144-fold in the cornea. Among all examined ALDHs, the mRNA level of lens ALDH4A1 was the lowest and set as the control (=1) to facilitate comparisons. Our results showed that the order of mRNA expression was ALDH3A1 > ALDH1B1 > ALDH2 > ALDH1A1 > ALDH7A1 >ALDH1A3> ALDH3A2 > ALDH1A2 > ALDH4A1 in the cornea (Fig. 1A) and ALDH1A1 > ALDH3A1 > ALDH7A1 > ALDH2 > ALDH1B1 > ALDH3A2 > ALDH1A2 > ALDH1A3 > ALDH4A1 in lens (Fig. 1B).
Fig. 1. Gene expression profile of nine Aldhs in mouse cornea and lens.
Total RNA was isolated from pooled corneas (A) and lenses (B) of 3~4 male C57BL/6J mice. mRNA levels were determined by Q-PCR and relative levels were expressed as fold of control after normalization to β-actin. The mRNA level of lens ALDH4A1 was artificially set as the control (=1) due to it having the lowest value.
Among all examined ALDHs, the high expression of ALDH3A1 mRNA (4300-fold) in the cornea and of ALDH1A1 mRNA (108-fold) in lens were expected and is in agreement with ALDH3A1 being a corneal crystallin and ALDH1A1 being a lens crystallin. On the other hand, a comparable level of ALDH3A1 mRNA (104-fold) to that of ALDH1A1 in lens was intriguing, since the ALDH3A1 protein does not accumulate in mouse lens [17]. A possible explanation for this could be that the cellular environment of mouse lens does not favour efficient translation or stability of ALDH3A1 protein. ALDH1B1 and ALDH7A1 also showed high levels of mRNA expression in the cornea and lens, respectively. ALDH1B1, formerly known as ALDHx or ALDH5, is a homotetramer present in mitochondrial matrix [26]. This mitochondrial ALDH has been reported to be expressed in bovine cornea and has also been found in non-ocular tissues including liver, kidney, heart, brain and skeletal muscle [26]. Little is known about its physiological role. In a recent study, we observed that human ALDH1B1 efficiently metabolized 4-HNE and MDA but not retinaldehydes (Stagos and Vasiliou, manuscript in preparation). Our results, combined with the upregulation of ALDH1B1 in human cells following UV treatment [28], may indicate an enzymatically-based protective role of ALDH1B1 against UVR-induced damage. ALDH7A1 has a primary role in lysine catabolism. Functional mutations in the ALDH7A1 gene contribute to the etiology of pyridoxine-dependent epilepsy in human patients [29]. It is thought that human ALDH7A1 may be involved in osmotic regulation in cells due to its 60% identity in amino acid sequence with the osmotic stress-induced pea turgor plant ALDH7B1 protein [30]. The human homolog of Aldh7a1 has also been found to be expressed in fetal human eyes [31]. The expression of Aldh2 gene, encoding another mitochondrial enzyme, was at a similar level as the Aldh1a1 gene (36-fold) in mouse cornea. ALDH2 protein shows 72% homology to ALDH1B1 and is believed to be important in the metabolism of 4-HNE and MDA [32]. The significant similarity between ALDH1B1 and ALDH2 may suggest similar functional roles of these isozymes in the cornea. ALDH1A2/1A3 are cytosolic proteins and, along with ALDH1A1, constitute the so called retinaldehyde dehydrogenases, which play important roles in retinoic acid signalling [26]. ALDH3A2 is a microsomal enzyme and is known as a fatty aldehyde dehydrogenase (FALDH) due to its ability to oxidize fatty alcohol to fatty acid [26]. ALDH4A1 is a mitochondrial protein involved in proline degradation by converting pyrroline-5-carboxylate to glutamate [26]. Interestingly, human ALDH4A1 gene was confined within a chromosomal region showing linkage to congenital cataracts in an Australian family, but no segregating mutations were identified suggesting that it did not account for the disease [33]. Despite their roles in diverse metabolic pathways, ALDH1A2, ALDH1A3, ALDH3A2 and ALDH4A1 had very low mRNA levels relative to ALDH1A1 in both mouse cornea and lens (<10-fold).
In summary our data showed that, in addition to abundant gene transcriptions of Aldh3a1 in the cornea and of Aldh1a1 in lens, other members of ALDH superfamily revealed moderately high levels of basal gene transcription in these ocular tissues, including Aldh1b1, Aldh2 and Aldh7a1. Given that human eyes are exposed to UVR at a daily base, future investigations on the gene transcription pattern of these ALDH isoenzymes in response to UVR exposure or other environmental challenges will provide important information on the physiological and/or toxicological relevance of these ALDHs in the eye.
Regulation of ALDH3A1/1A1 expression in the cornea
The expression of most, if not all, members of the ALDH superfamily is primarily regulated at the transcriptional level. In a recent study [34], ALDH3A1 protein and mRNA levels were found to increase substantially in mouse cornea by postnatal (PN) day 14; the preferential expression of ALDH3A1 in mouse cornea is regulated by Pax6, a transcription factor that plays a critical role in gene expression of lens crystallins as well as in the developmental process of lens [35;36]. It was also found that the transcription factor Oct1, a ubiquitously expressed POU domain-containing protein, also upregulates mouse ALDH3A1 protein [34]. So far, it has not been known if Pax6 and Oct1 act separately or synergistically to activate the promoter activity of the mouse Aldh3a1 gene. The coactivator p300 involved in lens crystallin gene expression [37] has also been shown to enhance the promoter activity of Aldh3a1 gene in mice by Pax6 [34]. Moreover, the Klf4 transcription factor, a member of the Krüpel-like transcription factors subfamily, was shown to enhance the promoter activity of mouse Aldh3a1 gene in SV40-transformed human corneal epithelial cells [38]. In agreement with this observation, Aldh3a1 was one of the genes that were down-regulated in the corneas of Klf4-conditional null (Klf4CN) mice [38]. When compared with corneas from wild-type mice, the expression of Pax6 was also reduced to about half in the corneas of Klf4-conditional null (Klf4CN) mice, suggesting that Klf4 may regulate the expression of ALDH3A1 indirectly through the Pax6 transcription factor. It is noteworthy that the Pax6, Oct1 and Klf4 transcription factors are expressed in mouse corneal epithelial cells in PN day 9, viz. before the upregulation of ALDH3A1 expression [34;38].
The preferential expression of ALDH1A1 in the rabbit cornea has been investigated at the molecular level using transfected cells and transgenic mice [25]. At least two xenobiotic response elements (XREs) and an E-box present in the promoter region (−3519 to +43) of rabbit Aldh1a1 gene are involved in this characteristic expression pattern of rabbit ALDH1A1. These cis-elements are also represented in promoter sequences of other genes that display high expression in mammalian corneas [25]. In addition, sequence analysis reveals that the 5′ flanking sequence of human ALDH1A1 gene has 60.2% homology to that of rabbit Aldh1a1 gene and ~50% to that of rodents [25]. This observation agrees with a closer phylogenetic relationship and expression pattern of ocular ALDH1A1 between humans and rabbits than humans and rodents. The cornea is challenged with environmental stresses and high oxygen tension and, as such, it is believed that environmental induction plays a key role in the regulation of corneal crystallins, which is in contrast to a major role of developmental regulation on lens crystallins. The involvement of XREs and E-boxes in the promoter activity of rabbit Aldh1a1 gene supports this idea. It is known that the bHLH-PAS transcription factors interact with these cis-elements to mediate responses to hypoxia and xenobiotics [39]. Indeed, messengers of a few genes encoding bHLH-PAS transcription factors, including Arnt1, AhR, Hif-1α, and Hif-3α, have been detected in rabbit corneas and HIF-3α activates the promoter activity of rabbit Aldh1a1 gene, suggesting a role of hypoxia-related pathway in the regulation of corneal ALDH1A1 [25].
Functions of corneal ALDH3A1/1A1
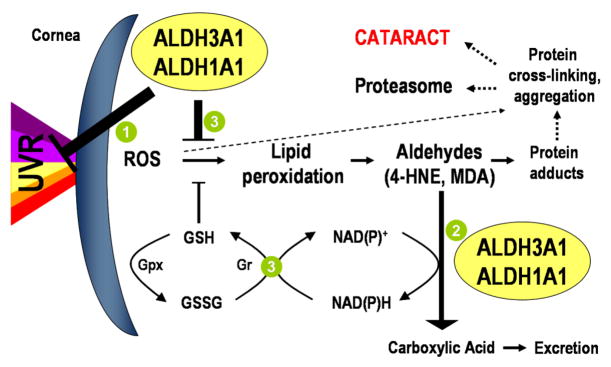
The expression of corneal crystallins, ALDH3A1 and ALDH1A1, at levels exceeding those needed for metabolism in the cornea has led to the proposal that these proteins may have additional roles and serve as an example of gene-sharing, a term invented to characterize the use of the same gene for more than one molecular functions [6]. An important role for ALDH3A1 in the cornea is implicated by the observation that decreased ALDH3A1 activity is associated with pathologic corneas [40]. A recent study by our laboratory reported the development of cataracts in Aldh3a1(−/−) knockout and Aldh1a1(−/−)/Aldh3a1(−/−) double knockout mice by one month of age, and in Aldh1a1(−/−) knockout mice later in life (6–9 months) [17]. These results are consistent with a protective function for ALDH3A1 and ALDH1A1 against cataract formation, which is associated with UVR exposure. It is believed that ALDH3A1 and ALDH1A1 play a key role in protecting the eye from UV-induced damage by the following mechanisms (Fig. 2): (1) direct absorption of UV light, (2) metabolism of toxic aldehydes produced by UV-induced lipid peroxidation, and (3) acting as antioxidants by scavenging directly UV-induced free radicals or by producing the antioxidant NAD(P)H. In addition to a protective role, it has been proposed that ALDH1A1 and ALDH3A1 also play a structural role in the cornea.
Fig. 2. Proposed roles of ALDH3A1 and ALDH1A1 in the cornea.
UVR produces reactive oxygen species (ROS) that can damage DNA and lipid membranes by initiating lipid peroxidation which, in turn, generates highly toxic aldehydes. ALDH3A1 and/or ALDH1A1 may (1) directly absorb UVR, (2) detoxify reactive aldehydes produced by lipid peroxidation to non-toxic metabolites and (3) act as antioxidants by scavenging ROS through active sites or by generating NAD(P)+H, which can be used in the regeneration of reduced glutathione (GSH) from its oxidized form (GSSG) via the glutathione reductase/peroxidase system (Gr/Gpx). GSH and NAD(P)+H may also eliminate ROS and thereby contribute to the cellular defense against UVR-induced oxidative damage.
ALDH3A1 and ALDH1A1 are potential major UVR filters
Direct absorption of UVR energy by cellular constituents can lead to the photo-oxidation of the absorbing species, including proteins, lipids and DNA. Consequences of UVR-induced modifications to proteins include enzyme inactivation, partial unfolding and non-native aggregation [41]. Such modifications may be partly responsible for the accumulation of aggregated proteins in the lens during cataract formation [42]. Several lines of evidence support a filtering role of UV light by ALDH3A1 and ALDH1A1, thereby protecting inner ocular tissues from UVR-induced damage. First, it was proposed early that ALDH3A1/BCP54 in the bovine cornea could directly absorb UVR [43]. This was supported by the observation that the water-soluble protein fraction of bovine cornea accounted for only 17% of the total cellular protein, but was responsible for almost 50% of the total absorption of UVB light (290–300 nm). This led to the suggestion for naming ALDH3A1 as ‘absorbin’ [44]. Second, the majority of UV light is absorbed by the corneal epithelium, the cellular layer in which ALDH3A1 is expressed abundantly [45]. Third, the UVR-absorbing capacity of ALDH3A1 is enhanced by its ability to bind NADPH [46]. Finally, studies indicate that the direct absorption of UVR by ALDH3A1, and possibly ALDH1A1, protect other corneal proteins at the expense of their own molecular inactivation. For example, purified recombinant ALDH3A1 is inactivated and covalently cross-linked by direct UVR exposure [47]. Similar inactivation of human recombinant ALDH1A1 by UVR is also observed (unpublished data). UV irradiation of mice leads to a decrease in corneal ALDH3A1 and ALDH1A1 activity by 85%, whereas the activities of other corneal enzymes remain unaltered [48]. We have recently shown that overexpression of ALDH3A1 in vitro diminishes UVR-induced inactivation of G6PD via direct absorption of UV energy [49].
ALDH3A1 and ALDH1A1 detoxify reactive aldehydes
ROS generated during UVR exposure can initiate lipid peroxidation by attacking polyunsaturated fatty acids in cell membrane phospholipids. The major aldehyde products of lipid peroxidation, such as 4-HNE and MDA, are highly reactive and potentially cytotoxic. Increased levels of 4-HNE or/and MDA have been observed in cataracts [50], corneal pathologies [51] and retinal disorders [52]. We have shown that human ALDH3A1 had high affinity for 4-HNE (Km &p; 40 μM) [18] and ALDH1A1 metabolized both 4-HNE and MDA (Km < 2 mM) [16]. Therefore, it is believed that enzymatic detoxification of these lipid-derived aldehydes is one mechanism by which ALDH3A1 and ALDH1A1 protect ocular tissues against UVR-induced damage. Furthermore, the presence of ALDH1A1 in the cornea may complement ALDH3A1 by oxidizing MDA, a poor substrate of ALDH3A1 [18]. Such “tandem” protective functions of ALDH3A1 and ALDH1A1 in the eye are supported by several studies that manipulate the cellular levels of these proteins. Overexpression of ALDH3A1 in human corneal epithelial cells inhibits the formation of 4-HNE-protein adducts and protects against 4-HNE-induced cytotoxicity and apoptosis [53]. In a rabbit corneal fibroblast cell line (TRK-43), stable transfection with human ALDH3A1 leads to reduced susceptibility to oxidative damage and levels of 4-HNE-protein adducts [54]. Inhibition of ALDH1A1 in human lens epithelial cells (HLECs) by siRNA is associated with decreased oxidation of 4-HNE and increased susceptibility of cells to oxidant-induced apoptosis [55]. Finally, the detection of increased levels of 4-HNE- and MDA-protein adducts in lens extracts from Aldh1a1(−/−)/Aldh3a1(−/−) null mice, who develop cataracts, further supports an in vivo role of ocular ALDH3A1 and ALDH1A1 in detoxifying 4-HNE and MDA [17].
ALDH3A1 and ALDH1A1 act as antioxidants
As mentioned above, the cornea is enriched in both enzymatic and non-enzymatic antioxidants to counteract oxidative insults, including UVR-generated ROS. There is evidence indicating that ALDH3A1 may also contribute to the antioxidant arsenal of the cornea. Bovine ALDH3A1 has been shown to protect RNase A from hydroxyl radical-induced modification, most likely by directly scavenging hydroxyl radical via its cysteine and methionine residues [56]. The high concentration of ALDH3A1 in the cornea may represent a target for ROS and reactive aldehydes, so that it provides a passive protective effect to other proteins [56;57]. Indeed, the level of ALDH3A1 carbonylation, a marker of oxidative damage, increases 2–3 fold in rabbit corneal fibroblasts treated with oxidative agents, but its enzymatic activity remains intact [54]. ALDH3A1 may also protect against UVR-induced oxidative damage by modifying proteasome activity, the major proteolytic system that removes damaged proteins [58]. This is supported by experiments in Aldh3a1(−/−) and Aldh3a1(−/−)/Aldh1a1(−/−) null mice who show decreased lens proteasome activity following UVR exposure relative to wild-type mice [17]. On the other hand, overexpression of ALDH3A1 inhibits UVR-induced loss of proteasome activity in rabbit corneal fibroblasts [54].
ALDH3A1 and ALDH1A1 may exhibit antioxidant activity indirectly by producing NAD(P)H during metabolism. NAD(P)H functions as an antioxidant in the cornea via multiple mechanisms. It is the reducing agent used by the GPX/GR system for the regeneration of GSH from its oxidized form GSSG [59]. It may act as a direct antioxidant by reducing glutathiyl and tyrosyl radicals generated during oxidative stress [60;61]. It can directly absorb UVR [46;62]. It may protect other antioxidant enzymes from ROS-induced inactivation [63]. Finally, it can maintain a reducing potential for pyridine nucleotide-dependent redox-active enzymes, such as isocitrate dehydrogenase, malic dehydrogenase and 6-phosphate dehydrogenase, which are involved in protecting eye tissues [64]. Consistent with a role for NAD(P)H in the actions of ALDH3A1 is the observation that overexpression of human ALDH3A1 prevents GSH depletion caused by oxidative agents in rabbit corneal fibroblast cells [54] and protects human corneal epithelial cells from oxidant-induced DNA damage [65].
ALDH3A1 and ALDH1A1 serve a possible structural role in the cornea
Aside from a crucial role in protecting the eye from UVR-induced damage, some evidence suggests that ALDH1A1 and ALDH3A1 may function as the structural elements of the cornea, a proposal manifesting largely from the abundance of these ALDH isozymes in the cornea. ALDH1A1 has been shown to contribute substantially to the transparency and refractory aspects of the cornea in the rabbit, consistent with a structural role as a corneal crystallin [20]. These studies demonstrated that injury-induced corneal haze was associated with the loss of ALDH1A1 expression in the corneal stroma. The development of postnatal corneal transparency was associated with stromal keratocyte quiescence and increased ALDH1A1 expression. By contrast, ALDH3A1 messenger and protein were undetectable in eyes from mouse embryos, but levels in the cornea increased dramatically between 9 and 14 days after birth, coincident with the eye opening [34]. In another study, decreased expression and enzymatic activity of ALDH3A1 were observed when human corneal keratocytes were transformed to a repair phenotype. This led to the speculation that reduced expression of ALDH3A1 may contribute to the loss of corneal transparency in humans after injuries [27]. Nevertheless, the role of ALDH1A1 and ALDH3A1 in mediating corneal transparency is not so certain given that mutant mice that do not express these isozymes possess structurally normal and transparent corneas [5;24;66].
Cellular localization of corneal ALDH3A1 and ALDH1A1
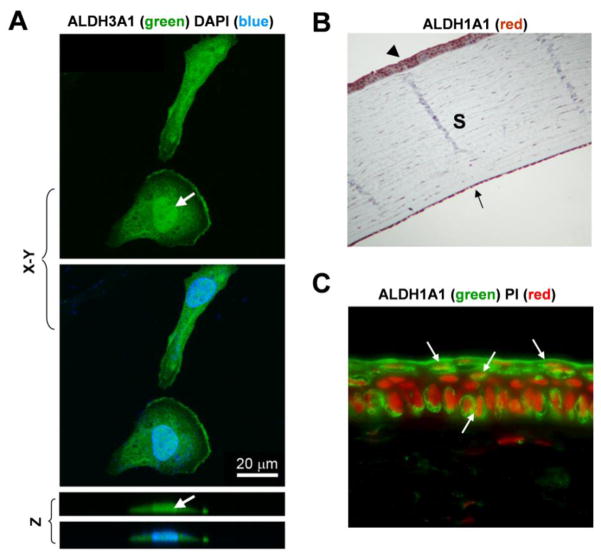
It has been known for decades that both ALDH1A1 and ALDH3A1 are cytoplasmic proteins. Recently this laboratory has reported the nuclear localization of ALDH3A1 in transfected human corneal epithelial cells [65]. We herein report the nuclear presence of ALDH3A1 in transfected rabbit corneal keratocytes. Confocal microscopy by X, Y and Z axes examination (Fig. 3A) showed that ALDH3A1 (green) was expressed in the cytoplasm and nucleus (arrows) of keratocytes. The physiological significance of the nuclear localization of ALDH3A1 in the cornea is of great interest and warrants further investigation. Based on the observation of an inverse relationship between ALDH3A1 expression and cell proliferation rate, a novel role for ALDH3A1 in cell cycle regulation has been proposed [65]. In this study, it was found that actively-proliferating primary human corneal epithelium displayed progressive loss of ALDH3A1 gene expression. When stably transfected with human ALDH3A1, human corneal epithelial (HCE) and human skin keratinocyte (NCTC2455) cell lines showed reduced growth rate and prolonged cell cycle length. Such growth inhibition by ALDH3A1 was associated with suppressed DNA synthesis, reduced cyclin A- and B-dependent kinase activities, decreased phosphorylation of retinoblastoma protein (Rb) and downregulation of several cell cycle regulators including cyclin A, B, and E, E2F and p21. The inhibitory effect of ALDH3A1 on cell cycle progression is proposed to be an additional mechanism by which this enzyme protects corneal cells from oxidative damage by facilitating DNA repair and cellular survival. The underlying mechanism(s) of this effect is unknown but may attribute to the catalytic and non-catalytic properties of ALDHs. From the catalytic perspective, ALDH3A1 metabolizes 4-HNE, an aldehyde that modulates cell proliferation and differentiation at low concentrations [67]. From the non-catalytic perspective, the physical localization of ALDH3A1 in the nucleus of corneal cells [65] supports a putative role of this isozyme in nuclear functions, such as mitotic control.
Fig. 3. Cellular localization of ALDH3A1 and ALDH1A1.
(A) Confocal microscopy of ALDH3A1-transfected corneal keratocytes, showing both cytoplasmic and nuclear presences of ALDH3A1 (green) by X, Y and Z axes examination. One set of samples were co-stained with DAPI (blue) to identify the nucleus. Bar = 20 μm. (B) Immunohistochemical staining for ALDH1A1 on paraffin-embedded rabbit eye sections (5 μm), showing positive staining for ALDH1A1 in corneal epithelium (arrow head), stroma (S) and endothelium (arrow). Magnification: 100x. (C) Immunofluorescent staining for ALDH1A1 (green) on frozen rabbit eye sections (8 μm), showing peri-nuclear staining for ALDH1A1 (arrows) in corneal epithelium. Sections were co-stained with PI (red) to identify the nucleus. Magnification: 1000x.
The expression pattern of ALDH1A1 in the cornea was examined in rabbits that express primarily this isozyme in the cornea. By immunohistochemical analysis (Fig. 3B) on rabbit eye sections, ALDH1A1 was detected in all three corneal cellular layers, namely epithelium, stroma and endothelium, a result consistent with previous studies [25]. Immunofluorescent analysis (Fig. 3C) revealed that in rabbit corneal epithelium, however, ALDH1A1 appeared predominantly as peri-nuclear. Up to this point, there is no direct evidence suggesting a role for ALDH1A1 in cellular growth regulation, However, a good correlation between the ALDH1A1 expression and cell cycle exit has been observed in rabbit corneal keratocytes [68].
Conclusion
With the characteristics of abundant expression and active metabolism of toxic aldehydes in the cornea, ALDH3A1 and ALDH1A1 are being recognized as playing important structural and functional roles in this ocular tissue. The novel nuclear localization of ALDH3A1 suggests additional roles, such as participating in the regulation of cell proliferation. Due to these features, these ALDH enzymes may serve as potential pharmacological targets for the prevention and treatment of UVR exposure-induced eye diseases, as such drug interventions would be directed at increasing the activity of ALDHs. Future studies aiming at a better understanding of ALDHs functions and regulations will provide a greater insight into the development of corneal diseases.
Acknowledgments
We thank Dr. David Thompson for critical reading and discussion of this manuscript. This work was supported by National Institutes of Health (NIH) grants EY11490, EY17963, and EY016665.
Footnotes
Conflict of Interest
The authors declare no conflict of interest or any commercial relationships.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Bergmanson JP, Soderberg PG. The significance of ultraviolet radiation for eye diseases. A review with comments on the efficacy of UV-blocking contact lenses. Ophthalmic Physiol Opt. 1995;15:83–91. [PubMed] [Google Scholar]
- 2.Shinohara T, Singh DP, Chylack LT., Jr Review: Age-related cataract: immunity and lens epithelium-derived growth factor (LEDGF) J Ocul Pharmacol Ther. 2000;16:181–191. doi: 10.1089/jop.2000.16.181. [DOI] [PubMed] [Google Scholar]
- 3.Sacca SC, Bolognesi C, Battistella A, Bagnis A, Izzotti A. Gene-environment interactions in ocular diseases. Mutat Res. 2008 doi: 10.1016/j.mrfmmm.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Piatigorsky J. Enigma of the abundant water-soluble cytoplasmic proteins of the cornea: the “refracton” hypothesis. Cornea. 2001;20:853–858. doi: 10.1097/00003226-200111000-00015. [DOI] [PubMed] [Google Scholar]
- 5.Lassen N, Black WJ, Estey T, Vasiliou V. The role of corneal crystallins in the cellular defense mechanisms against oxidative stress. Semin Cell Dev Biol. 2008;19:100–112. doi: 10.1016/j.semcdb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Piatigorsky J. Gene sharing in lens and cornea: facts and implications. Prog Retin Eye Res. 1998;17:145–174. doi: 10.1016/s1350-9462(97)00004-9. [DOI] [PubMed] [Google Scholar]
- 7.Jester JV. Corneal crystallins and the development of cellular transparency. Semin Cell Dev Biol. 2008;19:82–93. doi: 10.1016/j.semcdb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 9.Vasiliou V, Pappa A. Polymorphisms of human aldehyde dehydrogenases. Consequences for drug metabolism and disease. Pharmacology. 2000;61:192–198. doi: 10.1159/000028400. [DOI] [PubMed] [Google Scholar]
- 10.Verhagen C, Hoekzema R, Verjans GM, Kijlstra A. Identification of bovine corneal protein 54 (BCP 54) as an aldehyde dehydrogenase. Exp Eye Res. 1991;53:283–284. doi: 10.1016/0014-4835(91)90085-s. [DOI] [PubMed] [Google Scholar]
- 11.Holmes RS, Cheung B, VandeBerg JL. Isoelectric focusing studies of aldehyde dehydrogenases, alcohol dehydrogenases and oxidases from mammalian anterior eye tissues. Comp Biochem Physiol B. 1989;93:271–277. doi: 10.1016/0305-0491(89)90081-3. [DOI] [PubMed] [Google Scholar]
- 12.King G, Holmes RS. Human corneal aldehyde dehydrogenase: purification, kinetic characterisation and phenotypic variation. Biochem Mol Biol Int. 1993;31:49–63. [PubMed] [Google Scholar]
- 13.Pappa A, Sophos NA, Vasiliou V. Corneal and stomach expression of aldehyde dehydrogenases: from fish to mammals. Chem Biol Interact. 2001;130–132:181–191. doi: 10.1016/s0009-2797(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 14.Graham C, Hodin J, Wistow G. A retinaldehyde dehydrogenase as a structural protein in a mammalian eye lens. Gene recruitment of eta-crystallin. J Biol Chem. 1996;271:15623–15628. doi: 10.1074/jbc.271.26.15623. [DOI] [PubMed] [Google Scholar]
- 15.Zinovieva RD, Tomarev SI, Piatigorsky J. Aldehyde dehydrogenase-derived omega-crystallins of squid and octopus. Specialization for lens expression. J Biol Chem. 1993;268:11449–11455. [PubMed] [Google Scholar]
- 16.Manzer R, Qamar L, Estey T, Pappa A, Petersen DR, Vasiliou V. Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA. DNA Cell Biol. 2003;22:329–338. doi: 10.1089/104454903322216671. [DOI] [PubMed] [Google Scholar]
- 17.Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, Duester G, Day BJ, Huang J, Hines LM, Vasiliou V. Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem. 2007;282:25668–25676. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem J. 2003;376:615–623. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piatigorsky J. Multifunctional lens crystallins and corneal enzymes. More than meets the eye. Ann N Y Acad Sci. 1998;842:7–15. doi: 10.1111/j.1749-6632.1998.tb09626.x. [DOI] [PubMed] [Google Scholar]
- 20.Jester JV, Budge A, Fisher S, Huang J. Corneal keratocytes: phenotypic and species differences in abundant protein expression and in vitro light-scattering. Invest Ophthalmol Vis Sci. 2005;46:2369–2378. doi: 10.1167/iovs.04-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jester JV, Moller-Pedersen T, Huang J, Sax CM, Kays WT, Cavangh HD, Petroll WM, Piatigorsky J. The cellular basis of corneal transparency: evidence for ‘corneal crystallins’. J Cell Sci. 1999;112(Pt 5):613–622. doi: 10.1242/jcs.112.5.613. [DOI] [PubMed] [Google Scholar]
- 22.Shiao T, Tran P, Siegel D, Lee J, Vasiliou V. Four amino acid changes are associated with the Aldh3a1 locus polymorphism in mice which may be responsible for corneal sensitivity to ultraviolet light. Pharmacogenetics. 1999;9:145–153. [PubMed] [Google Scholar]
- 23.Cooper DL, Isola NR, Stevenson K, Baptist EW. Members of the ALDH gene family are lens and corneal crystallins. Adv Exp Med Biol. 1993;328:169–179. doi: 10.1007/978-1-4615-2904-0_19. [DOI] [PubMed] [Google Scholar]
- 24.Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hough RB, Piatigorsky J. Preferential transcription of rabbit Aldh1a1 in the cornea: implication of hypoxia-related pathways. Mol Cell Biol. 2004;24:1324–1340. doi: 10.1128/MCB.24.3.1324-1340.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pei Y, Reins RY, McDermott AM. Aldehyde dehydrogenase (ALDH) 3A1 expression by the human keratocyte and its repair phenotypes. Exp Eye Res. 2006;83:1063–1073. doi: 10.1016/j.exer.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Crabb DW, Stewart MJ, Xiao Q. Hormonal and chemical influences on the expression of class 2 aldehyde dehydrogenases in rat H4IIEC3 and human HuH7 hepatoma cells. Alcohol Clin Exp Res. 1995;19:1414–1419. doi: 10.1111/j.1530-0277.1995.tb01000.x. [DOI] [PubMed] [Google Scholar]
- 29.Striano P, Battaglia S, Giordano L, Capovilla G, Beccaria F, Struys EA, Salomons GS, Jakobs C. Two novel ALDH7A1 (antiquitin) splicing mutations associated with pyridoxine-dependent seizures. Epilepsia. 2008 doi: 10.1111/j.1528-1167.2008.01741.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee P, Kuhl W, Gelbart T, Kamimura T, West C, Beutler E. Homology between a human protein and a protein of the green garden pea. Genomics. 1994;21:371–378. doi: 10.1006/geno.1994.1279. [DOI] [PubMed] [Google Scholar]
- 31.Skvorak AB, Robertson NG, Yin Y, Weremowicz S, Her H, Bieber FR, Beisel KW, Lynch ED, Beier DR, Morton CC. An ancient conserved gene expressed in the human inner ear: identification, expression analysis, and chromosomal mapping of human and mouse antiquitin (ATQ1) Genomics. 1997;46:191–199. doi: 10.1006/geno.1997.5026. [DOI] [PubMed] [Google Scholar]
- 32.Vasiliou V, Pappa A, Petersen DR. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem Biol Interact. 2000;129:1–19. doi: 10.1016/s0009-2797(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 33.Burdon KP, Hattersley K, Lachke SA, Laurie KJ, Maas RL, Mackey DA, Craig JE. Investigation of eight candidate genes on chromosome 1p36 for autosomal dominant total congenital cataract. Mol Vis. 2008;14:1799–1804. [PMC free article] [PubMed] [Google Scholar]
- 34.Davis J, Davis D, Norman B, Piatigorsky J. Gene expression of the mouse corneal crystallin Aldh3a1: activation by Pax6, Oct1, and p300. Invest Ophthalmol Vis Sci. 2008;49:1814–1826. doi: 10.1167/iovs.07-1057. [DOI] [PubMed] [Google Scholar]
- 35.Cvekl A, Piatigorsky J. Lens development and crystallin gene expression: many roles for Pax-6. Bioessays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 36.Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- 37.Chen Q, Dowhan DH, Liang D, Moore DD, Overbeek PA. CREB-binding protein/p300 co-activation of crystallin gene expression. J Biol Chem. 2002;277:24081–24089. doi: 10.1074/jbc.M201821200. [DOI] [PubMed] [Google Scholar]
- 38.Swamynathan SK, Davis J, Piatigorsky J. Identification of candidate Klf4 target genes reveals the molecular basis of the diverse regulatory roles of Klf4 in the mouse cornea. Invest Ophthalmol Vis Sci. 2008;49:3360–3370. doi: 10.1167/iovs.08-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 40.Gondhowiardjo TD, van Haeringen NJ, Volker-Dieben HJ, Beekhuis HW, Kok JH, van RG, Pels L, Kijlstra A. Analysis of corneal aldehyde dehydrogenase patterns in pathologic corneas. Cornea. 1993;12:146–154. doi: 10.1097/00003226-199303000-00010. [DOI] [PubMed] [Google Scholar]
- 41.Dean RT, Fu S, Stocker R, Davies MJ. Biochemistry and pathology of radical-mediated protein oxidation. Biochem J. 1997;324(Pt 1):1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Abedinia M, Pain T, Algar EM, Holmes RS. Bovine corneal aldehyde dehydrogenase: the major soluble corneal protein with a possible dual protective role for the eye. Exp Eye Res. 1990;51:419–426. doi: 10.1016/0014-4835(90)90154-m. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell J, Cenedella RJ. Quantitation of ultraviolet light-absorbing fractions of the cornea. Cornea. 1995;14:266–272. doi: 10.1097/00003226-199505000-00007. [DOI] [PubMed] [Google Scholar]
- 45.Kolozsvari L, Nogradi A, Hopp B, Bor Z. UV absorbance of the human cornea in the 240- to 400-nm range. Invest Ophthalmol Vis Sci. 2002;43:2165–2168. [PubMed] [Google Scholar]
- 46.Atherton SJ, Lambert C, Schultz J, Williams N, Zigman S. Fluorescence studies of lens epithelial cells and their constituents. Photochem Photobiol. 1999;70:823–828. [PubMed] [Google Scholar]
- 47.Manzer R, Pappa A, Estey T, Sladek N, Carpenter JF, Vasiliou V. Ultraviolet radiation decreases expression and induces aggregation of corneal ALDH3A1. Chem Biol Interact. 2003;143–144:45–53. doi: 10.1016/s0009-2797(02)00171-0. [DOI] [PubMed] [Google Scholar]
- 48.Downes JE, Swann PG, Holmes RS. Ultraviolet light-induced pathology in the eye: associated changes in ocular aldehyde dehydrogenase and alcohol dehydrogenase activities. Cornea. 1993;12:241–248. doi: 10.1097/00003226-199305000-00010. [DOI] [PubMed] [Google Scholar]
- 49.Estey T, Cantore M, Weston PA, Carpenter JF, Petrash JM, Vasiliou V. Mechanisms involved in the protection of UV-induced protein inactivation by the corneal crystallin ALDH3A1. J Biol Chem. 2007;282:4382–4392. doi: 10.1074/jbc.M607546200. [DOI] [PubMed] [Google Scholar]
- 50.Bhuyan KC, Bhuyan DK, Santos O, Podos SM. Antioxidant and anticataractogenic effects of topical captopril in diquat-induced cataract in rabbits. Free Radic Biol Med. 1992;12:251–261. doi: 10.1016/0891-5849(92)90112-t. [DOI] [PubMed] [Google Scholar]
- 51.Buddi R, Lin B, Atilano SR, Zorapapel NC, Kenney MC, Brown DJ. Evidence of oxidative stress in human corneal diseases. J Histochem Cytochem. 2002;50:341–351. doi: 10.1177/002215540205000306. [DOI] [PubMed] [Google Scholar]
- 52.Verdejo C, Marco P, Renau-Piqueras J, Pinazo-Duran MD. Lipid peroxidation in proliferative vitreoretinopathies. Eye. 1999;13(Pt 2):183–188. doi: 10.1038/eye.1999.48. [DOI] [PubMed] [Google Scholar]
- 53.Pappa A, Chen C, Koutalos Y, Townsend AJ, Vasiliou V. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic Biol Med. 2003;34:1178–1189. doi: 10.1016/s0891-5849(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 54.Lassen N, Pappa A, Black WJ, Jester JV, Day BJ, Min E, Vasiliou V. Antioxidant function of corneal ALDH3A1 in cultured stromal fibroblasts. Free Radic Biol Med. 2006;41:1459–1469. doi: 10.1016/j.freeradbiomed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 55.Choudhary S, Xiao T, Vergara LA, Srivastava S, Nees D, Piatigorsky J, Ansari NH. Role of aldehyde dehydrogenase isozymes in the defense of rat lens and human lens epithelial cells against oxidative stress. Invest Ophthalmol Vis Sci. 2005;46:259–267. doi: 10.1167/iovs.04-0120. [DOI] [PubMed] [Google Scholar]
- 56.Uma L, Hariharan J, Sharma Y, Balasubramanian D. Corneal aldehyde dehydrogenase displays antioxidant properties. Exp Eye Res. 1996;63:117–120. doi: 10.1006/exer.1996.0098. [DOI] [PubMed] [Google Scholar]
- 57.Estey T, Cantore M, Weston PA, Carpenter JF, Petrash JM, Vasiliou V. Mechanisms involved in the protection of UV-induced protein inactivation by the corneal crystallin ALDH3A1. J Biol Chem. 2007;282:4382–4392. doi: 10.1074/jbc.M607546200. [DOI] [PubMed] [Google Scholar]
- 58.Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 59.Dickinson DA, Forman HJ. Glutathione in defense and signaling: lessons from a small thiol. Ann N Y Acad Sci. 2002;973:488–504. doi: 10.1111/j.1749-6632.2002.tb04690.x. [DOI] [PubMed] [Google Scholar]
- 60.Forni LG, Willson RL. Thiyl free radicals and the oxidation of ferrocytochrome c. Direct observation of coupled hydrogen-atom- and electron-transfer reactions. Biochem J. 1986;240:905–907. doi: 10.1042/bj2400905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirsch M, De Groot H. NAD(P)H, a directly operating antioxidant? FASEB J. 2001;15:1569–1574. doi: 10.1096/fj.00-0823hyp. [DOI] [PubMed] [Google Scholar]
- 62.Davies MJ, Truscott RJ. Photo-oxidation of proteins and its role in cataractogenesis. J Photochem Photobiol B. 2001;63:114–125. doi: 10.1016/s1011-1344(01)00208-1. [DOI] [PubMed] [Google Scholar]
- 63.Kirkman HN, Rolfo M, Ferraris AM, Gaetani GF. Mechanisms of protection of catalase by NADPH. Kinetics and stoichiometry. J Biol Chem. 1999;274:13908–13914. doi: 10.1074/jbc.274.20.13908. [DOI] [PubMed] [Google Scholar]
- 64.Pappa A, Chen C, Koutalos Y, Townsend AJ, Vasiliou V. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic Biol Med. 2003;34:1178–1189. doi: 10.1016/s0891-5849(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 65.Pappa A, Brown D, Koutalos Y, DeGregori J, White C, Vasiliou V. Human aldehyde dehydrogenase 3A1 inhibits proliferation and promotes survival of human corneal epithelial cells. J Biol Chem. 2005;280:27998–28006. doi: 10.1074/jbc.M503698200. [DOI] [PubMed] [Google Scholar]
- 66.Nees DW, Wawrousek EF, Robison WG, Jr, Piatigorsky J. Structurally normal corneas in aldehyde dehydrogenase 3a1-deficient mice. Mol Cell Biol. 2002;22:849–855. doi: 10.1128/MCB.22.3.849-855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barrera G, Pizzimenti S, Dianzani MU. 4-hydroxynonenal and regulation of cell cycle: effects on the pRb/E2F pathway. Free Radic Biol Med. 2004;37:597–606. doi: 10.1016/j.freeradbiomed.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 68.Jester JV, Lee YG, Huang J, Houston J, Adams B, Cavanagh HD, Petroll WM. Postnatal corneal transparency, keratocyte cell cycle exit and expression of ALDH1A1. Invest Ophthalmol Vis Sci. 2007;48:4061–4069. doi: 10.1167/iovs.07-0431. [DOI] [PubMed] [Google Scholar]