Abstract
Many bioactive peptides are featured by their unique amino acid compositions such as argine/lysine-rich peptides. However, histidine-rich bioactive peptides are hardly found. In this study, histidine-containing peptides were constructed by selectively replacing the corresponded lysine residues in a lytic peptide LL-1 with histidines. Interestingly, all resulting peptides demonstrated pH-dependent activities. The cell lysis activities of these peptides could be increased up to four times as the solution pHs dropped from pH=7.4 to pH=5.5. The pH sensitivity of a histidine-containing peptide was determined by histidine substitution numbers. Peptide derivatives with more histidines were associated with increased pH sensitivity. Results showed that not the secondary structures but pH-affected cell affinity changes were responsible for the pH-dependent activities of histidine-containing peptides. The histidine substitution approach demonstrated here may present a general strategy to construct bioactive peptides with desired pH sensitivity for various applications.
Keywords: Peptide, pH sensitivity, histidine, bioactive, cell lysis
Introduction
Bioactive peptides are featured by their unique amino acid compositions such as proline-rich [1], cysteine-rich [2], and argine/lysine-rich [3]. Lysine and arginine have proven to be essential amino acids for many bioactive peptides including anticancer and antibacterial peptides. These two basic amino acids play a key role in peptide membrane partitioning and contribute greatly to the cell lysis activity of most bioactive peptides. However, another basic amino acid, histidine, is hardly found in bioactive peptides. Histidine-containing antibacterial peptides have been reported in few peptides including Histatin, Clavanin and Chrysophsin [4–9]. Interestingly, these histidine-containing peptides have proven to be pH-sensitive and become more active at acidic pHs.
Bioactive peptides with pH-dependent activities are of great importance for biomedical research and disease treatments. For examples, it has been found that the extracellular pH of some solid tumor are consistently lower as compared to the pH = 7.2~7.4 in normal tissues/organs [10, 11]. The origins of this extracellular acidity lie in the chaotic nature of tumor vasculature, increased glycolytic flux in tumor cells, increased export of protons from tumor cells, diminished buffering capacity of tumor interstitial fluid, and diffusion-limited rates of transport of lactic acid from the interstitium into the vasculature. To this regard, if bioactive peptides with appropriate pH sensitive toxicity that fit the extracellular pH difference between tumor and normal tissues, they should be able to kill tumors in acidic environments selectively but spare normal tissues with physiological pH.
Lytic peptide is a group of peptides which have their primary targeting sites on cell membranes and demonstrates strong anti-bacterial and anti-cancer activities [12–14]. In this study, we had tried to construct histidine-containing lytic peptides by replacing the corresponded lysine residues in lytic peptide LL-1 with histidines. All obtained histidine-containing peptides demonstrated pH-dependent cell lysis activities. This study may present a general strategy to create bioactive peptides with desired pH sensitivity to meet the needs of various medical applications including cancer and bacterial infection treatments [9, 15].
Materials and methods
Materials
Human alveolar basal epithelial cells (A549), Dulbecco’s modified Eagle’s medium (DMEM), and Kaighn’s Modification of Ham’s F-12 medium were purchased from American Type Culture Collection (ATCC). Calcein, Cholesterol, and Cell Growth Determination Kit (MTT kit) were from Sigma–Aldrich Company. The 1, 2-Dipalmitoyl-sn-Glycero-3-Phospho-L-Serine (DPPS) and 1, 2-Dipalmitoyl-sn-Glycero-3-Phosphocholine (DPPC) were purchased from Avanti polar lipids, Inc. Peptides were synthesized by GenScript. The purity of peptides was confirmed by HPLC and electrospray ionization mass spectrometry (see supplementary Table S1). All other reagents used in this experiment were analytic grade.
Cytotoxicity measurement
The activity of peptides was determined by MTT assay as described previously [16]. Briefly, A549 cells in complete medium were added into 96-well plates (5 ×103 cells/well) and cultured at 37 °C for 14–16 hrs. After being washed, cells were fed with serum-free DMEM medium containing various concentrations of peptides and incubated 37 °C for 2 hours. After that, 10 µl of MTT (5 mg/ml) were added into each well and cell viability was tested after additional four-hour incubation at 37 °C.
Preparation of lipid vesicles
Lipid vesicles were prepared by the extrusion method [17]. Briefly, 50 µmol DPPC, 2.5 µmol DPPS and 10 µmol cholesterol were dissolved in 4.0 ml chloroform. The organic solvent was removed by vacuum evaporation and, the dried lipid film was suspended by vortexing it in PBS buffer. After freezing (−20 °C) and thawing (45 °C) cycles (5 ~10 times), the lipid suspensions were extruded through polycarbonate filters with pore size of 1.0 µm. The final lipid concentration in the lipid vesicles was determined by the ferrithiocyanate method [18]. The surface charges of lipid vesicles were measured using Nanosizer.
Peptide binding to lipid vesicles
The bindings of peptides to lipid vesicles were studied by monitoring the intrinsic tryptophan fluorescence changes of LL-1 derivatives. A fixed amount of peptide (10 µM) was incubated with lipid vesicles (20 ~ 200 µM) for 10 mins at room temperature. Fluorescence spectra were recorded on Jasco FT-6500 fluorescence spectrofluorimeter using 1.0 cm path quartz cuvettes with excitation at 280 nm and emission from 310 to 380 nm. The spectra were corrected for light scattering by subtracting the corresponding blank spectra without peptides.
Fluorescence quenching by KI and acrylamide
The peptide was added to a lipid vesicle suspension (2µM, final concentration) in 50 mM potassium phosphate buffer. Small aliquots of the stock solutions of quenchers (8 M acrylamide; 5 M potassium iodide) were added to vesicle suspensions. The concentrations of quenching reagents used in the tests were set at 20 to 200 mM for KI and 10 to 80 mM for acrylamide. We have found that compared to 280 nm, excitation at 295 nm gave more reliable results with less interferences in acrylamide quenching experiments. Therefore, the peptide samples were excited at 280 nm and 295 nm in KI and acrylamide quenching experiments, respectively. In KI related quenching experiments, KCl was used to compensate KI concentration change to maintain constant solution ionic strength. The emission wavelength was recorded in a range from 310 to 380 nm. Results were fitted to the Stern-Volmer equation (F0/F=1+Ksv[Q]), where F and F0 are the fluorescence intensities with and without quencher, Ksv is the Stern-Volmer constant, and [Q] is the quencher concentration.
Secondary structure assay using circular dichroism (CD) [19]
The CD spectra of peptides were recorded with a Jasco J-710 spectropolarimeter. The samples contained peptide (30 µM) in different solutions of various pHs. The CD spectra were scanned at 25 °C in a capped, quartz optical cell with a 1.0 mm path length. Data were collected from 250 to 190 nm at an interval of 1.0 nm with an integration time of two seconds at each wavelength. Five to ten scans were averaged, smoothed, background-subtracted, and converted to mean residue molar ellipticity [θ] (degrees cm2 dmol−1) for each measurement. CDPRO software was used to analyze the data obtained from the CD spectrophotometer
Results
The pH-sensitivity of histidine-containing peptides
A lytic peptide, LL-1, was selected as the template peptide for histidine-containing peptide construction. Peptide LL-1 is a derivative of peptide PTP7 in which the phenylalanine residue at position 5 is substituted by tryptophan. Peptide LL-1 has strong anticancer and antibacterial activities (data not shown). The two lysine residues in peptide LL-1 at position 7 and position 11 were selectively replaced by histidines to yield three histidine-containing derivatives, LL-1a, LL-1b, and LL-1c. Unlike the amino group (pKa = 10.5) in lysine, the pKa value of imidazole group in histidine is about 6.0. At the physiological pH (pH ~7.4), the side chains of histidine is only partially charged and thus each histidine residue in peptides carries less than one unit of positive charge. Therefore, a direct consequence of lysine replacement by histidines was the low isoelectric point (pI) values and significantly decreased positive charges in resulting histidine-containing lytic peptides (Table 1). As a reflection of these chemical property changes, all three histidine-containing derivatives from peptide LL-1 showed reduced cytotoxicity (Fig. 1A). The activity loss in histidine-containing peptides was determined by histidine substitution numbers. Peptide LL-1a and LL-1b with one histidine had moderate and also almost the same activity while peptide LL-1c in which both of the two lysines were replaced by histidines showed very low cytotoxicity. The activity order of peptides at pH=7.4 is LL-1>LL-1a~LL-1b >LL-1c.
Table 1.
Properties of histidine-containing peptides from LL-1
| Peptide | Sequence | Number of histidine |
Number of lysine |
a Net charge | b Charge ratio | a PI | |
|---|---|---|---|---|---|---|---|
| pH=7.4 | pH=5.5 | ||||||
| LL-1 | FLGALWKALSKLL | 0 | 2 | 2.0 | 2.0 | 1.0 | 10.6 |
| LL-1a | FLGALWHALSKLL | 1 | 1 | 1.0 | 1.7 | 1.7 | 10.0 |
| LL-1b | FLGALWKALSHLL | 1 | 1 | 1.0 | 1.7 | 1.7 | 10.0 |
| LL-1c | FLGALWHALSHLL | 2 | 0 | 0.1 | 1.5 | 15 | 7.6 |
Theoretical calculation [20]
Positive charge ratios of peptides at pH=5.5 verse at pH= 7.4
Figure 1.
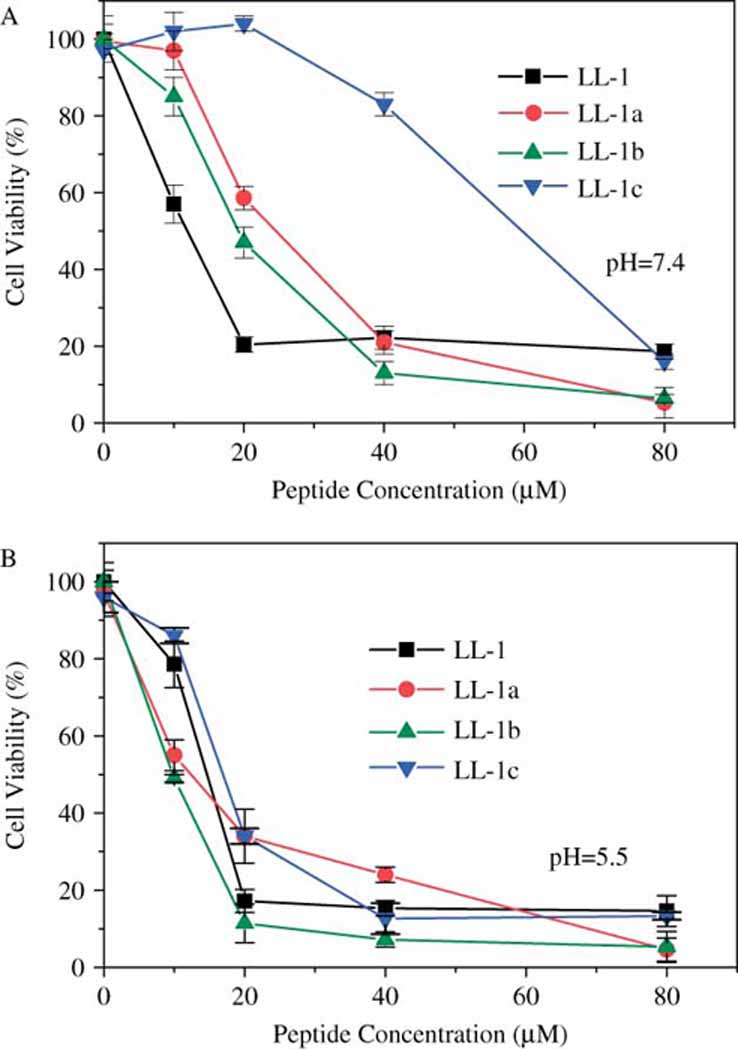
The cytotoxicity of peptide LL-1 and its histidine-containing derivatives. The log-phase cell growth inhibition assay (MTT assay) was performed on 96-well plates seeded with A549 cells in the presence of peptides at pH=7.4 (A) and pH=5.5 (B), respectively. Cell viability results were obtained after 2 hours of incubation.
Interestingly, although the cytotoxicity of peptide LL-1 was hardly affected by solution pHs, all three histidine-containing peptides demonstrated pH-dependent cytotoxicity and became more active under acidic conditions. At pH=5.5, histidine-containing peptides had the same or even more potent activity as peptide LL-1 (Fig. 1B). Since the solution pHs in the range of pH=7.4 ~ 5.5 had no effects on cytotoxicity results from MTT assay [21], the pH sensitivity of histidine-containing peptides was not an artifact. Furthermore, the same pH sensitivity of histidine-containing peptides were also observed on other cell lines (data not shown), suggesting that the pH sensitivity is not cell specific but an intrinsic feature of these histidine-containing peptides.
The acting mechanism of histidine-containing peptides
As we know, the secondary structure of a peptide is greatly affected by the hydrophobicity and the intra-chain interactions of all amino acids in this peptide. Because of distinguished chemical and physical properties (Table 1), histidine-containing derivatives were expected to have varied and different secondary structures from peptide LL-1. Surprisingly, three histidine-containing derivatives from LL-1 gave almost the same secondary structures as their parent peptide (Fig. 2A). More interestingly, the secondary structure of these peptides proved to be very stable and did not change in the wide pH range from pH=7.4 to pH=5.5 (Fig. 2B). Although increased helical contents (associated with decreased random coil structures) were observed for these peptides in a hydrophobic environment (10 mM SDS, Fig. 2D) compared to in an aqueous solution (20 mM NaCl, Fig. 2C), the effects of solution pH were still negligible in both cases (see supplementary Table S2).
Figure 2.
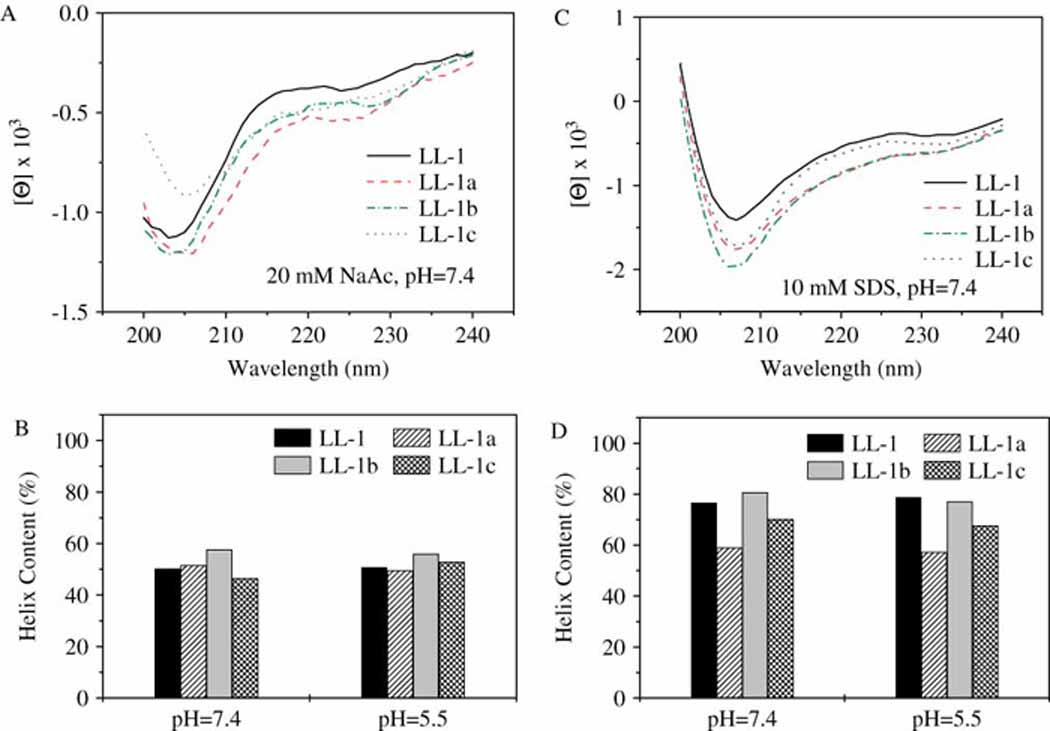
Secondary structure comparison. CD spectra of LL-1 and its histidine-containing derivative (10 µM) were recorded in two different solution systems: 20 mM NaAc (A) and 10 mM SDS (C). The pH affected helical content changes in peptides in these two solution systems (B, 20 mM NaAc; D, 10 mM SDS) were compared.
Because of the presence of tryptophan, pH-induced possible conformation changes in peptide LL-1 and its histidine-containing derivatives were further examined using well established fluorescence quenching assays [22]. The microenvironment differences around the tryptophan residue at position six in peptides were probed by two quenching agents, acrylamide and iodide (Table 2). Acrylamide quenching was more efficient than iodide, as the Stern-Volmer constant (Ksv) of acrylamide and iodide for the four peptides amounted up to 80% and 50% of that of free tryptophan, respectively, suggesting that tryptophan residues in peptides were shielded from solutions. Solution pH was found to have very limited effects on acrylamide and iodide mediated tryptophan fluorescence quenching. We had demonstrated that solution pH in the range of pH = 7.4 to pH = 5.5 did not induce significant structure or conformation changes in all LL-1 peptides (Fig. 2). The tryptophan quenching data we obtained here provide further evidences to support CD analysis and confirm that peptide LL-1 and its derivatives form stable secondary structures which are hardly affected by solution pHs. These results imply that secondary structures have little to do with the pH sensitivity of histidine-containing peptides.
Table 2.
Stern-Volmer constant (Ksv) of the tryptophan residue in peptides by KI and acrylamide
| Ksv (M−1) at pH=7.4 | Ksv (M−1) at pH=5.5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Free Trp | LL-1 | LL-1a | LL-1b | LL-1c | LL-1 | LL-1a | LL-1b | LL-1c | |
| KI | 11.3 | 5.7 | 5.05 | 5.1 | 4.9 | 5.15 | 4.05 | 4.55 | 4.4 |
| Acrylamide | 20.7 | 16.3 | 16.17 | 14.8 | 17.5 | 16.5 | 12.4 | 12.2 | 13.2 |
It is known that the electrostatic attraction of peptides to cell membrane is the critical step for lytic peptide interactions with cell membranes. According to theoretical calculation, the positive charges in histidine-containing peptide LL-1a, LL-1b, and LL-1c will be increased dramatically as the solution pH change from pH=7.4 to pH=5.5 (see supplementary Fig. S1). On the contrary, the positive charges on peptide LL-1 keep constant in the same pH range (Table 1). The pH-sensitivity of histidine-containing peptides from LL-1 matched perfectly with pH-induced positive charge changes in these peptides (Fig 3). Mammalian cell surface carries net negative charges although they are not significantly negative. Therefore, it is very likely that the pH sensitivity of histidine-containing peptides is from pH-affected electrostatic interactions between cells and peptides.
Figure 3.
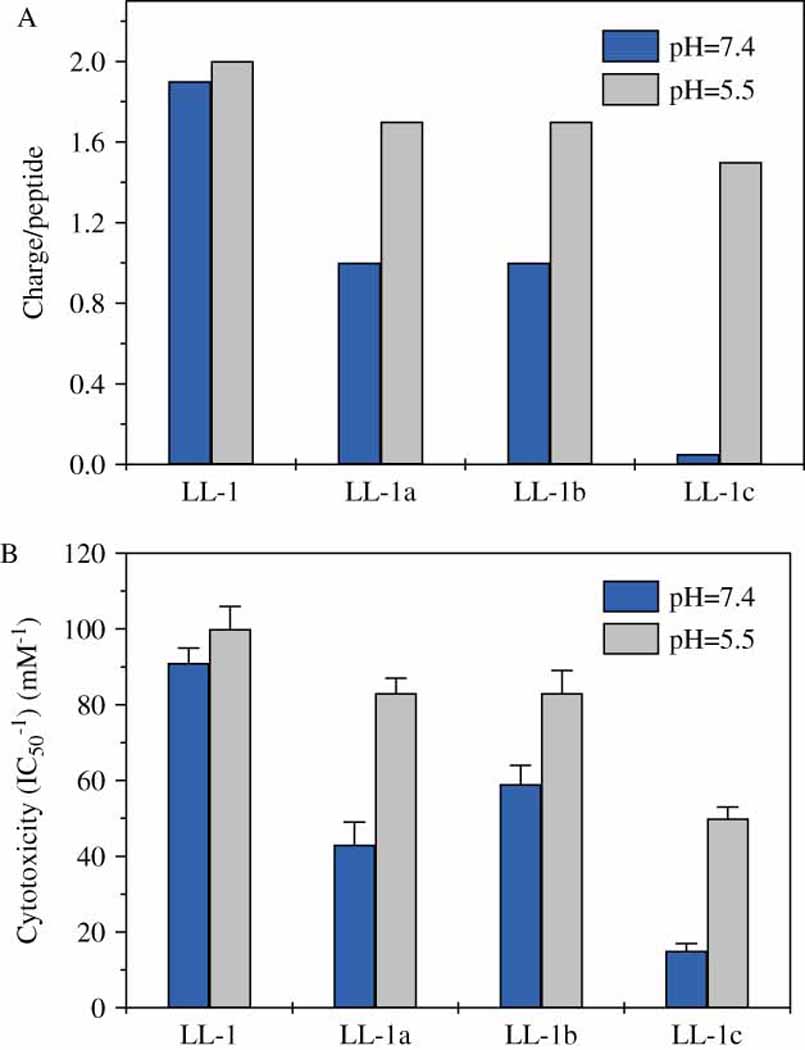
Comparison of pH-induced charge (A) and cytotoxicity (B) changes in peptides. The net positive charges of peptides were estimated using the isoelectric plot application from JaMBW 1.1.
To prove this conclusion, we examined cell-peptide interactions on an artificial system by measuring peptide binding to lipid vesicles induced tryptophan fluorescence peak shifts (Fig. 4A). In order to avoid the effects from pH-induced charge variations in lipid vesicles, we chose a lipid system (DPPC:DPPS:cholesterol = 20:1:4) which mimicked the lipid compositions of cell membranes but maintained constant surface charges (measured as surface Zeta potentials) in the pH range from 7.4 to 5.5 (Fig. 4B). Tryptophan fluorescence peak shifts resulted from peptide binding to lipid vesicles were plotted against lipid concentrations (Fig. 4C & Fig 4D). The plot slope represents the lipid membrane affinity of a specific peptide. As had expected, varied lipid membrane binding abilities were observed for peptide LL-1 and its histidine-containing derivatives. Peptide LL-1 showed the strongest lipid membrane binding ability at pH=7.4, followed by peptide LL-1a and LL-1b. Peptide LL-1c with two histidines gave the lowest lipid membrane affinity (Fig. 4C). At acidic pHs, all histidine-containing peptides showed improved lipid membrane binding ability. Since solution pH hardly affected the lipid membrane affinity of peptide LL-1, histidine-containing derivatives gave almost the same lipid membrane binding ability as peptide LL-1 at pH=5.5. This pH-affected membrane affinity changes fitted well with the pH sensitivity of histidine-containing peptides (Fig. 1), implying that pH-induced charge and thus cell affinity changes in peptides are responsible for the pH-dependent cytotoxicity of histidine-containing lytic peptides.
Figure 4.
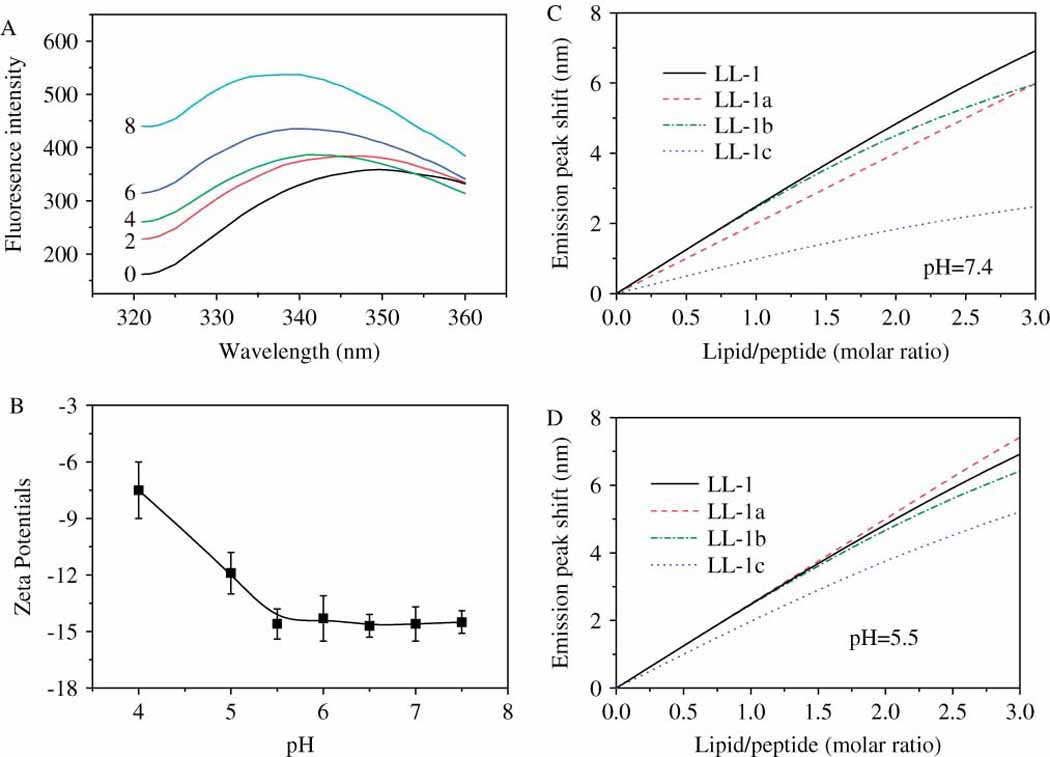
The pH-affected lipid membrane binding abilities of peptides. A, Peptide LL-1 binding to lipid vesicles associated fluorescence spectrum changes. Each curve was labeled by a number which represents the lipid/peptide (molar) ratio; B, Surface Zeta potentials (surface charges) of lipid vesicles at different pHs; C & D, Peptide binding to lipid vesicles as indicated tryptophan fluorescence peak (350nm) shifts at pH=7.4 and pH=5.5, respectively.
Discussions
In this study, we have demonstrated that we can construct pH-sensitive peptides by selectively replacing the corresponded lysines residues in a lytic peptide with histidines. There is a positive correlation between the pH sensitivity and the histidine substitution numbers in resulted histidine-containing peptides (Table 1 & Fig. 1). Despite the significant chemical property differences between peptide LL-1 and its histidine-containing derivatives, all these peptides have almost identical secondary structures in a wide pH range from pH=7.4 to pH=5.5 (Fig. 2 & Table 2). On the contrary, a good correlation between the cytotoxicity and pH-induced positive charge changes in histidine-containing peptides has been found (Fig.3). Results from lipid membrane binding studies suggest that the pH-dependent activities of histidine-containing peptides come from pH-affected histidine protonization and thus the cell affinity changes in peptides (Fig. 4). Indeed, the electrostatic attraction of peptides to cell membrane has proven to be the critical step in lytic peptides caused cell lysis. The concentration of a peptide of charge Z= +3 at the membrane surface could be 350-fold larger than that in bulk solution [23]. This is why almost all lytic peptides are positively charged (lysine or arginine rich) with very few exceptions. Replacement of lysine residues in lytic peptides with histidines will greatly decrease the cells affinity of these peptides at physiological pH of approximately 7.4 and thus explains significantly reduced cytotoxicity of histidine-containing lytic peptides (Table 1). As the imidazole groups in hisitidines are protonated under acidic conditions, hisitidine-containing lytic peptides resume their cell binding abilities and thus demonstrate pH-dependent activities. However, it should be pointed out that even at pH=5.5 and when the solution pH is lower than pKa (pKa =6.0) of imidazole groups, histidine residue will carry less than one unit of positive charge. Therefore, histidine-containing peptides have less net positive charges (1.7 units for LL1a and LL1b, and 1.5 units for LL-1c, respectively) compared to their parent peptide LL-1 (Table 1). This may explain the progressive activity loss of peptides with the increased histidine substitution numbers.
It has been demonstrated that the transition of the lytic peptides into the plane of binding and insertion into the lipid bilayer of cell membranes depends greatly on the hydrophobic/hydrophilic balance of the molecule groups and forces involved [24]. Under acidic conditions and when the imidazole group is protonated, histidine has very close interfacial free energy (ΔG = 0.96 kcal/mol) as lysine (ΔG = 0.99 kcal/mol). Therefore, histidine-containing peptides yielded in this study through lysine substitution may have the same or very close membrane partitioning property as peptide LL-1 at acidic pH. This explains why only lysine replacement by histidines leads to pH-sensitive peptides with reasonable cell lysis activities. Histidine substitution at positions other than lysine residues resulted in permanent activity loss in peptides easily (data not shown). To this regard, replacement of corresponded lysine residues with histidines may present a general strategy to create bioactive peptides with pH-dependent activities to meet the needs of various applications including cancer and bacterial infection treatments [9, 15]
Supplementary Material
Acknowledgements
This work was supported by the grant (GM081874) from National Institute of Health (NIH).
References
- 1.Otvos L., Jr The short proline-rich antibacterial peptide family. Cell Mol Life Sci. 2002;59:38–50. doi: 10.1007/s00018-002-8493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimarcq JL, Bulet P, Hetru C, Hoffmann J. Cysteine-rich antimicrobial peptides in invertebrates. Biopolymers. 1998;47:465–477. doi: 10.1002/(SICI)1097-0282(1998)47:6<465::AID-BIP5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 3.Brown KL, Hancock RE. Cationic host defense (antimicrobial) peptides. Curr Opin Immunol. 2006;18:24–30. doi: 10.1016/j.coi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Den Hertog AL, Wong Fong Sang HW, Kraayenhof R, Bolscher JG, Van't Hof W, Veerman EC, Nieuw Amerongen AV. Interactions of histatin 5 and histatin 5-derived peptides with liposome membranes: surface effects, translocation and permeabilization. Biochem J. 2004;379:665–672. doi: 10.1042/BJ20031785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Kan EJ, Demel RA, Breukink E, van der Bent A, de Kruijff B. Clavanin permeabilizes target membranes via two distinctly different pH-dependent mechanisms. Biochemistry. 2002;41:7529–7539. doi: 10.1021/bi012162t. [DOI] [PubMed] [Google Scholar]
- 6.Domingues MM, Lopes SC, Santos NC, Quintas A, Castanho MA. Fold-unfold transitions in the selectivity and mechanism of action of the N-terminal fragment of the bactericidal/permeability-increasing protein (rBPI(21) Biophys J. 2009;96(3):987–996. doi: 10.1016/j.bpj.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason AJ, Bertani P, Moulay G, Marquette A, Perrone B, Drake AF, Kichler A, Bechinger B. Membrane interaction of chrysophsin-1, a histidine-rich antimicrobial peptide from red sea bream. Biochemistry. 2007;46(51):15175–15187. doi: 10.1021/bi701344m. [DOI] [PubMed] [Google Scholar]
- 8.Mason AJ, Gasnier C, Kichler A, Prévost G, Aunis D, Metz-Boutigue MH, Bechinger B. Enhanced membrane disruption and antibiotic action against pathogenic bacteria by designed histidine-rich peptides at acidic pH. Antimicrob Agents Chemother. 2006;50(10):3305–3311. doi: 10.1128/AAC.00490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hait WN, Hambley TW. Targeted cancer therapeutics. Cancer Res. 2009;69(4):1263–1267. doi: 10.1158/0008-5472.CAN-08-3836. [DOI] [PubMed] [Google Scholar]
- 10.Engin K, Leeper DB, Cater JR, Thistlethwaite AJ, Tupchong L, McFarlane JD. Extracellular pH distribution in human tumours. Int J Hyperthermia. 1995;11:211–216. doi: 10.3109/02656739509022457. [DOI] [PubMed] [Google Scholar]
- 11.Raghunand N, He X, van Sluis R, Mahoney B, Baggett B, Taylor CW, Paine-Murrieta G, Roe D, Bhujwalla ZM, Gillies RJ. Enhancement of chemotherapy by manipulation of tumour pH. Br J Cancer. 1999;80:1005–1011. doi: 10.1038/sj.bjc.6690455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang N, Lejon T, Rekdal O. Antitumour activity and specificity as a function of substitutions in the lipophilic sector of helical lactoferrin-derived peptide. J Pept Sci. 2003;9(5):300–311. doi: 10.1002/psc.457. [DOI] [PubMed] [Google Scholar]
- 13.Wegener KL, Wabnitz PA, Carver JA, Bowie JH, Chia BC, Wallace JC, Tyler MJ. Host defense peptides from the skin glands of the Australian Blue Mountains tree-frog Litoria Citropa. Eur J Biochem. 1999;265:627–637. doi: 10.1046/j.1432-1327.1999.00750.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Kim SS, Bang YJ, Kim SJ, Lee BJ. In vitro activities of native and designed peptide antibiotics against drug sensitive and resistant tumor cell lines. Peptides. 2003;24(7):945–953. doi: 10.1016/s0196-9781(03)00194-3. [DOI] [PubMed] [Google Scholar]
- 15.Islam B, Khan SN, Khan AU. Dental caries: from infection to prevention. Med Sci Monit. 2007;13(11):RA196–RA203. [PubMed] [Google Scholar]
- 16.Liang JF, Yang VC. Synthesis of doxorubicin-peptide conjugates with multidrug resistant tumor cell killing activity. Bioorg Med Chem Lett. 2005;15:5071–5075. doi: 10.1016/j.bmcl.2005.07.087. [DOI] [PubMed] [Google Scholar]
- 17.Wei SY, Wu JM, Kuo YY, Chen HL, Yip BS, Tzeng SR, Cheng JW. Solution structure of a novel tryptophan-rich peptide with bidirectional antimicrobial activity. J Bacteriol. 2006;188:328–334. doi: 10.1128/JB.188.1.328-334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirkpatrick Daniel T, Guth Daniel J, Mavis Richard D. Detection of in vivo lipid peroxidation using the thiobarbituric acid assay for lipid hydroperoxides. J Biochem Toxicol. 2006;1:93–104. doi: 10.1002/jbt.2570010110. [DOI] [PubMed] [Google Scholar]
- 19.Liang JF, Yang VC, Vaynshteyn Y. The minimal functional sequence of protamine. Biochem Biophys Res Commun. 2005;336(2):653–659. doi: 10.1016/j.bbrc.2005.08.151. [DOI] [PubMed] [Google Scholar]
- 20.IUPAC-IUB Joint Commission on Biochemical Nomenclature. Nomenclature and symbolism for Amino Acids and Peptides. Eur. J. Biochem. 1984;138:9–37. doi: 10.1111/j.1432-1033.1984.tb07877.x. [DOI] [PubMed] [Google Scholar]
- 21.Tu Z, Hao J, Kharidia R, Meng XG, Liang JF. Improved stability and selectivity of lytic peptides through self-assembly. Biochem Biophys Res Commun. 2007;361(3):712–717. doi: 10.1016/j.bbrc.2007.06.178. [DOI] [PubMed] [Google Scholar]
- 22.Lakowicz JR. Principles of Fluorescence Spectroscopy (2006) 3rd ed. New York: Springer; 2006. pp. 278–317. [Google Scholar]
- 23.Terzi E, Hölzemann G, Seelig J. Interaction of Alzheimer betaamyloid peptide(1–40) with lipid membranes. Biochem. 1997;38:14845–14852. doi: 10.1021/bi971843e. [DOI] [PubMed] [Google Scholar]
- 24.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


