Abstract
Nanoparticles are finding utility in myriad biotechnological applications, including gene regulation, intracellular imaging, and medical diagnostics. Thus, evaluating the biocompatibility of these nanomaterials is imperative. Here we use genome-wide expression profiling to study the biological response of HeLa cells to gold nanoparticles functionalized with nucleic acids. Our study finds that the biological response to gold nanoparticles stabilized by weakly bound surface ligands is significant (cells recognize and react to the presence of the particles) yet when these same nanoparticles are stably functionalized with covalently attached nucleic acids, the cell shows no measurable response. This finding is important for researchers studying and using nanomaterials in biological settings, as it demonstrates how slight changes in surface chemistry and particle stability can lead to significant differences in cellular responses.
Keywords: gold, nanoparticle, nucleic acid, oligonucleotide, DNA, RNA, gene regulation, biocompatibility, toxicity
Introduction
Nanogold has been employed for various therapeutic applications for many years.1-4 Moreover, a wide range of nanomaterials have been recently developed and the unique properties exhibited by these novel materials often make them ideal for therapeutic and diagnostic applications. With the advent of this new class of materials, it is important to carefully characterize the biocompatibility and safety of the nanomaterials if they are to be used for medical purposes.5-8 One nanomaterial that is particularly promising for therapeutic and diagnostic applications is the polyvalent oligonucleotide-gold nanoparticle. Consisting of a gold core densely functionalized with DNA or RNA surface ligands, this nanomaterial exhibits many surprising properties as a result of its high surface density. These properties include increased binding affinities for complementary nucleic acids,9 avid cellular uptake,10 nuclease resistance,11 and a limited immune response.12, 13 Nanoparticles allow for the design and synthesis of complex, multifunctional materials.14, 15 In comparison to small molecule drugs, the multifunctional potential and tailorability of nanomaterials have made them extremely attractive for pharmaceutical development.16, 17 Indeed, nanoparticles conjugated with various biomolecules have proven useful in applications of gene regulation12, 18, 19 and intracellular detection20-22 and the highly selective and sensitive detection of biomarkers in complex biological fluids, further demonstrating their value in therapeutic and diagnostic applications.16, 17 To date, we have not observed any detectable toxicity associated with cultured mammalian cells or with mouse models following introduction of densely functionalized oligonucleotide-gold nanoparticle conjugates. However, research investigating the biological potential of various nanomaterials has shown that the chemistry on the surface of the nanoparticle can have significant effects on the biological response of the nanoparticles,23-26 The results of these studies have led us to specifically investigate the biocompatibility of this exceptional nanomaterial. Genome-wide expression profiling studies are often used to examine the cellular effects of therapeutic agents as they provide a detailed analysis of the complex interactions occurring inside the cell.27, 28 As such, we use gene expression profiling in this study to compare how HeLa cells respond to citrate stabilized nanoparticles, nanoparticles densely functionalized with nucleic acids, and protein coated nanoparticles. We further demonstrate the impact a nanomaterials surface chemistry can have on its stability and subsequent effect on biocompatibility and identify a striking example of how a subtle difference in composition can have a substantial impact on the biological response of nanomaterials.
Results and Discussion
In our experiments we use 15 nm gold nanoparticles that are either stabilized with electrostatically bound citrate molecules or functionalized with covalently bound single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), double-stranded RNA (dsRNA) or coated with bovine serum albumin (BSA). Each of the nanoparticle types used in this study were characterized in terms of size and zeta potential (Table S1). Dynamic light scattering determined the size of the citrate stabilized particles to be approximately 17 nm in diameter, and following adsorption of the BSA the particles increased in size to approximately 74 nm. The nucleic acid functionalized particles increased in size to approximately 26 nm, which is to be expected based on the length of the oligonucleotides. Zeta potential measurements indicate the surface potential of the citrate stabilized particles to be -32.3 ± 1.6 mV. Following adsorption of the BSA the surface potential became slightly more negative (-34.3 ± 0.8 mV) and the ssDNA (-30.7 ± 1.2 mV), dsDNA (28.3 ± 1.6 mV), and dsRNA (-28.8 ± 2.6 mV) functionalized particles became slightly less negative due to a high concentration of sodium ions surrounding the particles which screen the negative charges between the nucleic acids and allow dense loading on the particle surface. The nucleic acid functionalized nanoparticles were specifically chosen as we have previously demonstrated that these nanoconjugates are biologically relevant and able to function in a cellular environment in terms of gene regulation12, 19 and intracellular detection.20 The citrate stabilized nanoparticles were tested as they serve as the starting material for the nanoconjugates before the functional groups are attached, and the BSA coated nanoparticles were chosen as a control as they are stable under cell culture conditions but are internalized by cells to a lesser extent than oligonucleotide functionalized nanoparticles. In three separate experiments, HeLa cells were treated with each of the nanoparticle types for 24 hours at a concentration of 10 nM. This concentration of nanoparticles was chosen based on previous studies in which the nanoparticles were functional at 10 nM concentrations or less. To avoid confusion between downstream changes in gene expression as a result of knocking down a target gene and changes in expression caused by nanoparticle treatment, non-targeting DNA and RNA sequences were used in this study. Following nanoparticle treatment, genome-wide expression analysis of the three replicate experiments revealed that 15 nm citrate stabilized gold nanoparticles induce significant changes in the gene expression profile of HeLa cells that have been exposed to a 10 nM concentration of nanoparticles (Figure 1). 127 genes (119 down-regulated; 8 up-regulated) were identified as being differentially expressed using a 1.5 fold change in gene expression and a p-value less than 0.05 as a threshold (Table 1 and S2). When these same 15 nm gold nanoparticles are functionalized with ssDNA, dsDNA, dsRNA, or BSA (thereby displacing the weakly bound citrate) there were no significant changes observed in the gene expression profile using the same threshold conditions.
Figure 1.
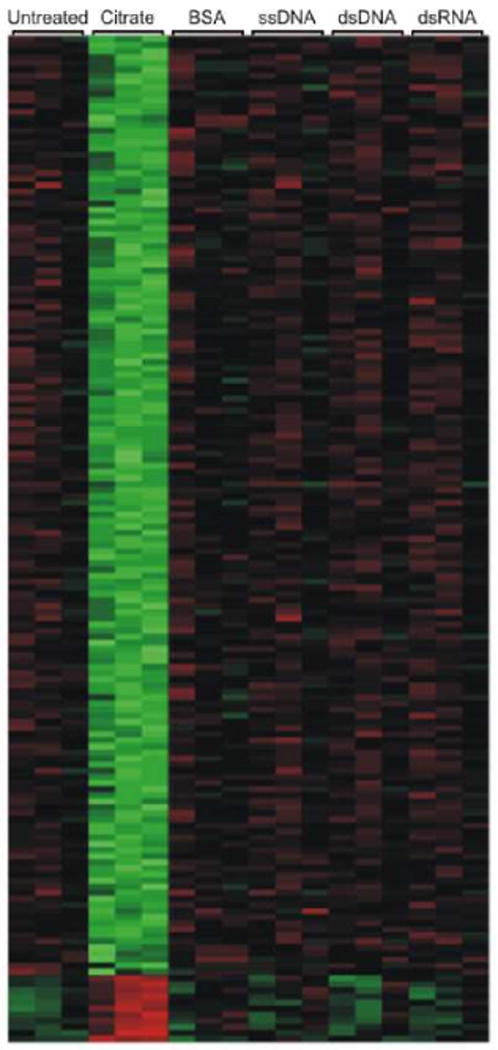
A whole-genome expression analysis of HeLa cells following a 24 hour treatment period with 10 nM gold nanoparticles that were functionalized with various surface ligands (citrate, bovine serum albumin (BSA), single-stranded DNA, double-stranded DNA, double-stranded RNA). Labeled columns represent untreated and nanoparticle treated cells for three replicate experiments. This heat map depicts the similarity between the expression profiles for the different treatment conditions. The relative gene expression levels are denoted by green (low copy number), red (high copy number), and black (equal copy number).
Table 1.
Categories of genes identified in Table S1 following 24 hour treatment with 10 nM citrate stabilized gold nanoparticles.
| Gene Category | Number of Genes |
|---|---|
| Small Molecule Biochemistry | 52 |
| Molecular Transport | 14 |
| Cell Morphology | 12 |
| Drug Metabolism | 8 |
| RNA Post-Transcriptional Modification | 4 |
| Other | 37 |
We performed cell-cycle analysis and quantified the induction of apoptosis to supplement the results of the gene expression profiling. An analysis of cell-cycle progression demonstrates that the HeLa cell population begins to shift into the G1 and S phase with limited progression into the M phase after 24 hours of treatment with 10 nM citrate stabilized nanoparticles.29 Nucleic acid functionalized and BSA coated particles, however, show no effect on cell-cycle progression (Figure 2). Consistent with both the gene expression profiling and the cell-cycle analysis, the results of an annexin assay (measuring the degree of apoptosis) also show an increased cell population in an early apoptotic stage following treatment with citrate stabilized nanoparticles while no changes were observed following treatment with nucleic acid functionalized nanoparticles or BSA coated nanoparticles (Figure 3).
Figure 2.
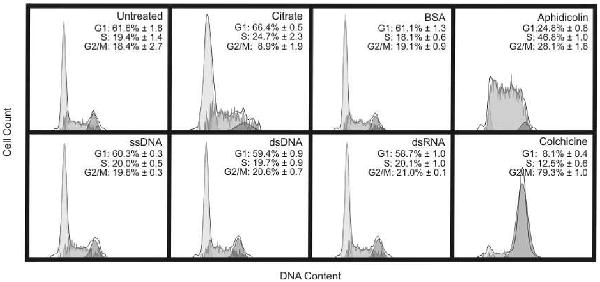
Cell-cycle analysis of HeLa cells following a 24 hour treatment period with 10 nM gold nanoparticles that were functionalized with various surface ligands (citrate, bovine serum albumin, single-stranded DNA, double-stranded DNA, double-stranded RNA). As a control, the cells were also treated with 0.1 μM colchicine to arrest the cells in G2/M phase, or with 0.5 μM aphidicolin to arrest the cells in S phase. Data from three separate experiments ± standard deviation are reported and a representative histogram from one of the three experiments is depicted in the figure.
Figure 3.
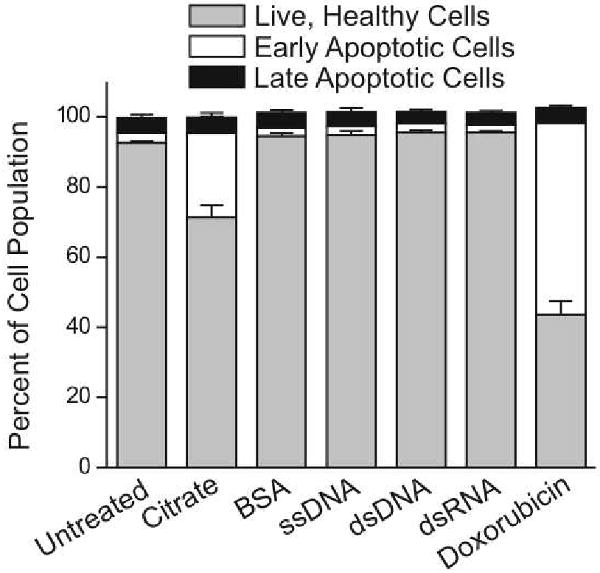
Quantification of apoptosis induction of HeLa cells following a 24 hour treatment period with 10 nM gold nanoparticles that were functionalized with various surface ligands (citrate, bovine serum albumin, single-stranded DNA, double-stranded DNA, double-stranded RNA). As a control, the cells were also treated with 0.5 μM doxorubicin to induce apoptosis. Data from three separate experiments ± standard deviation are reported.
The surface chemistry on the nanoparticle can greatly affect the rate of cellular uptake for the nanoconjugates.10 We measured the cellular uptake for each type of nanoparticle to ensure that the changes induced by the citrate stabilized gold nanoparticles were not due to differences in the ability of the various nanoparticles to enter cells (Figure 4). The results of this experiment indicate that the BSA coated nanoparticles had the lowest level of uptake, which may explain the lack of changes in gene expression for this condition, but the nucleic acid functionalized particles enter the cells to a greater extent than the citrate stabilized particles. We originally hypothesized that nucleic acid functionalized nanoparticles would cause the greatest changes in gene expression as we predicted they would have the highest cellular uptake. However, the citrate stabilized nanoparticles caused the most changes in gene expression, which we attribute to the instability of these particles and the interaction of the aggregated particles with the cells. It is unclear why the dsDNA functionalized nanoparticles to do not enter the cells as readily as the ssDNA or the dsRNA, however, determining the mechanism by which the nanoparticles are internalized is currently an area of active research. Nonetheless, the ssDNA, dsDNA and dsRNA functionalized nanoparticles entered the cells to the same or greater extent as the citrate stabilized particles, yet in contrast to the citrate stabilized particles, both the nucleic acid functionalized particles and the BSA coated nanoparticles caused no significant changes in gene expression, cell-cycle progression or apoptosis induction.
Figure 4.
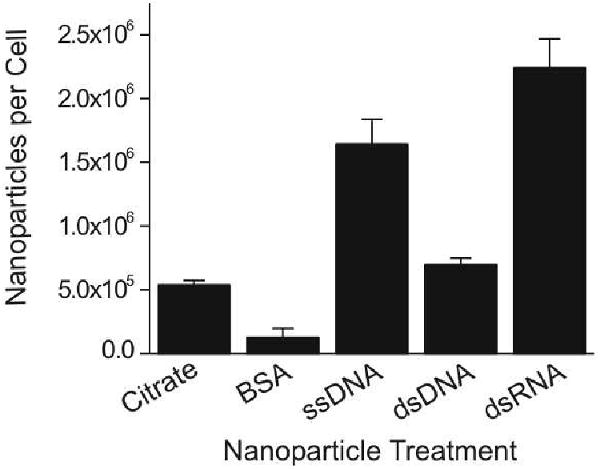
Quantification of nanoparticle uptake by HeLa cells following a 24 hour treatment period with 10 nM gold nanoparticles that were functionalized with various surface ligands (citrate, bovine serum albumin, single-stranded DNA, double-stranded DNA, double-stranded RNA). Data from three separate experiments ± standard deviation are reported.
The citrate stabilized nanoparticles, which act as the starting material for nucleic acid functionalized nanoparticles, show a significant cellular response, while cellular treatment with nucleic acid or BSA functionalized nanoparticles caused no detectable changes in gene expression, cell-cycle progression, or apoptosis induction. The changes associated with the citrate stabilized nanoparticles are likely due to the citrate molecule's weak association with the gold nanoparticle core. Indeed, in cell culture medium the citrate stabilized nanoparticles quickly aggregate, indicated by a red shift in their plasmon resonance (Figure S1) and a concomitant color change from red to purple, and settle to the bottom of the culture flask over time (Figure S2, S3). As a result, the citrate stabilized nanoparticles coat the surface of the cells with a layer of aggregated nanoparticles and do not interact with the cells as discreet particles. In contrast, nucleic acid or BSA functionalized nanoparticles remain stable over the course of the 24 hour treatment period. Thus, the chemistry of surface ligand attachment to the nanoparticle surface and the ability of the ligand to maintain nanomaterial stability seem to be principal factors in determining biocompatibility.
Conclusions
Although these results may not be consistent across different cell types or in an animal model where different cell types are able to interact with and signal to each other, they suggest there are limited off-target effects resulting when nucleic acid functionalized gold nanoparticles are used in a cell culture model under treatment conditions that have been demonstrated to be functionally relevant.12, 19, 20 This study provides encouraging results for the continued development of densely functionalized polyvalent oligonucleotide-gold nanoparticle conjugates for therapeutic and diagnostic applications. These results are especially promising in regards to nucleic acid based therapies as traditional methods of introducing nucleic acids to the intracellular environment are limited by their cytotoxic and off-target effects.30-32 Additionally, in agreement with other studies23-26 investigating the biocompatibility of various nanomaterials for therapeutic and diagnostic applications, this work further highlights in a dramatic manner how making a “small change”, to a nanoparticle, such as the use of a different surface ligand, can significantly impact its biological response. Researchers developing nanomaterials for biological applications will need to carefully examine the materials not just in terms of their size and shape, but also their surface functionalization making it difficult to draw general conclusions about a single class of materials.
Methods
Nucleic Acid Synthesis
DNA was synthesized using an Expedite 8909 Nucleotide Synthesis System (ABI) using solid-phase phosphoramidite chemistry. RNA was synthesized using a MerMade 6 (Bioautomation) and 2-O-TriisopropylsilylOxyMethyl (TOM) -protected RNA bases. Bases and reagents were purchased from Glen Research. Oligonucleotides were purified using published methods.33 After purification, oligonucleotides were lyophilized and stored at -80°C until use. DNA Sequence: 5′ - GAG CTG CAC GCT GCC GTC AAA AAA AAA A(thiol) – 3′, RNA sequence: 5′ - GAG CUG CAC GCU GCC GUC AAA AAA AAA A(thiol) – 3′.
Nanoparticle Synthesis and Functionalization
Citrate-stabilized gold nanoparticles (∼15 nm) were prepared using published methods.33 First, the colloid was adjusted to 0.3% SDS (sodium dodecyl sulfate) and 0.01 M phosphate buffer, pH 7.4. A 1:1 ratio of each sequence and its complement was allowed to hybridize in phosphate buffered saline (0.5 M NaCl) at 70°C for one hour and then slowly cooled to room temperature. Thiol-modified single-stranded, duplex DNA or duplex RNA was added to the 15 nm citrate-stabilized nanoparticles (approximately 1.5 nmol oligonucleotide per 1 mL of 10 nM gold colloid). After 30 minutes of gentle mixing, 2.0 M NaCl in nanopure water was added to bring the NaCl concentration to 0.05 M and the mixture was sonicated for 20 seconds. Two more additions of 2.0 M NaCl were added in 30 minute intervals, each followed by sonication, to bring the mixture to a final concentration to 0.15 M NaCl. For RNA particles, following the third salt addition, OEG-thiol, (1-mercaptoundec-11-yl)tri(ethylene glycol), was added to create a 30 μM final concentration. The final mixture was gently shaken for 24 hours to complete the functionalization process. The particles were centrifuged (13000 rpm, 20 minutes; 3×) and resuspended in phosphate buffered saline (PBS).
Cell Culture and Transfection
HeLa cells were grown in 5% CO2 at 37°C in Minimal Essential Medium (EMEM) that was supplemented with 10% heat-inactivated fetal bovine serum (FBS) and penicillin and streptomycin. Cells were plated and grown to a density of approximately 80% confluence, cell culture media was removed and replaced with nanoparticle containing medium for a 24 hour treatment period.
Gene Expression Analysis
RNA expression analysis was performed using the Illumina Human HT-12 BeadChip, which provides coverage of over 48,802 genes and expressed sequence tags. HeLa cells were treated with 10 nM concentrations of specified nanoparticle types for 24 hours. Total RNA was extracted using TRIzol (Invitrogen) following manufacturer's recommended protocol. Extracted RNA was processed using an RNeasy MinElute Cleanup Kit (Qiagen). High quality RNA was labeled using a commercial kit (TargetAmp 1-Round Aminoallyl-aRNA Kit; Epicentre, Madison, WI, USA). Labeled RNA was next hybridized to microarrays (Human HT-12 BeadChip; Illumina, San Diego, CA). Raw signal intensities of each probe were obtained using data analysis software (Beadstudio; Illumina) and imported to the Lumi package of Bioconductor for data transformation and normalization.34-36 Differentially expressed genes were identified using an Analysis of Variance (ANOVA) model with empirical Bayesian variance estimation.37 The problem of multiple comparisons was corrected using the false discovery rate (FDR). Initially, genes were identified as being differentially expressed on the basis of a statistically significant (raw p-value < 0.05), FDR < 5% and 1.5-fold change (up or down) in expression level in different nanoparticle treatment samples.
Cellular Assays
HeLa cells were treated for 24 hours with 10 nM concentrations of specified nanoparticle types or with 0.1 μM colchicine,38 0.5 μM aphidicolin,39 or 0.5 μM doxorubicin as controls. After treatment, the medium was removed and a solution of 150 mM KI and 25 mM I2 in PBS was added to the cells for approximately 2 minutes to dissolve any non-internalized, cell surface-bound, nanoparticles.40 The cells were then washed with PBS, collected, and fixed and stained using Guava's Cell Cycle Reagent following manufacturer's recommended protocol, or stained to quantify induction of apoptosis using Guava's Nexin Reagent following manufacturer's recommended protocol. A Guava EasyCite Mini flow cytometer was used to run cellular assays (Guava Technologies).
Nanoparticle Uptake
HeLa cells were treated with 10 nM concentrations of specified nanoparticle types for 24 hours. After treatment, the medium was removed and a solution of 150 mM KI and 25 mM I2 in PBS was added to the cells for approximately 2 minutes to dissolve any non-internalized nanoparticles. The cells were then washed with 1× PBS, collected, and counted using a Guava EasyCyte Mini flow cytometer (Guava Technologies). Uptake quantification was accomplished using inductively coupled plasma mass spectrometry (ICP-MS (Thermo-Fisher). To prepare samples for ICP-MS, the cells were dissolved with nitric acid at 60° C overnight, diluted in a matrix consisting of 3% HNO3 and 1 ppb Indium (internal standard). The number of nanoparticles in each sample was calculated based on the concentration of Au found in the sample.10
Supplementary Material
Acknowledgments
We thank the Genomics Core at Northwestern University's Center for Genomic Medicine for analyzing the RNA quality, synthesizing and labeling the cRNA, hybridizing the labeled cRNA, and scanning the microarray bead chip. We also thank the Bioinformatics Core at Northwestern University for performing the statistical analysis for the gene expression profiling. C. A. M. acknowledges a Cancer Center for Nanotechnology Excellence (NCI CCNE) award for support of this research. C.A.M. is also grateful for the NIH funded Skin Disease Research Center (NU-SDRC – Award No. 1P30AR057216). He is also grateful for an NSSEF Fellowship from the DoD.
Footnotes
Supporting Information Available. Comprehensive list of gene names, gene ID numbers, fold change and associated p-values for genes identified as being differentially expressed, UV-Vis and TEM characterization of nanoparticles, and images of cells following nanoparticle treatment. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Tsai CY, Shiau AL, Chen SY, Chen YH, Cheng PC, Chang MY, Chen DH, Chou CH, Wang CR, Wu CL. Amelioration of Collagen-Induced Arthritis in Rats by Nanogold. Arthritis and Rheumatism. 2007;56(2):544–554. doi: 10.1002/art.22401. [DOI] [PubMed] [Google Scholar]
- 2.Jaeger GT, Larsen S, Soli N, Moe L. Two Years Follow-up Study of the Pain-Relieving Effect of Gold Bead Implantation in Dogs with Hip-Joint Arthritis. Acta Vet Scand. 2007;49:9. doi: 10.1186/1751-0147-49-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown CL, Whitehouse MW, Tiekink ER, Bushell GR. Colloidal Metallic Gold Is Not Bio-Inert. Inflammopharmacology. 2008;16(3):133–7. doi: 10.1007/s10787-007-0017-6. [DOI] [PubMed] [Google Scholar]
- 4.Freyberg RH, Block WD, Metabolism Levey S. Toxicity and Manner of Action of Gold Compounds Used in the Treatment of Arthritis. I. Human Plasma and Synovial Fluid Concentration and Urinary Excretion of Gold During and Following Treatment with Gold Sodium Thiomalate, Gold Sodium Thiosulfate, and Colloidal Gold Sulfide. J Clin Invest. 1941;20(4):401–12. doi: 10.1172/JCI101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall JB, Dobrovolskaia MA, Patri AK, McNeil SE. Characterization of Nanoparticles for Therapeutics. Nanomedicine. 2007;2(6):789–803. doi: 10.2217/17435889.2.6.789. [DOI] [PubMed] [Google Scholar]
- 6.Jan E, Byrne SJ, Cuddihy M, Davies AM, Volkov Y, Gun'ko YK, Kotov NA. High-Content Screening as a Universal Tool for Fingerprinting of Cytotoxicity of Nanoparticles. ACS Nano. 2008;2(5):928–938. doi: 10.1021/nn7004393. [DOI] [PubMed] [Google Scholar]
- 7.Shaw SY, Westly EC, Pittet MJ, Subramanian A, Schreiber SL, Weissleder R. Perturbational Profiling of Nanomaterial Biologic Activity. Proc Natl Acad Sci U S A. 2008;105(21):7387–92. doi: 10.1073/pnas.0802878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nel A, Xia T, Madler L, Li N. Toxic Potential of Materials at the Nanolevel. Science. 2006;311(5761):622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 9.Jin R, Wu G, Li Z, Mirkin CA, Schatz GC. What Controls the Melting Properties of DNA-Linked Gold Nanoparticle Assemblies? J Am Chem Soc. 2003;125(6):1643–54. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- 10.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Oligonucleotide Loading Determines Cellular Uptake of DNA-Modified Gold Nanoparticles. Nano Lett. 2007;7(12):3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Lett. 2009;9(1):308–11. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science. 2006;312(5776):1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 13.Massich MD, Giljohann DA, Seferos DS, Ludlow LE, Horvath CM, Mirkin CA. Regulating Immune Response Using Polyvalent Nucleic Acid-Gold Nanoparticle Conjugates. Mol Pharm. 2009 doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature. 1996;382(6592):607–9. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 15.Park SY, Lytton-Jean AK, Lee B, Weigand S, Schatz GC, Mirkin CA. DNA-Programmable Nanoparticle Crystallization. Nature. 2008;451(7178):553–6. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 16.Rosi NL, Mirkin CA. Nanostructures in Biodiagnostics. Chem Rev. 2005;105(4):1547–62. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 17.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Gold Nanoparticles for Biology and Medicine. Angew Chem Int Ed Engl. 2010;49(19):3280–94. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel PC, Giljohann DA, Seferos DS, Mirkin CA. Peptide Antisense Nanoparticles. Proc Natl Acad Sci U S A. 2008;105(45):17222–6. doi: 10.1073/pnas.0801609105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene Regulation with Polyvalent siRNA-Nanoparticle Conjugates. J Am Chem Soc. 2009;131(6):2072–3. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-Flares: Probes for Transfection and Mrna Detection in Living Cells. J Am Chem Soc. 2007;129(50):15477–+. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. Nano-Flares for Mrna Regulation and Detection. ACS Nano. 2009;3(8):2147–52. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Aptamer Nano-Flares for Molecular Detection in Living Cells. Nano Lett. 2009;9(9):3258–61. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clift MJD, Rothen-Rutishauser B, Brown DM, Duffin R, Donaldson K, Proudfoot L, Guy K, Stone V. The Impact of Different Nanoparticle Surface Chemistry and Size on Uptake and Toxicity in a Murine Macrophage Cell Line. Toxicol Appl Pharmacol. 2008;232(3):418–427. doi: 10.1016/j.taap.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Goodman CM, McCusker CD, Yilmaz T, Rotello VM. Toxicity of Gold Nanoparticles Functionalized with Cationic and Anionic Side Chains. Bioconjugate Chem. 2004;15(4):897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- 25.Hauck TS, Ghazani AA, Chan WCW. Assessing the Effect of Surface Chemistry on Gold Nanorod Uptake, Toxicity, and Gene Expression in Mammalian Cells. Small. 2008;4(1):153–159. doi: 10.1002/smll.200700217. [DOI] [PubMed] [Google Scholar]
- 26.Alkilany AM, Nagaria PK, Hexel CR, Shaw TJ, Murphy CJ, Wyatt MD. Cellular Uptake and Cytotoxicity of Gold Nanorods: Molecular Origin of Cytotoxicity and Surface Effects. Small. 2009;5(6):701–8. doi: 10.1002/smll.200801546. [DOI] [PubMed] [Google Scholar]
- 27.Clarke PA, te Poele R, Workman P. Gene Expression Microarray Technologies in the Development of New Therapeutic Agents. Eur J Cancer. 2004;40(17):2560–91. doi: 10.1016/j.ejca.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham MJ, Liang S, Fuhrman S, Seilhamer JJ, Somogyi R. Gene Expression Microarray Data Analysis for Toxicology Profiling. Ann N Y Acad Sci. 2000;919:52–67. doi: 10.1111/j.1749-6632.2000.tb06867.x. [DOI] [PubMed] [Google Scholar]
- 29.Meikrantz W, Schlegel R. Apoptosis and the Cell Cycle. J Cell Biochem. 1995;58(2):160–74. doi: 10.1002/jcb.240580205. [DOI] [PubMed] [Google Scholar]
- 30.Omidi Y, Hollins AJ, Benboubetra M, Drayton R, Benter IF, Akhtar S. Toxicogenomics of Non-Viral Vectors for Gene Therapy: A Microarray Study of Lipofectin- and Oligofectamine-Induced Gene Expression Changes in Human Epithelial Cells. Journal of Drug Targeting. 2003;11(6):311–323. doi: 10.1080/10611860310001636908. [DOI] [PubMed] [Google Scholar]
- 31.Omidi Y, Barar J, Heidari HR, Ahmadian S, Yazdi HA, Akhtar S. Microarray Analysis of the Toxicogenomics and the Genotoxic Potential of a Cationic Lipid-Based Gene Delivery Nanosystem in Human Alveolar Epithelial A549 Cells. Toxicol Mech Methods. 2008;18(4):369–378. doi: 10.1080/15376510801891286. [DOI] [PubMed] [Google Scholar]
- 32.Akhtar S, Benter I. Toxicogenomics of Non-Viral Drug Delivery Systems for Rnai: Potential Impact on Sirna-Mediated Gene Silencing Activity and Specificity. Adv Drug Delivery Rev. 2007;59(2-3):164–182. doi: 10.1016/j.addr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-Pot Colorimetric Differentiation of Polynucleotides with Single Base Imperfections Using Gold Nanoparticle Probes. J Am Chem Soc. 1998;120(9):1959–1964. [Google Scholar]
- 34.Du P, Kibbe WA, Lin SM. Lumi: A Pipeline for Processing Illumina Microarray. Bioinformatics (Oxford, England) 2008;24(13):1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 35.Lin SM, Du P, Huber W, Kibbe WA. Model-Based Variance-Stabilizing Transformation for Illumina Microarray Data. Nucleic acids research. 2008;36(2):e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du P, Kibbe WA, Lin SM. Nuid: A Universal Naming Scheme of Oligonucleotides for Illumina, Affymetrix, and Other Microarrays. Biology direct. 2007;2:16. doi: 10.1186/1745-6150-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wettenhall JM, Smyth GK. Limmagui: A Graphical User Interface for Linear Modeling of Microarray Data. Bioinformatics. 2004;20(18):3705–6. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 38.Prather RS, Boquest AC, Day BN. Cell Cycle Analysis of Cultured Porcine Mammary Cells. Cloning. 1999;1(1):17–24. doi: 10.1089/15204559950020067. [DOI] [PubMed] [Google Scholar]
- 39.Pedrali-Noy G, Spadari S, Miller-Faures A, Miller AO, Kruppa J, Koch G. Synchronization of Hela Cell Cultures by Inhibition of DNA Polymerase Alpha with Aphidicolin. Nucleic Acids Res. 1980;8(2):377–87. doi: 10.1093/nar/8.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho EC, Xie J, Wurm PA, Xia Y. Understanding the Role of Surface Charges in Cellular Adsorption Versus Internalization by Selectively Removing Gold Nanoparticles on the Cell Surface with a I2/Ki Etchant. Nano Lett. 2009;9(3):1080–4. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


