Crystal structure of a prokaryotic homologue of the mammalian oligopeptide–proton symporters, PepT1 and PepT2
This study presents the crystal structure of a prokaryotic oligopeptide–proton symporter, homologous to mammalian peptide transporters, giving insights into the molecular mechanism by which this important family of proteins act.
Keywords: major facilitator superfamily transporter, occluded state, peptide transport
Abstract
PepT1 and PepT2 are major facilitator superfamily (MFS) transporters that utilize a proton gradient to drive the uptake of di- and tri-peptides in the small intestine and kidney, respectively. They are the major routes by which we absorb dietary nitrogen and many orally administered drugs. Here, we present the crystal structure of PepTSo, a functionally similar prokaryotic homologue of the mammalian peptide transporters from Shewanella oneidensis. This structure, refined using data up to 3.6 Å resolution, reveals a ligand-bound occluded state for the MFS and provides new insights into a general transport mechanism. We have located the peptide-binding site in a central hydrophilic cavity, which occludes a bound ligand from both sides of the membrane. Residues thought to be involved in proton coupling have also been identified near the extracellular gate of the cavity. Based on these findings and associated kinetic data, we propose that PepTSo represents a sound model system for understanding mammalian peptide transport as catalysed by PepT1 and PepT2.
Introduction
The absorption of dietary nitrogen in the form of peptides by plasma membrane transporters belonging to the solute carrier (SLC) 15 family is essential for human health. Evolutionarily, these transporters form part of the widely distributed proton-dependent oligopeptide transporter (POT) family (TC 2.A.17), also referred to as the peptide transporter or PTR2 family (Paulsen and Skurray, 1994; Steiner et al, 1995), members of which transport peptides, amino acids and nitrate (Huang et al, 1999). They are proton-driven symporters, and in both eukaryotes and prokaryotes use the inwardly directed proton (H+) electrochemical gradient to drive the uptake of peptides across cell membranes (Ganapathy and Leibach, 1983; Daniel et al, 2006). Human PepT1 (SLC15A1) is found predominantly in the small intestine, whereas PepT2 (SLC15A2) is found in the kidney, the lungs and central nervous system (Daniel and Kottra, 2004). PepT1 and PepT2 are predicted to contain 12 transmembrane (TM) helices with both N- and C-termini facing the cytoplasm, as is typical for major facilitator superfamily (MFS) members (Fei et al, 1994; Covitz et al, 1998). PepT1 is a high-capacity, low-affinity transporter and is the main route for dietary peptide uptake, whereas PepT2 operates as a low-capacity, high-affinity transporter, thought to mediate more selective transport in the kidney and other tissues (Terada et al, 1997; Doring et al, 2002; Daniel and Kottra, 2004; Biegel et al, 2006). In addition to peptides, the human proteins transport a broad spectrum of orally administered drugs, including the β-lactam antibiotics (Wenzel et al, 1995; Tamai et al, 1997; Faria et al, 2004), and are under active clinical investigation to improve the pharmacokinetic properties of antivirals such as valacyclovir (Ganapathy et al, 1998) and the vasopressor midodrine (Tsuda et al, 2006).
To study the function of the mammalian transporters, a number of distantly related prokaryotic homologues with similar substrate specificities have been employed as model systems (Hagting et al, 1994; Harder et al, 2008; Ernst et al, 2009). Here, we report the crystal structure of a peptide transporter from the bacterium Shewanella oneidensis, PepTSo, which shows a high degree of sequence conservation within the TM region (∼30% identity) to the mammalian PepT1 and PepT2 proteins. All previously identified residues proposed to be functionally important in the mammalian transporters are conserved, including a critical histidine residue (Uchiyama et al, 2003) (His57 in human PepT1) (Supplementary Figure S1). The structure of PepTSo reveals important information concerning the spatial arrangement of residues involved in peptide and drug transport as catalysed by mammalian peptide transporters. In addition, it represents a ligand-bound occluded conformation for an MFS symporter, providing fresh insight into the alternating access model of membrane transport.
Results
Structure of PepTSo
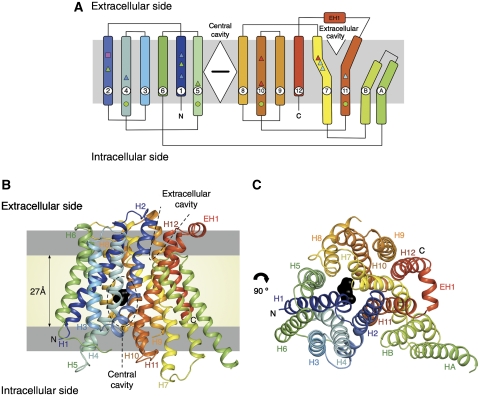
PepTSo contains 14 TM helices (Figure 1A), of which helices H1–H12 adopt the overall fold observed previously for the MFS transporters LacY, GlpT and EmrD (Figure 1B) (Abramson et al, 2003; Huang et al, 2003; Yin et al, 2006). The arrangement of the helices is also consistent with the EM projection structure of a previously studied POT protein, DtpD from Escherichia coli (Casagrande et al, 2009). Like previous MFS transporter structures, the N- and C-terminal six-helix bundles, formed by helices H1–H6 and H7–H12, come together to form a ‘V'-shaped transporter, related by a pseudo two-fold symmetry axis running perpendicular to the membrane plane. PepTSo has two additional TM helices, HA and HB, which are inserted into the cytoplasmic loop connecting the N- and C-terminal bundles. These form a hairpin-like structure in the membrane that packs against the periphery of the protein (Figure 1C). Their role is currently unclear. The apparent absence of these helices in the fungal, plant and metazoan protein sequences, however, suggests they do not contribute to any conserved transport mechanism. The position of PepTSo within the membrane has been examined using coarse-grained lipid bilayer self-assembly simulations (Scott et al, 2008). These demonstrate that PepTSo, including the hairpin helices HA and HB, reproducibly inserts into a modelled bilayer (Supplementary Figure S2).
Figure 1.
Structure of PepTSo. (A) PepTSo topology. The central and extracellular cavities are shown as a closed diamond and open triangle, respectively. A bound ligand in the central cavity is represented as a black horizontal bar. Functionally important residues conserved between PepTSo and metazoan peptide transporters are highlighted by shapes in Supplementary Figure S1 and mapped onto the topology diagram. (B) PepTSo structure viewed in the plane of the membrane. The two hydrophilic cavities present in the structure are outlined in dashed lines. The hydrophobic core of the membrane (pale yellow) is distinguished from the interfacial region (light grey). N and C represent the N- and C-termini, respectively. Bound ligand is shown in black. Helices are labelled. (C) View from the extracellular side of the membrane.
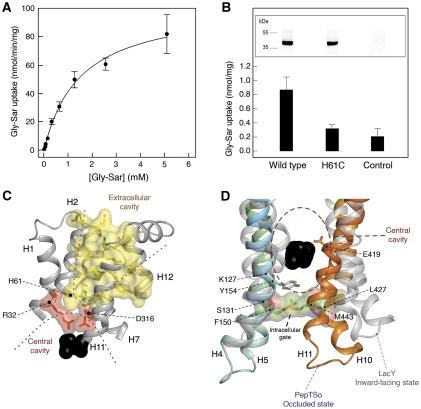
The apparent KM for transport of the hydrolysis resistant di-peptide glycylsarcosine is 1.5±0.15 mM, similar to the value reported for human PepT1 of 1.1±0.1 mM (Brandsch et al, 1994) (Figure 2A). Uptake of a fluorescent di-peptide, β-Ala-Lys-Ne-7-amino-4-methylcoumarin-3-acetic acid (β-Ala-Lys-(AMCA)) in cells overexpressing the PepTSo gene was reduced upon addition of either di- or tri-alanine peptides to the media (Supplementary Figure S3). Addition of L-alanine or the larger tetra-alanine peptide, however, had little effect, suggesting a similar preference for di- and tri-peptides as reported for the mammalian transporters (Fei et al, 1994). Uptake was also abolished by the proton ionophore carbonyl cyanide p-chlorophenylhydrazone, consistent with a dependence on the proton electrochemical gradient (ΔμH+) to drive transport. The crystal structure was solved by multiple isomorphous replacement with anomalous scattering using mercury derivative crystals and seleno-L-methionine incorporated protein (Table I and Supplementary Tables I and II). Assignment of the amino-acid sequence to the density map was aided through identification of the 22 selenium and three mercury sites present in the molecule (Supplementary Figure S4). The model was built and refined using data with anisotropic truncation of the observed structure factors (Strong et al, 2006) to 4.3 Å along the A and B axes, while keeping the C axis at 3.6 Å (Supplementary Figure S5). The final model was refined to an Rfactor of 27.8% and a corresponding Rfree of 29.6% (Table I). There are three PepTSo molecules in the crystallographic asymmetric unit and their structures are identical in the context of this analysis.
Figure 2.
Transport of peptides by PepTSo. (A). Concentration dependence of PepTSo-mediated glycylsarcosine (Gly-Sar) uptake in E. coli. Results shown, expressed per milligram of His-tagged PepTSo protein, are mean values±s.d. (n=4). (B) Effect on transport activity of mutating His61 to cysteine. Uptake of [3H]-glycylsarcosine (Gly-Sar) over a period of 10 min was measured in E. coli cells expressing the indicated forms of His-tagged PepTSo or in control cells lacking the transporter. Results shown are mean values±s.d. (n=3) and are expressed per milligram dry weight of bacteria. The inset shows western blots of equivalent samples from each culture, stained with a monoclonal antibody against oligohistidine. (C) Extracellular cavity viewed in the membrane plane. The central and extracellular cavities are isolated from each other by a putative extracellular gate. Residues in the central and extracellular cavities are highlighted in red and yellow, respectively. His61, part of the proposed proton–substrate coupling machinery is shown in green. Bound ligand is shown as a black CPK model of a di-alanine peptide. (D) Intracellular gate viewed in the membrane plane. Residues forming the gate are shown as stick models with transparent CPK surfaces. LacY helices (grey) are superposed onto PepTSo. Bound ligand is shown as a black CPK model as in C.
Table 1. Data collection and refinement statistics.
| Native | MMC-1a | MMC-2 | MMC-3 | HgAc | Se | |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P32 | P32 | P32 | P32 | P32 | P32 |
| Cell dimensions | ||||||
| a, b, c (Å) | 159.4, 159.4, 153.0 | 159.7, 159.7, 153.9 | 157.6, 157.6, 153.1 | 158.1, 158.1, 153.6 | 159.7, 159.7, 153.9 | 157.6, 157.6, 153.1 |
| α, β, γ (deg) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 40–3.6 (3.8–3.6)b | 40–4.6 (4.8–4.6) | 40–4.0 (4.1–4.0) | 40–4.5 (4.7–4.5) | 40–4.6 (4.8–4.6) | 40–5.0 (5.2–5.0) |
| Rmergec | 8.9 (67.5) | 9.3 (82.4) | 10.4 (76.1) | 16.0 (82.3) | 9.4 (50.2) | 10.9 (50.0) |
| I/σI | 9.0 (1.1) | 7.1 (1.1) | 8.8 (1.4) | 7.7 (1.0) | 8.5 (1.68) | 7.5 (2.0) |
| Completeness (%) | 93.8 (88.7) | 90.3 (90) | 97.7 (96.4) | 99.7 (99.0) | 96.4 (95.7) | 99.0 (99.0) |
| Redundancy | 2.0 (1.8) | 1.7 (1.7) | 3.2 (2.8) | 3.3 (3.0) | 2.4 (2.2) | 3.3 (3.1) |
| Refinement | ||||||
| Resolution (Å) | 19–3.6 (3.8–3.6) | |||||
| No of reflections | 47 021 | |||||
| Rwork/Rfree | 27.8 (35.8)/ 29.6 (40.4) | |||||
| No of protein atoms | 10 533 | |||||
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.009 | |||||
| Bond angles (deg) | 1.12 | |||||
| aFor details on derivatisation see Supplementary Materials and methods. | ||||||
| bValues in parenthesis are for the highest-resolution shell. | ||||||
| cThe last shell Rmerge is high for some of the derivative data because of severe anisotropy in the diffraction images. | ||||||
Hydrophilic cavities
In the structure, we observe a central cavity and a smaller extracellular cavity, both of which are hydrophilic (Figure 1B). The central cavity is situated within the centre of the membrane and closed to the extracellular space by a gate made of helices H1, H2, H7 and H8, which pack closely together (Figures 1C and 2C). Previous secondary active transporter structures have all revealed a ligand-binding site located within the centre of the membrane, essentially equidistant between extracellular and intracellular sides (reviewed in Boudker and Verdon, 2010). Indeed, the residues extending into the central cavity in PepTSo are all known to affect peptide binding and/or transport in the mammalian proteins (Terada et al, 1996, 2004; Fei et al, 1997; Bolger et al, 1998; Yeung et al, 1998; Chen et al, 2000; Uchiyama et al, 2003; Hauser et al, 2005; Pieri et al, 2009; Xu et al, 2009). This cavity is therefore an obvious location for the peptide-binding site, as we discuss below. As further confirmation, we also observe clear electron density within this cavity for a bound ligand. The apparent Km for the substrate peptide glycylsarcosine is low, 1.5 mM; therefore, this density is unlikely to represent a co-purified natural peptide. The density is more likely to represent a non-natural ligand or a high-affinity inhibitor acquired from either the purification or the crystallization conditions. The position of this density corresponds to the same location of the bound sugar analogue β-D-galactopyranosyl-thio-β-D-galactopyranoside observed in the binding site of LacY (Abramson et al, 2003) (Supplementary Figure S6). Access to the cytoplasm from this cavity is restricted by an intracellular gate formed by side-chain interactions between two-helix hairpins, helices H4 and H5 on the N-terminal side, and H10 and H11 on the opposing C-terminal side (Figure 2D). The interaction between these helices occurs through residues that are conserved across the vertebrate peptide transporters (Figure 1A; Supplementary Figure S1). The most prominent of these interactions involves Leu427(603) on helix H10 packing against Tyr154(167) and Phe150(163) on helix H4, both of which form part of the highly conserved POT family PTR2_2 motif (FYxxINxG), suggesting a possible role in regulating the exit of peptides from the central cavity. Numbers in brackets correspond to the equivalent residues in human PepT1. Indeed, several point mutations within the PTR2_2 motif have been found to inactivate or greatly impair transport (Hauser et al, 2005). Considering the position of a bound ligand within the central cavity and its confinement through the closure of both extracellular and intracellular gates (Figure 3A and B), we have described the present state of PepTSo as substrate occluded, in analogy with the LeuT superfamily transporters (Krishnamurthy et al, 2009).
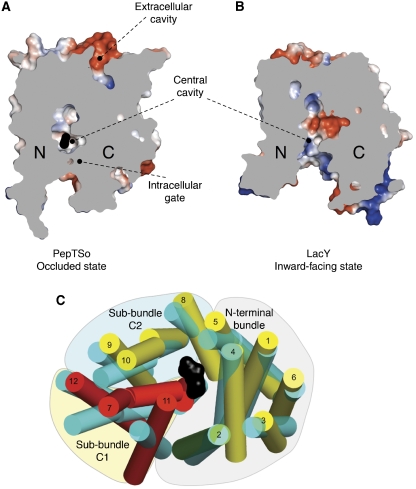
Figure 3.
Comparison of PepTso and LacY structures. Electrostatic surface representation showing the location of the hydrophilic cavities in a section through the protein volumes of (A) PepTSo and (B) LacY. The N- and C-terminal six-helix bundles are labelled. (C) Superposed transmembrane helices of PepTSo and LacY viewed from the intracellular side of the membrane. PepTSo helices are labelled and shown in yellow except for helix H2 (green) and helices H7, H11 and H12 (red), which form sub-bundle C1. The N-terminal six-helix bundle and the C-terminal sub-bundles C1 and C2 are highlighted. LacY helices are shown in cyan. Bound ligand is shown as a black CPK model of a di-alanine peptide. Helices HA and HB have been omitted for clarity.
Comparing this conformation with previous MFS transporter structures has highlighted a potentially important structural feature of transport within the MFS. The various LacY structures are all in inward-facing open conformations (Abramson et al, 2003). The EmrD structure on the other hand likely corresponds to an occluded state, although no evidence of bound ligand could be observed in the electron density maps (Yin et al, 2006). Using a secondary structure matching algorithm (Krissinel and Henrick, 2004) to overlay these evolutionarily distinct proteins, it is clear that the structure of PepTSo is more similar to that of EmrD than to LacY, with an average distance of 2.4 Å for 155 equivalent Cα atoms (Supplementary Figure S7). This observation supports the conclusion that PepTSo also represents the occluded conformation for the POT family.
To further understand the differences between PepTSo and LacY, which represent two different conformational states for MFS transporters, we also compared their structures by calculating the change in position between Cα atoms of related helical segments within the N- and C-terminal six-helix bundles, respectively (see Supplementary data). The main difference was clearly identified as residing within the C-terminal domain, the average distances between atoms that make up the N- and C-terminal helix bundles being 2.8 and 4.4 Å, respectively (Supplementary Table III). Further insight was drawn from comparing Cα displacements of individual helices within the C-terminal domain. In this case, helix H7 of PepTSo and LacY showed the largest average Cα displacement between the two structures of 5.7 Å. The C-terminal six-helix bundle of MFS transporters is made up of inverted repeats constructed from helices H7–H9 and H10–H12. Structurally, however, they also form two sub-bundles of helices consisting of H7, H11 and H12 (sub-bundle C1) and H8, H9 and H10 (sub-bundle C2) (Figure 3C). When the N-terminal domains of PepTSo and LacY are superimposed, the largest deviation is observed in sub-bundle C1, which is displaced by an approximate 11° rotation (Supplementary Figure S8). These helices seem to be the ones mainly responsible for the asymmetry between the N- and C-terminal helix bundles between the occluded conformation of PepTSo and inward open conformation of LacY. These observations suggest that a large conformational change takes place predominantly within the C-terminal domain of MFS transporters during the transition from ligand-bound occluded to inward open state.
The observed extracellular cavity is also located at the interface between the N- and C-terminal domains and is roughly cone shaped, with the apex at the bottom near the central cavity, opening outward (Figures 1B and 2C). The overall dimensions of the cavity are ∼16 × 8 × 8 Å. Atomistic molecular dynamics (MD) simulations reproducibly show that both this extracellular cavity and the central cavity are fully solvated and not blocked by lipid molecules in the present conformation (Supplementary Figure S9). The EmrD structure exhibits a similar cavity (Supplementary Figure S7), which differs mainly in that its surface is composed primarily of hydrophobic residues. This observation supports the possibility that this cavity represents the vestiges of either an entrance (for PepTSo) or an exit (for EmrD) pathway for substrates, these being hydrophilic peptides for PepTSo and hydrophobic compounds for EmrD, when the central cavity is open towards the extracellular side.
The peptide-binding site
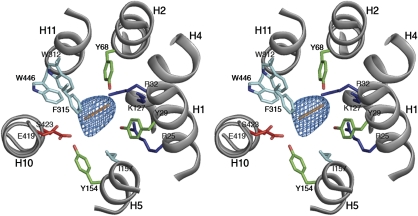
As previously noted, many of the residues conserved between PepTSo and the mammalian peptide transporters cluster around the central hydrophilic cavity, with approximate dimensions of 13 × 12 × 11 Å. These dimensions are sufficient to accommodate both di- and tri-peptides, although would be sterically restrictive for larger tetra-peptide ligands. This may explain the competition we observe in the in vivo transport assay between peptides of this size and the β-Ala-Lys-(AMCA) (Supplementary Figure S3). The dimensions of the cavity could also explain the lack of affinity for single amino acids, as these would presumably be incapable of interacting sufficiently with both the N- and C-terminal domains of the transporter. Sitting within the centre of the cavity we observe strong (>4σ) electron density for an unidentified ligand (Figure 4; Supplementary Figure S10) of approximately the same dimensions as a di-peptide. In the figure, a Cα model of a di-alanine peptide has been placed into the density as a reference to evaluate the size of the cavity, although no peptide was modelled during refinement.
Figure 4.
The peptide-binding site. Stereo view of the central cavity as viewed from above on the extracellular side of the membrane. Conserved residues between PepTSo and the mammalian peptide transporters are labelled and coloured according to side-chain type, Arg and Lys (blue), Glu and Ser (red), Tyr (green) and Trp, Phe and Leu (cyan). A di-peptide sized Cα baton (orange) is fitted as a size reference into the mFo-DFc electron density observed in the central cavity (blue mesh), contoured at 4 σ.
The binding site is formed by residues from helices H1, H2, H4 and H5 from the N-terminal six-helix bundle and from helices H7, H8, H10 and H11 from the C-terminal bundle. On the N-terminal side of the binding site, three conserved positively charged residues, Arg25(27), Arg32(34) and Lys127(140) extend into the cavity. It has been reported that mutation of Arg25(27) in human PepT2 to a histidine completely inactivates transport (Terada et al, 2004). Two conserved tyrosine residues, Tyr29(31) and Tyr68(64), are positioned close to this positively charged cluster. On the C-terminal side of the binding site, at a distance of ∼13 Å from Lys127(140), are two further strictly conserved residues, Glu419(595) and Ser423(599), located in close proximity to Tyr154(167). Various mutants of Glu419(595) in PepT1 have been reported to drastically reduce transport activity, except where mutation was to an aspartic acid (Xu et al, 2009), indicating the importance of a negatively charged residue at this position. The arrangement of opposite charges within the binding site may have an important role in the recognition and orientation of peptides through the creation of a dipole moment. The presence of several possible hydrogen-bond donors and acceptors could be advantageous in adapting to peptides of various lengths, sequences and charges. For example, the tyrosine residues described above could have important roles in forming hydrogen bonds and for providing a hydrophobic environment for side chains, as observed in many antigen-recognition sites (Fellouse et al, 2004).
Most of the other residues in the binding site are conserved hydrophobic residues, including Ile157(170), Trp312(294), Phe315(297) and Trp446(622). These residues are likely to provide a suitable environment for peptide side chains that in general are more hydrophobic than the peptide backbone. This idea is supported by the fact that mutation of Trp312(294) of PepT1 to alanine reduces substrate uptake in HEK293 cells (Bolger et al, 1998). The presence of hydrophobic pockets formed by these residues in the peptide-binding site of PepT1 has also been predicted through biochemical studies (Bailey et al, 2000). The water molecules that were identified by the MD simulation as filling the central cavity may also have an important role in allowing the binding site to accommodate various types of amino-acid side chain, as shown in the study of OppA, a periplasmic protein that also binds a diverse library of oligopeptides (Tame et al, 1996).
Interestingly, some conserved residues are not located within the central cavity. The PTR2_1 motif, also conserved throughout the POT family (Daniel et al, 2006), spans the cytoplasmic linker connecting helices H2 and H3 (Supplementary Figure S1). Residues within this motif do not make any contribution to the interior of the protein and most likely sit in the interfacial region of the lipid bilayer, a location also suggested by the coarse-grained MD simulations (Supplementary Figure S2). Glu21(23) and Glu24(26) on helix H1 are also well conserved among the POTs. These residues are not located in the peptide-binding site but are in close proximity to Arg25(27) and Lys127(140), and may have an important role in positioning these residues.
Discussion
Implications for proton-driven peptide symport
Although MFS transporters have been extensively studied using biochemical and biophysical methods, the current lack of additional structural information pertaining to the multiple states has hampered further understanding of the transport mechanism in this large and important transporter family. The PepTSo structure presents a ligand-bound occluded conformation for an MFS symporter with a clearly identified substrate-binding site. Its comparison with the inward-facing LacY structure in this study provides new insight into the transition states of the alternating access mechanism as operating within the MFS. Figure 5 summarizes a possible model for transport, which is further discussed below.
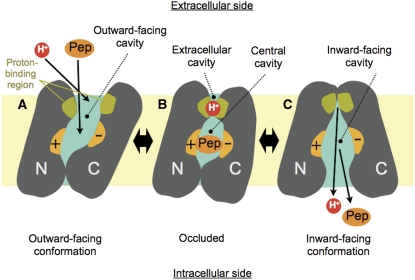
Figure 5.
A possible mechanism for peptide–proton symport. (A) Outward-facing state: peptide (Pep) and proton (H+) can access respective binding sites through the outward-facing cavity that is open towards the extracellular side of the membrane. The peptide-binding site is made from the surfaces of both the N- and C-terminal helix bundles (indicated by + and − signs), whereas the proton-binding site is located in the area close to the extracellular gate. (B) Occluded state: both ends of the central cavity are closed with peptide occluded into the central cavity. The proton-binding site is still exposed to the extracellular side through the extracellular cavity. (C) Inward-facing state: peptide and proton are released on the intracellular side of the membrane through the inward-facing cavity. Note that the proton-binding site is exposed to the intracellular side in this conformation.
In the present structure, both ends of the central cavity containing residues involved in peptide binding are closed (Figure 5B). For uptake of peptide, the central cavity needs to be connected to the extracellular space, potentially through the extracellular cavity observed in PepTSo (Figure 5A). The central and extracellular cavities are separated by a putative extracellular gate, which is made of helix H7 packed against helix H2 at the interface between the N- and C-terminal helix bundles (Figure 2C). Kaback and colleagues have identified this region in LacY as forming a periplasmic gate, the opening of which is essential for substrate uptake from the extracellular space (Zhou et al, 2008; Nie et al, 2009). Exit of bound peptide to the intracellular side is currently restricted by side-chain interactions between two helix hairpins formed by helices H4 and H5 and H10 and H11 on the opposing N- and C-terminal helix bundles, respectively (Figure 2D). When we compare the occluded structure of PepTSo with the inward-facing LacY and occluded EmrD structures, the N-terminal helix sub-bundles, including the helix hairpin formed by H4 and H5, maintain similar positions as discussed above (Figure 3C; Supplementary Figure S7). Opening of the intracellular gate, therefore, seems to be controlled by the movement of helix hairpin H10–H11 in the C-terminal helix bundle. This hairpin would move together with the sub-bundle C1, composed of helices H7, H11 and H12, which show the largest difference between the LacY and PepTSo structures (Supplementary Figure S8).
Considering the proton–substrate symport mechanism within the peptide transporter family, one of the more interesting residues is His61(57) buried within the H2–H7 helix interface that forms the extracellular gate (Figures 2C and 5B). The region containing His61(57) is completely exposed to the intracellular side in the LacY structures, suggesting a significant conformational change in moving from the occluded to inward-facing state (Figure 5C). The His61(57) residue has been identified as the primary protonation site in human PepT1 and PepT2 (Fei et al, 1997; Uchiyama et al, 2003). Indeed, a His61(57)Arg mutant of human PepT1 is active only at higher pH, indicating that protonation and deprotonation of this side chain is essential for peptide transport (Fei et al, 1997; Uchiyama et al, 2003). Mutation of this residue in PepTSo to cysteine also inactivates the transporter in an in vivo peptide uptake assay (Figure 2B). In the vicinity of His61(57), there is Asp316(298) that sits at the top of the central cavity. This residue is currently interacting with Arg32(34) in the ligand-binding site (Figure 2C). Although neither His61(57) nor Asp316(298) is strictly conserved in POTs from bacteria and lower eukaryotes, this region is clearly important in proton binding and/or opening of the extracellular gate in PepTSo and its mammalian homologues. In EmrD and related homologues, a conserved glutamate, Glu227, in helix H7, which is located close to the equivalent position of Asp316(298) in PepTSo, has been suggested as a possible proton-binding site (Yin et al, 2006) (Supplementary Figure S7). An EmrD homologue, LmrP from Lactococcus lactis, also conserves a carboxylate group (Asp235) in a similar region of the protein, which is suggested to form part of a flexible proton/ligand-binding site (Bapna et al, 2007; Schaedler and van Veen, 2010) and is responsible in part for conformational changes observed in LmrP upon proton binding (Gbaguidi et al, 2007).
The structural model employed to explain transport within the MFS has to date been a symmetrical rigid body rocking motion between the N- and C-terminal six-helix bundles (Abramson et al, 2003; Huang et al, 2003). The occluded PepTSo structure, however, indicates that there is a potentially important role sharing arrangement between these bundles. Our structure suggests the N-terminal helix bundle should be less dynamic, and in the POT family more involved in peptide binding, whereas the C-terminal helix bundle contains the mobile gates, quite possibly driven by the proton electrochemical gradient. Such a functional division between the two halves of MFS transporters is supported by previous biochemical studies in LacY and other distantly related transporters (Guan and Kaback, 2006; Kasho et al, 2006; Boudker and Verdon, 2010). This mechanism is conceptually similar to the one reported for the benzyl-hydantoin transporter Mhp1, wherein the central four-helix bundle provides a stable platform and the ‘hash motif', made by the other four-helix bundle, acts as a mobile gate (Shimamura et al, 2010).
To understand the mechanism in full however, it will be essential to determine the PepTSo structure in different conformational states and determine the sites of protonation and peptide binding. A central question still remaining is how the human peptide transporters PepT1 and PepT2 are able to transport not only peptides and peptide analogues such as β-lactam antibiotics, but also much larger peptide prodrugs, such as valacyclovir and val-val-lopinavir (Brandsch, 2009). The structure of PepTSo provides the first structural model with which to further investigate this important aspect of mammalian peptide transporter biochemistry.
Materials and methods
Protein expression and purification
The gene encoding PepTSo (SO_0002, Uniprot identifier Q8EKT7) was amplified from a previously constructed expression plasmid pMPSIL0079A and cloned into the pWaldo-GFPe plasmid (Drew et al, 2001) for all subsequent overexpression, purification and crystallization procedures (for details see Supplementary data).
Crystallography
Crystals of PepTSo were obtained in 30% PEG 300, 0.1 M MES pH 6.50 and 0.1 M NaCl using the hanging drop vapour diffusion technique at 4°C. For cryoprotection, the crystals were transferred to a solution containing 36–40% PEG 300, 0.1 M MES pH 6.50, 0.1 M NaCl and 0.03% DDM, before being flash vitrified in liquid nitrogen. The crystals always showed strong anisotropic diffraction, with the best crystals diffracting between 3.6 and 3.8 Å in the best direction. Mercury derivatives were prepared in two ways, either before crystallization or through soaking of native crystals. Data sets MMC-1, HgAc and MMC-3 were obtained from crystals grown using protein pre-incubated with mercury compounds, while data set MMC-2 was obtained through soaking a seleno-L-methionine incorporated crystal. Modification of free cysteines and seleno-L-methionine incorporation of the protein before crystal growth was carried out as described in Extended experimental procedures. Data were collected on beamlines ID23eh1 and ID29 at the European synchrotron radiation facility and on IO2 and IO3 at the Diamond Light Source Ltd, UK. Data were processed and scaled using the HKL suite of programs (Otwinowski and Minor, 1997). The space group was determined to be P32, with three molecules in the asymmetric unit. Two mercury sites were initially located in each of the three molecules. These were found manually using RSPS (Knight, 2000). The mercury positions were refined and initial phases were calculated using SHARP (de La Fortelle and Bricogne, 1997). The resulting phases were used to locate a third mercury site. Phases from the mercury sites were used to locate the selenium sites in difference Fourier maps and these were added to the nine mercury sites to improve the phase information. Over 30 data sets were collected during the structure determination from >1000 crystals screened at the synchrotron. These data sets were grouped according to isomorphism and anomalous signal strength, as judged by Xtriage (Adams et al, 2002) (Supplementary Tables I and II). Each set of isomorphous data was used to calculate phases in SHARP and the calculated maps were analysed. This process resulted in two sets of non-isomorphous data that gave good initial maps that showed clear solvent boundaries around the three molecules. Set 1 contained the high-resolution native and two mercury derivative data sets, MMC-1 and HgAc. Set 2 contained two mercury derivative data sets, MMC-2, which was a seleno-L-methionine incorporated crystal soaked overnight in 2 mM MMC and MMC-3. Set 2 also contained a separate seleno-L-methionine crystal collected at the anomalous edge for selenium. The three-fold non-crystallographic symmetry present in the packing of the crystal was used to improve the phase information. A rough mask was calculated around one of the three molecules using O (Jones and Kjeldgaard, 1997) and the NCS operators calculated and improved using programs from the RAVE (Jones and Kjeldgaard, 1997) package. These were then used in DM (Cowtan, 1994) to average over the three molecules in the asymmetric unit. The two sets of phases were also combined using cross-crystal averaging in DMMulti (Cowtan, 1994). The resulting maps were then of sufficient quality to see all 14 helices from each of the three molecules. The optimal solvent content for density modification was found to be 79%.
Model building and refinement
An initial Cα model was built into the density from all three sources of phases described above, from SHARP, DM and DMMulti, using O (Jones and Kjeldgaard, 1997). The partial models were further cycled back into phase calculation in SHARP to improve the initial solvent envelope used for the solvent flipping procedure. This cycle was iterated many times until a reasonably complete model could be built. The amino-acid side chains were then built into the partial model using the selenium and mercury sites to determine the correct register. Refinement of the model was carried out in BUSTER (BUSTER-TNT 2.X) against the highest resolution data set with inclusion of the experimental phase information. Refinement was improved by anisotropic truncation of the structure factors (Strong et al, 2006) along the A and B axes to 4.3 Å, with the C axis kept at 3.6 Å. To increase the contribution of the high-resolution terms in the resulting 2mFo-DFc electron density maps, a B-factor sharpening term was introduced during map calculation in FFT of between −50 and −80 Å2. Model validation was carried out using the Molprobity server (Davis et al, 2007) (Supplementary Table IV). The quality of the model compares favourably with structures of a similar resolution in the Protein Data Bank. Images were prepared using PyMol (The PyMOL Molecular Graphics System) and VMD (Humphrey et al, 1996).
In vivo [3H]-glycylsarcosine transport assay
Uptake of the radioactive di-peptide [3H]-glycylsarcosine (Gly-Sar) was measured at 37°C in cells overexpressing the PepTSo gene as described in Supplementary data. After an uptake period of 15 s, employed to approximate initial velocities of transport, cells were filtered to terminate transport and washed twice with ice-cold transport buffer (Henderson and Macpherson, 1986). For estimation of the apparent Vmax and Km values for transport, uptake was measured over a substrate concentration range of 10 μM to 5 mM. PepTSo-mediated transport was calculated by subtraction of rates seen in induced cells harbouring a control expression vector encoding the E. coli nucleoside transporter NupG. The resultant rates were expressed per milligram of PepTSo protein, as measured by quantitative immunoblotting. Data were fitted to the Michaelis–Menten equation using the non-linear curve-fitting program KaleidaGraph (version 4.0, Synergy software) in order to estimate kinetic parameters.
In vivo β-Ala-Lys-(AMCA) competition transport assay
Uptake of the fluorescent di-peptide β-Ala-Lys-(AMCA) was measured at 37°C in cells overexpressing the PepTSo gene essentially as described by Weitz et al (2007) and in Supplementary data. Transport of β-Ala-Lys-(AMCA) was measured using fluorescence in a Fluostar Optima plate reader using an excitation wavelength of 340 nm and an emission filter with a wavelength of 460 nm.
Supplementary Material
Acknowledgments
We thank all the members and advisors of the Membrane Protein Structure Initiative for advice and support. We also thank Manami Kanazawa, Mutsuko Grant and Gill Murray for administrative support; and James Mansfield for technical assistance with the bioreactor fermentations and Jocelyn Baldwin for bioinformatic analyses. This research was funded primarily by the Biotechnology and Biological Sciences Research Council (BBSRC), Membrane Protein Structure Initiative (MPSi) (grant BBS/B/14418), with important contributions from the Wellcome Trust (grant 062164/Z/00/Z) and the Japan Science and Technology Agency. We thank the Membrane Protein Laboratory (MPL) staff at the Diamond Light Source Limited for their support. We also thank the beamline staff at the European Synchrotron Radiation Facility (ID23, ID29 and ID14eh4) and Diamond Light Source, UK (IO2, IO3 and IO4). DD was the recipient of an EMBO LT Fellowship. SN and DD acknowledge the current support of the Medical Research Council and Royal Society through the University Research Fellowship schemes, respectively. A part of this work was also supported by the Targeted Proteins Research Program of MEXT, Japan. The atomic coordinates have been deposited in the Protein Data Bank (accession code 2XUT).
Author contributions: SN, XX, DD, VP, MJM and SAB cloned the PepTSo gene and tested expression in the different vectors. VP, XX, JCI and SAB designed, conducted and analysed the transport assay data. SN, DD, AC and SI purified, crystallized and collected X-ray diffraction data. SN and AC solved the heavy atom substructure, built the model and refined the structure. SN, DD, AC, VP, SAB and SI analysed the structure, functional data and wrote the paper. PWF and MSPS performed and analysed the molecular dynamics calculations.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301: 610–615 [DOI] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58(Pt 11): 1948–1954 [DOI] [PubMed] [Google Scholar]
- Bailey PD, Boyd CA, Bronk JR, Collier ID, Meredith D, Morgan KM, Temple CS (2000) How to make drugs orally active: a substrate template for peptide transporter PepT1. Angew Chem Int Ed Engl 39: 505–508 [DOI] [PubMed] [Google Scholar]
- Bapna A, Federici L, Venter H, Velamakanni S, Luisi B, Fan TP, van Veen HW (2007) Two proton translocation pathways in a secondary active multidrug transporter. J Mol Microbiol Biotechnol 12: 197–209 [DOI] [PubMed] [Google Scholar]
- Biegel A, Knütter I, Hartrodt B, Gebauer S, Theis S, Luckner P, Kottra G, Rastetter M, Zebisch K, Thondorf I, Daniel H, Neubert K, Brandsch M (2006) The renal type H+/peptide symporter PEPT2: structure-affinity relationships. Amino Acids 31: 137–156 [DOI] [PubMed] [Google Scholar]
- Bolger MB, Haworth IS, Yeung AK, Ann D, von Grafenstein H, Hamm-Alvarez S, Okamoto CT, Kim KJ, Basu SK, Wu S, Lee VH (1998) Structure, function, and molecular modeling approaches to the study of the intestinal dipeptide transporter PepT1. J Pharm Sci 87: 1286–1291 [DOI] [PubMed] [Google Scholar]
- Boudker O, Verdon G (2010) Structural perspectives on secondary active transporters. Trends Pharmacol Sci 31: 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsch M (2009) Transport of drugs by proton-coupled peptide transporters: pearls and pitfalls. Expert Opin Drug Metab Toxicol 5: 887–905 [DOI] [PubMed] [Google Scholar]
- Brandsch M, Miyamoto Y, Ganapathy V, Leibach FH (1994) Expression and protein kinase C-dependent regulation of peptide/H+ co-transport system in the Caco-2 human colon carcinoma cell line. Biochem J 299(Pt 1): 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSTER-TNT 2.X GPL, Sheraton House, Cambridge CB3 0AX, UK
- Casagrande F, Harder D, Schenk A, Meury M, Ucurum Z, Engel A, Weitz D, Daniel H, Fotiadis D (2009) Projection structure of DtpD (YbgH), a prokaryotic member of the peptide transporter family. J Mol Biol 394: 708–717 [DOI] [PubMed] [Google Scholar]
- Chen XZ, Steel A, Hediger MA (2000) Functional roles of histidine and tyrosine residues in the H(+)-peptide transporter PepT1. Biochem Biophys Res Commun 272: 726–730 [DOI] [PubMed] [Google Scholar]
- Covitz KM, Amidon GL, Sadée H (1998) Membrane topology of the human dipeptide transporter, hPEPT1, determined by epitope insertions. Biochemistry 37: 15214–15221 [DOI] [PubMed] [Google Scholar]
- Cowtan K (1994) Joint CCP4 and ESF-EACBM newsletter on protein crystallography. 31: 34–38
- Daniel H, Kottra G (2004) The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pflugers Arch 447: 610–618 [DOI] [PubMed] [Google Scholar]
- Daniel H, Spanier B, Kottra G, Weitz D (2006) From bacteria to man: archaic proton-dependent peptide transporters at work. Physiology (Bethesda) 21: 93–102 [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35 (Web Server issue): W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol 276: 472–494 [DOI] [PubMed] [Google Scholar]
- Doring F, Martini C, Walter J, Daniel H (2002) Importance of a small N-terminal region in mammalian peptide transporters for substrate affinity and function. J Membr Biol 186: 55–62 [DOI] [PubMed] [Google Scholar]
- Drew DE, von Heijne G, Nordlund P, de Gier JW (2001) Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett 507: 220–224 [DOI] [PubMed] [Google Scholar]
- Ernst HA, Pham A, Hald H, Kastrup JS, Rahman M, Mirza O (2009) Ligand binding analyses of the putative peptide transporter YjdL from E. coli display a significant selectivity towards dipeptides. Biochem Biophys Res Commun 389: 112–116 [DOI] [PubMed] [Google Scholar]
- Faria TN, Timoszyk JK, Stouch TR, Vig BS, Landowski CP, Amidon GL, Weaver CD, Wall DA, Smith RL (2004) A novel high-throughput pepT1 transporter assay differentiates between substrates and antagonists. Mol Pharm 1: 67–76 [DOI] [PubMed] [Google Scholar]
- Fei YJ, Kanai Y, Nussberger S, Ganapathy V, Leibach FH, Romero MF, Singh SK, Boron WF, Hediger MA (1994) Expression cloning of a mammalian proton-coupled oligopeptide transporter. Nature 368: 563–566 [DOI] [PubMed] [Google Scholar]
- Fei YJ, Liu W, Prasad PD, Kekuda R, Oblak TG, Ganapathy V, Leibach FH (1997) Identification of the histidyl residue obligatory for the catalytic activity of the human H+/peptide cotransporters PEPT1 and PEPT2. Biochemistry 36: 452–460 [DOI] [PubMed] [Google Scholar]
- Fellouse FA, Wiesmann C, Sidhu SS (2004) Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci USA 101: 12467–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy ME, Huang W, Wang H, Ganapathy V, Leibach FH (1998) Valacyclovir: a substrate for the intestinal and renal peptide transporters PEPT1 and PEPT2. Biochem Biophys Res Commun 246: 470–475 [DOI] [PubMed] [Google Scholar]
- Ganapathy V, Leibach FH (1983) Role of pH gradient and membrane potential in dipeptide transport in intestinal and renal brush-border membrane vesicles from the rabbit. Studies with L-carnosine and glycyl-L-proline. J Biol Chem 258: 14189–14192 [PubMed] [Google Scholar]
- Gbaguidi B, Hakizimana P, Vandenbussche G, Ruysschaert JM (2007) Conformational changes in a bacterial multidrug transporter are phosphatidylethanolamine-dependent. Cell Mol Life Sci 64: 1571–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Kaback HR (2006) Lessons from lactose permease. Annu Rev Biophys Biomol Struct 35: 67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagting A, Kunji ER, Leenhouts KJ, Poolman B, Konings WN (1994) The di- and tripeptide transport protein of Lactococcus lactis. A new type of bacterial peptide transporter. J Biol Chem 269: 11391–11399 [PubMed] [Google Scholar]
- Harder D, Stolz J, Casagrande F, Obrdlik P, Weitz D, Fotiadis D, Daniel H (2008) DtpB (YhiP) and DtpA (TppB, YdgR) are prototypical proton-dependent peptide transporters of Escherichia coli. FEBS J 275: 3290–3298 [DOI] [PubMed] [Google Scholar]
- Hauser M, Kauffman S, Naider F, Becker JM (2005) Substrate preference is altered by mutations in the fifth transmembrane domain of Ptr2p, the di/tri-peptide transporter of Saccharomyces cerevisiae. Mol Membr Biol 22: 215–227 [DOI] [PubMed] [Google Scholar]
- Henderson PJF, Macpherson AJ (1986) Assay, genetics, proteins and reconstitution of proton-linked galactose, arabinose and xylose transport systems of Escherichia coli. Methods Enzymol 125: 387–429 [DOI] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF (1999) Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Lemieux MJ, Song J, Auer M, Wang DN (2003) Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301: 616–620 [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14: 33–38, 27–38 [DOI] [PubMed] [Google Scholar]
- Jones TA, Kjeldgaard M (1997) Electron-density map interpretation. Methods Enzymol 277: 173–208 [DOI] [PubMed] [Google Scholar]
- Kasho VN, Smirnova IN, Kaback HR (2006) Sequence alignment and homology threading reveals prokaryotic and eukaryotic proteins similar to lactose permease. J Mol Biol 358: 1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SD (2000) RSPS version 4.0: a semi-interactive vector-search program for solving heavy-atom derivatives. Acta Crystallogr D Biol Crystallogr 56(Pt 1): 42–47 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy H, Piscitelli CL, Gouaux E (2009) Unlocking the molecular secrets of sodium-coupled transporters. Nature 459: 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1): 2256–2268 [DOI] [PubMed] [Google Scholar]
- Nie Y, Zhou Y, Kaback HR (2009) Clogging the periplasmic pathway in LacY. Biochemistry 48: 738–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Skurray RA (1994) The POT family of transport proteins. Trends Biochem Sci 19: 404. [DOI] [PubMed] [Google Scholar]
- Pieri M, Gan C, Bailey P, Meredith D (2009) The transmembrane tyrosines Y56, Y91 and Y167 play important roles in determining the affinity and transport rate of the rabbit proton-coupled peptide transporter PepT1. Int J Biochem Cell Biol 41: 2204–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler TA, van Veen HW (2010) A flexible cation binding site in the multidrug major facilitator superfamily transporter LmrP is associated with variable proton coupling. FASEB J 24: 3653–3661 [DOI] [PubMed] [Google Scholar]
- Scott KA, Bond PJ, Ivetac A, Chetwynd AP, Khalid S, Sansom MS (2008) Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure 16: 621–630 [DOI] [PubMed] [Google Scholar]
- Shimamura T, Weyand S, Beckstein O, Rutherford NG, Hadden JM, Sharples D, Sansom MS, Iwata S, Henderson PJ, Cameron AD (2010) Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science 328: 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner HY, Naider F, Becker JM (1995) The PTR family: a new group of peptide transporters. Mol Microbiol 16: 825–834 [DOI] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D (2006) Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA 103: 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai I, Nakanishi T, Hayashi K, Terao T, Sai Y, Shiraga T, Miyamoto K, Takeda E, Higashida H, Tsuji A (1997) The predominant contribution of oligopeptide transporter PepT1 to intestinal absorption of beta-lactam antibiotics in the rat small intestine. J Pharm Pharmacol 49: 796–801 [DOI] [PubMed] [Google Scholar]
- Tame JR, Sleigh SH, Wilkinson AJ, Ladbury JE (1996) The role of water in sequence-independent ligand binding by an oligopeptide transporter protein. Nat Struct Biol 3: 998–1001 [DOI] [PubMed] [Google Scholar]
- Terada T, Irie M, Okuda M, Inui K (2004) Genetic variant Arg57His in human H+/peptide cotransporter 2 causes a complete loss of transport function. Biochem Biophys Res Commun 316: 416–420 [DOI] [PubMed] [Google Scholar]
- Terada T, Saito H, Mukai M, Inui KI (1996) Identification of the histidine residues involved in substrate recognition by a rat H+/peptide cotransporter, PEPT1. FEBS Lett 394: 196–200 [DOI] [PubMed] [Google Scholar]
- Terada T, Saito H, Mukai M, Inui K (1997) Recognition of beta-lactam antibiotics by rat peptide transporters, PEPT1 and PEPT2, in LLC-PK1 cells. Am J Physiol 273(5 Pt 2): F706–F711 [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System Version 1.2r3pre, Schrödinger LLC
- Tsuda M, Terada T, Irie M, Katsura T, Niida A, Tomita K, Fujii N, Inui K (2006) Transport characteristics of a novel peptide transporter 1 substrate, antihypotensive drug midodrine, and its amino acid derivatives. J Pharmacol Exp Ther 318: 455–460 [DOI] [PubMed] [Google Scholar]
- Uchiyama T, Kulkarni AA, Davies DL, Lee VHL (2003) Biophysical evidence for His57 as a proton-binding site in the mammalian intestinal transporter hPepT1. Pharm Res 20: 1911–1916 [DOI] [PubMed] [Google Scholar]
- Weitz D, Harder D, Casagrande F, Fotiadis D, Obrdlik P, Kelety B, Daniel H (2007) Functional and structural characterization of a prokaryotic peptide transporter with features similar to mammalian PEPT1. J Biol Chem 282: 2832–2839 [DOI] [PubMed] [Google Scholar]
- Wenzel U, Thwaites DT, Daniel H (1995) Stereoselective uptake of beta-lactam antibiotics by the intestinal peptide transporter. Br J Pharmacol 116: 3021–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Haworth IS, Kulkarni AA, Bolger MB, Davies DL (2009) Mutagenesis and cysteine scanning of transmembrane domain 10 of the human dipeptide transporter. Pharm Res 26: 2358–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung AK, Basu SK, Wu SK, Chu C, Okamoto CT, Hamm-Alvarez SF, von Grafenstein H, Shen WC, Kim KJ, Bolger MB, Haworth IS, Ann DK, Lee VH (1998) Molecular identification of a role for tyrosine 167 in the function of the human intestinal proton-coupled dipeptide transporter (hPepT1). Biochem Biophys Res Commun 250: 103–107 [DOI] [PubMed] [Google Scholar]
- Yin Y, He X, Szewczyk P, Nguyen T, Chang G (2006) Structure of the multidrug transporter EmrD from Escherichia coli. Science 312: 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Guan L, Freites JA, Kaback HR (2008) Opening and closing of the periplasmic gate in lactose permease. Proc Natl Acad Sci USA 105: 3774–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.