Structure of the archaeal Na+/H+ antiporter NhaP1 and functional role of transmembrane helix 1
Structural information on the pharmacologically important cation–proton antiporter 1 family members is scarce. Here, the first such insight into the organization of the archaeal NhaP1 antiporter suggests a conserved ion-translocation motif but differences in the overall helical fold compared the distantly related bacterial NhaA structure.
Keywords: electron cryo-microscopy, membrane protein structure, membrane transport, molecular model, monovalent cation/proton antiporters
Abstract
We have determined the structure of the archaeal sodium/proton antiporter NhaP1 at 7 Å resolution by electron crystallography of 2D crystals. NhaP1 is a dimer in the membrane, with 13 membrane-spanning α-helices per protomer, whereas the distantly related bacterial NhaA has 12. Dimer contacts in the two antiporters are very different, but the structure of a six-helix bundle at the tip of the protomer is conserved. The six-helix bundle of NhaA contains two partially unwound α-helices thought to harbour the ion-translocation site, which is thus similar in NhaP1. A model of NhaP1 based on detailed sequence comparison and the NhaA structure was fitted to the 7 Å map. The additional N-terminal helix 1 of NhaP1, which appears to be an uncleaved signal sequence, is located near the dimer interface. Similar sequences are present in many eukaryotic homologues of NhaP1, including NHE1. Although fully folded and able to dimerize, NhaP1 constructs without helix 1 are inactive. Possible reasons are investigated and discussed.
Introduction
The monovalent cation/proton antiporters (CPAs) are an extensive family of essential secondary transporters that control cytosolic pH and intracellular Na+ concentration in all living organisms. The CPA superfamily is divided into two main branches, namely, CPA1 and CPA2 (Brett et al, 2005). The CPA1 branch includes Methanococcus NhaP1, Nhx from yeast, the plant NHX transporters, and the ubiquitous and pharmacologically important mammalian NHE sodium–proton exchangers. The CPA2 branch includes NhaA from Escherichia coli and related bacteria, the plant CHX transporters, and the more recently discovered mammalian NHA transporters (Brett et al, 2005). The CPA1 and CPA2 branches differ not only by phylogeny but also in their transport stoichiometry. Whereas the CPA1 transporters seem to be electroneutral, those of the CPA2 type appear to be electrogenic, exchanging two protons for one monovalent cation.
In terms of three-dimensional (3D) structure, E. coli NhaA is so far the only well-characterized member of the CPA superfamily. A 4 Å projection map obtained by electron cryo-microscopy (cryo-EM) of 2D crystals indicated that NhaA is a dimer in the membrane (Williams et al, 1999). A 3D map of the NhaA dimer at 6 Å resolution obtained by electron crystallography revealed 12 transmembrane α-helices (TMHs) per protomer (Williams, 2000) for the first time. The 12 helices were arranged in two groups, namely, one row of six more or less tilted TMHs along the dimer interface and a six-helix bundle. The 3.45 Å X-ray structure of the NhaA protomer (Hunte et al, 2005) indicated that helices IV and XI in the six-helix bundle are partly unwound at the point where they cross one another in the hydrophobic centre of the membrane. Polar and charged residues in these unwound regions are thought to harbour the ion-translocation site.
Both the 6 Å EM map and the 3.45 Å X-ray structure of NhaA show the same locked conformation at pH 4, where this transporter is inactive (Taglicht et al, 1991). A recent study of 2D crystals of NhaA at higher pH in the absence or presence of substrate ions revealed subtle, but well-defined conformational changes that mark the transition from the inactive to the pH-activated state as the pH rises from 6 to 7, and a distinct, substrate ion-induced movement of the periplasmic half of helix IV in the fully active, ion-translocating state at pH 8 (Appel et al, 2009).
Apart from NhaA, NhaP1 of Methanococcus jannaschii is currently the only other member of the CPA superfamily for which structural information is available. Cryo-EM of 2D crystals has shown that NhaP1 is also a dimer in the membrane, broadly similar but different in detail from NhaA (Vinothkumar et al, 2005). Like NhaA, NhaP1 undergoes a conformational change between pH 4 and 8, as revealed by projection maps of 2D crystals at 8 Å resolution (Vinothkumar et al, 2005). The pH-induced change in NhaP1 is both different and more pronounced than in NhaA. Interestingly, the activity profile of NhaP1 is the opposite of that of NhaA: the latter is fully active at pH 8 but inactive at pH 6, whereas NhaP1 is inactive at pH 8 and active at pH 6 (Hellmer et al, 2002; Vinothkumar et al, 2005). Sequence comparison indicates a sequence identity of ∼20% identity between M. jannaschii NhaP1 and the membrane domain of its eukaryotic homologues NHX7 (Arabidopsis thaliana) and NHE1 (Homo sapiens), whereas the sequence identity of E. coli NhaA is only 15%. Hydrophobicity analysis has suggested 13 rather than 12 TMHs in NhaP1 (Hellmer et al, 2003).
We report here the 3D structure of M. jannaschii NhaP1 at 7 Å resolution, determined by electron crystallography of 2D crystals. A detailed comparison to the 3D EM map (Williams, 2000) and the 3.45 Å X-ray structure (Hunte et al, 2005) of NhaA reveals that the structure of the six-helix bundle thought to harbour the ion-translocation site is similar in both proteins. The six-helix bundle includes the two unwound regions in NhaA helices IV and XI, corresponding to TMH 5 and 12 in NhaP1. The motif of unwound regions in the two crossed helices is thus likely to be common to all CPAs. The 3D map shows that the E. coli-expressed NhaP1 indeed has 13 TMHs, 12 of which can be related to those of NhaA. The extra helix is located at the N-terminus and has the characteristics of an uncleaved signal sequence. Sequence comparison indicates that similar N-terminal helices are also present in several eukaryotic homologues of NhaP1. The deletion of this helix has a profound effect on antiporter function.
Results and discussion
Electron crystallography
Dialysis of NhaP1 solubilized in 0.1% dodecyl maltoside (DDM) against acetate buffer (pH 4) produced tubular crystals that were 0.3–0.6 μm wide (Vinothkumar et al, 2005). Both the size and the order of the crystals improved upon detergent exchange at the stage of protein purification (Supplementary Figure S1). NhaP1 solubilized in DDM and purified in 1% n-octyl glucoside (OG) yielded wider crystals that were more suitable for 3D data collection by cryo-EM. Crystal quality improved further when the lipids added for 2D crystallization were dissolved in 1% DDM rather than DM. The low CMC of DDM means that this detergent is removed more slowly by dialysis than either OG or DM, and this may help the formation of better 2D crystals.
Cryo-EM and image processing of tubular crystals flattened on the support film showed two easily distinguishable 2D lattices from the two sides of the vesicle, including an angle of 12–15° with one another. The crystals had the same p22121 symmetry and virtually the same unit cell (Table I) as those previously obtained (Vinothkumar et al, 2005). In all, 74 images of 2D crystals tilted up to 45° were processed (Supplementary Figure S2). In most cases, the two lattices yielded two sets of projection data per image. Amplitudes and phases were merged into a 3D data set (Table I) and plotted along each lattice line to 7 Å resolution. Lattice lines were fitted with smooth curves (Supplementary Figure S3) and sampled to generate a set of structure factors from which a 3D map of NhaP1 was calculated.
Table 1. Electron crystallographic data.
| Plane group | p22121 |
| Unit-cell dimensions | a=81 Å, b=103 Å, γ=90° |
| Number of imagesa | 74 |
| Resolution in planeb | 7 Å |
| Resolution in z directionb | 15 Å |
| Total no. of observationsc | 7475 |
| No. of unique observations | 1494 |
| Overall weighted R-factor (%)c | 0.297 |
| Overall weighted phase errorc | 17.8° |
| aDistribution of tilt angles: 15 (0°), 14 (20°), 17 (30°), 28 (45°). | |
| bAs calculated from the point-spread function of the experimental data. | |
| cFrom program LATLINEK. |
3D map of the NhaP1 dimer
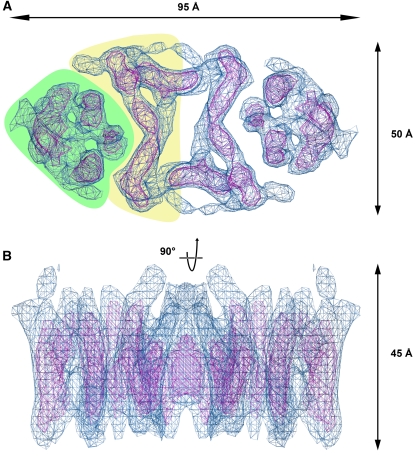
The 7 Å 3D map of NhaP1 indicates that the archaeal antiporter is a dimer of two identical protomers, related by crystallographic two-fold symmetry. The NhaP1 dimer is oval shaped, with a long axis of 95 Å and a short axis of 50 Å (Figure 1). We identified 13 rod-shaped densities in the protomer, characteristic of membrane-spanning α-helices at this resolution, which we refer to by Arabic numerals as helices 1–13, to distinguish them from helices I–XII in NhaA (Hunte et al, 2005). At the position of the two-fold axis, there is a central cavity measuring 10 × 20 Å that is most likely filled with lipid. On either side of this cavity, the protomers are in close contact. In each protomer, seven α-helices line the dimer interface (yellow background in Figure 1A). The helices in this region are tilted by up to 40° relative to the membrane normal. At either end of the dimer, there is a bundle of six α-helices (green background in Figure 1A), with three helices facing the TMHs at the interface, while the other three face the lipid bilayer. The six helices in this bundle are less highly tilted, including angles of 5–25° with the membrane normal.
Figure 1.
Top view (A) and side view (B) of the 3D map of NhaP1 from M. jannaschii obtained by electron crystallography of 2D crystals. The six-helix bundle and the group of transmembrane helices near the dimer interface in (A) are shown on a green or yellow background, respectively.
Comparison with NhaA
In overall shape and dimensions, the NhaP1 dimer is similar to NhaA (Williams, 2000). The z dimension of both antiporters is 45 Å, which is about 10 Å more than the thickness of the E. coli inner membrane, so that they protrude by no more than a few Å from each membrane surface. Closer inspection reveals important differences, as had already been apparent from the projection maps (Williams et al, 1999; Vinothkumar et al, 2005). In NhaA, the two protomers are connected by a β-sheet at the periplasmic membrane surface (Appel et al, 2009), inserted between TMH I and II. Otherwise, the NhaA protomers are well separated by a lipid-filled gap, so that there are minimal hydrophobic contacts between them. By contrast, the NhaP1 protomers are held together by tight helix–helix interactions across an extensive, most likely hydrophobic contact surface at the dimer interface. The map shows no evidence of a periplasmic β-sheet, or any other contact above or below the membrane (Figure 1B).
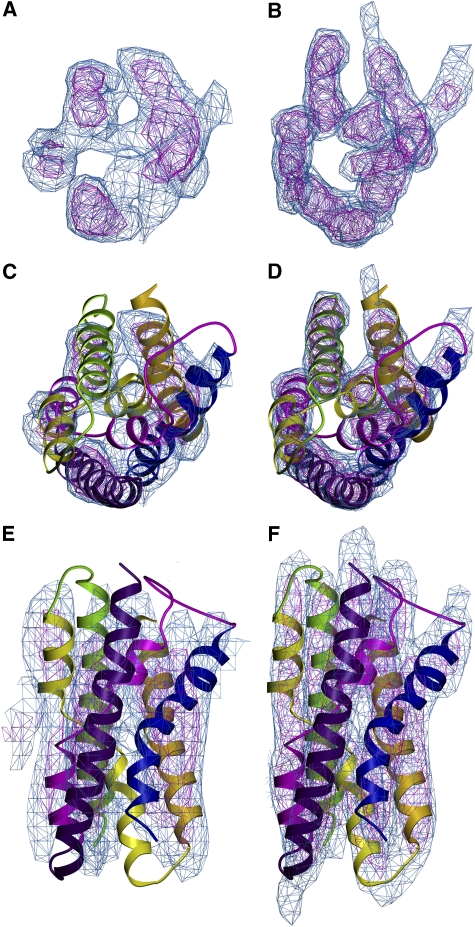
Whereas the protomers of the two antiporters clearly interact in different ways to form dimers, the six-helix bundles at either end of the NhaA and NhaP1 dimers look remarkably similar (Figure 2). This helix bundle contains the two pairs of half helices referred to as IVc/IVp and XIc/XIp in NhaA. Each pair is connected by a short stretch of unwound polypeptide, with a number of charged or polar residues that are thought to form the ion binding and translocation site (Hunte et al, 2005). Comparison of the corresponding map regions of NhaA (Hunte et al, 2005; Appel et al, 2009) and NhaP1 (Figure 2C–F) suggests that the six-helix bundle, including the two pairs of half helices and their unwound stretches are conserved in NhaP1. However, due to the missing cone of data in electron crystallography, and the resulting limited z-resolution of ∼15 Å for NhaP1 (Table I), or ∼14 Å for NhaA (Williams, 2000), the peptide stretches linking the half helices are not resolved in the current EM maps.
Figure 2.
The 7 Å map of six-helix bundle in NhaP1 (A); 6 Å map of NhaA (B; Williams (2000)), both contoured at 1 s (blue) and 2 s (magenta). X-ray structure of six-helix bundle in NhaA (Hunte et al, 2005) fitted to the corresponding regions of the NhaP1 (C, E) and NhaA maps (D, F; Appel et al (2009)). (C, D) The six-helix bundle is seen from the top and in (E, F) from the membrane.
Sequence comparison
Prompted by the observed similarities and differences in the 3D maps of NhaP1 and NhaA, we compiled a sequence alignment of 11 representative members of the CPA superfamily, including 3 bacterial NhaAs, 3 archaeal NhaPs, plus 5 eukaryotic sodium/proton antiporters from yeast, plants, and mammals. The alignment (Figure 3) confirms that M. jannaschii NhaP1 is indeed more closely related to the eukaryotic antiporters of the CPA1 clade than NhaA. The periplasmic β-sheet between TMH I and II, which links the two protomers in E. coli NhaA is not present in the CPA1 clade, and apparently is not present in all bacterial NhaA homologues.
Figure 3.
Sequence alignment of 11 members of the CPA superfamily. Transmembrane helices in the NhaA X-ray structure (Hunte et al, 2005) as well as the predicted transmembrane regions are shown in cyan and as secondary structure elements above the alignment. Blue boxes mark stretches of unwound helix in the NhaA structure. Conserved aspartates involved in ion binding or translocation are marked with red asterisks. The N-termini of NhaPΔ22 and NhaPΔ27 are highlighted by red circles.
NhaP1 had been predicted to have 13 rather than 12 TMHs (Hellmer et al, 2002, 2003; Kedrov et al, 2007), and indeed our 7 Å structure shows 13 membrane-spanning helix densities in the NhaP1 protomer. Sequence alignment (Figure 3) indicates that each of the 13 TMHs of NhaP1 has a correspondence in NhaA, except for one extra helix at the N-terminus of NhaP1. This extra N-terminal helix is common to all archaeal NhaPs. In membrane proteins with several TMHs, the first hydrophobic sequence generally acts as a signal sequence that targets the nascent peptide to the membrane (Hegde and Bernstein, 2006). These signal sequences can be either cleaved off or remain as part of the mature protein (Higy et al, 2004). Indeed, the N-terminal helix of NhaP1 has the characteristics of a signal sequence (Emanuelsson et al, 2007)
A similar hydrophobic signal sequence is found in most, although not all, of the eukaryotic CPAs in our alignment. At least one of them (human NHE6a) seems to have two hydrophobic stretches before the equivalent of NhaA helix I. A. thaliana NHX1 lacks such a sequence (Sato and Sakaguchi, 2005). In the case of human NHE1, cystein mutagenesis in combination with biotinylation experiments has suggested that this hydrophobic signal sequence is present in the mature protein, and that its N-terminus is located in the cytosol (Wakabayashi et al, 2000). However, subsequent studies showed that it is cleaved off in vitro (Miyazaki et al, 2001), and this also seems to be true of the N-terminal signal sequence of NHE3 (Zizak et al, 2000) and NHE6 (Miyazaki et al, 2001).
It is worth noting that all the eukaryotic members of the CPA1 family in our alignment (Figure 3) have another hydrophobic sequence inserted between the equivalents of TMH 4 and 5 in NhaP1, TMH III and IV in NhaA, respectively. Perhaps this hydrophobic stretch forms another TMH that conserves the membrane topography and transport directionality in those of the eukaryotic antiporters, which contain an uncleaved signal sequence, and hence an extra N-terminal TMH, bringing the total number of helices up to 14. The 12 TMHs found in the NhaA structure thus seem to be a minimum, rather than the rule in the family.
Conserved six-helix bundle
With the knowledge that the helices in the six-helix bundle, and in particular the two partially unwound helices IV and XI of E. coli NhaA are structurally conserved in NhaP1, we can assign the 13 helix densities in the NhaP1 structure to their corresponding polypeptide sequences. The unwound region of NhaA helix IV extends from A131 to I134 and has the sequence ATDI. This corresponds to ATDP in NhaP1 (A130–P133), human NHE6a and Saccharomyces cerevisiae Nhx1 (Figure 3). The aspartate in this motif is fully conserved throughout the family, and essential for function in NhaP1 (Hellmer et al, 2003). Similar considerations hold for the unwound peptide of helix XI in NhaA, which stretches from G336 to T340 and comprises the sequence motif GIGFT. This motif corresponds to PRGVV in NhaP1 (P346–V350), with the RG pair being conserved in the archaeal NhaP and the eukaryotic CPA1 families (Figure 3). The proline, arginine, and glycine in the PRGVV sequence of NhaP1 would certainly break a hydrophobic membrane-spanning helix, and presumably this is also true in the eukaryotic homologues, even if they lack the proline residue. The glycine in this sequence is the only fully conserved residue in the second unwound polypeptide stretch. We conclude that helices IVc/IVp and XIc/XIp in NhaA correspond to two pairs of half helices in NhaP1, referred to as 5a/5b and 12a/12b.
Two other buried charges that have essential roles in the transport mechanism are D163 and D164 in helix V of NhaA. The first of these two aspartates (D161 in NhaP) is fully conserved in the CPA superfamily. The IIDDLG motif in bacterial NhaA (I161–G166) occurs as FNDPLG in the archaeal NhaPs (F159–G164 in NhaP) (Figure 3). The N160/D161 pair in this sequence is conserved throughout the CPA1 family. In NhaP1, D161 is essential for function (Hellmer et al, 2003). It may be significant that NhaA, which exchanges two protons for one Na+, has two consecutive aspartates in this position, whereas the CPA1 family members, which apparently exchange only one proton per Na+, have one.
Molecular model of NhaP1
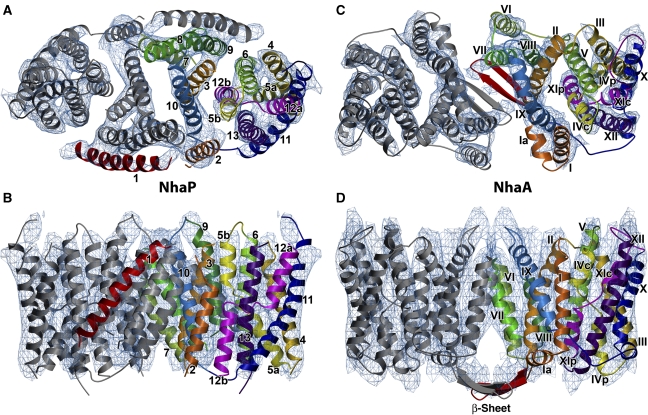
Based on the 3.45 Å structure of NhaA (Hunte et al, 2005) and the sequence alignment of Figure 3, we built a molecular model of NhaP1 (Figure 4). Starting with the structurally conserved six-helix bundle (Figure 2), and using the conserved residues identified above as reference points, we fitted NhaA helices III, IV, V as well as X, XI, and XII manually to the corresponding map regions of the NhaP1 six-helix bundle (helices 4, 5, 6, and 11, 12, 13, respectively). Only minor readjustments in length and orientation of the NhaA helices were necessary to achieve an excellent fit.
Figure 4.
Model of NhaP1 based on the sequence alignment in Figure 4 and the NhaA X-ray structure (Hunte et al, 2005), fitted to the 7 Å map of NhaP1 (A, B). Model of the NhaA dimer (pdb code 3FI1; Appel et al (2009)) fitted to the 6 Å map of NhaA (C, D; Williams (2000)). (A, C) Viewed from the top. (B, D) Viewed from the side. Arabic numbers in (A, B) refer to the 13 helices of NhaP1 and Roman numbers in (C, D) refer to the 12 helices of NhaA. Corresponding helices in one protomer of each antiporter are shown in the same colour. The uncleaved signal peptide helix 1 in NhaP1 and the β-hairpin in NhaA are shown in red.
The helix densities at the dimer interface were less similar in the two structures, and required some rearrangement to fit the NhaP1 map in this region. Only small movements were needed to fit NhaA helix I and the N-terminus of helix II into the corresponding NhaP1 map densities. The C-terminus of helix II was moved by ∼10 Å, and the N-terminus of helix IX, located in the same region as the C-terminus of helix II, was shifted by ∼8 Å. The N-terminal end of NhaA helix VI was shifted by 12 Å and in its C-terminal end by 18 Å to fit the corresponding region of the NhaP1 map. The densities corresponding to helices VII and VIII were clearly longer than in NhaA. Helices 8 and 9 of the NhaP1 model were therefore extended by 8 and 10 residues, respectively, to span the entire thickness of the protomer.
After fitting the 12 helices of NhaA into the NhaP1 map, an unassigned helix density remained, which was accounted for by the extra helix of NhaP1. The C-terminal end of this helix was close to the equivalent of NhaA helix I, and easily linked to it by a short loop. Accordingly, this map region corresponds to the N-terminal helix 1 of NhaP1, identified above as an uncleaved signal sequence. With this assignment, the equivalent of helix I in NhaA becomes helix 2 in NhaP1, helix II in NhaA becomes helix 3 in NhaP1, and so on to helix XII of NhaA that becomes helix 13 in NhaP1.
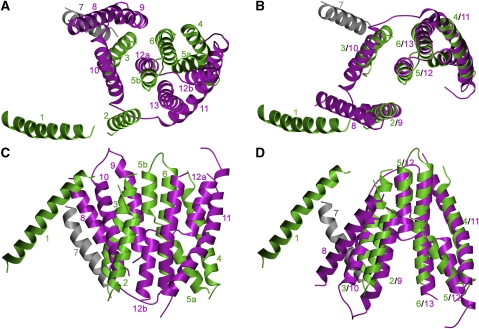
Inverted helix repeat in NhaP1
Helices IV and XI of NhaA that contain the two functionally important stretches of unwound polypeptide are at the centre of an inverted repeat. This repeat relates TMHs III, IV, and V in one half of the six-helix bundle to X, XI, and XII in the other half of the bundle (Hunte et al, 2005). The same inverted repeat is also observed in the corresponding structures of NhaP1. However, in the archaeal antiporter, it extends further and applies to 12 out of the 13 TMHs, including all except one at the dimer interface. This striking relationship is seen most clearly when helices 8 to 13 of NhaP1 are rotated by ∼180° in-plane around the long axis of the dimer (Figure 5) and superposed onto helices 1 to 6. The NhaP antiporters thus consist of two groups of six TMHs each inverted relative to one another and connected by a single helix (helix 7 in NhaP1). The repeat motif includes helix 1, which suggests that this uncleaved signal sequence is part of the mature protein. Assuming that the inverted repeat reflects a single event in the early evolution of the CPA family, NhaP1 must be the more ancient of the two antiporters.
Figure 5.
Inverted helix repeat in NhaP1. (A, C) Helices 1–6 (green) and 8–13 (purple) of the inverted repeat. The connecting helix 7 is grey. (B, D) The superposition of the repeated motifs after rotation of helices 8–13 by ∼180° roughly along the long axis of the NhaP1 dimer. (A, B) Seen from the top. (C, D) Seen from the membrane.
Functional role of N-terminal helix 1
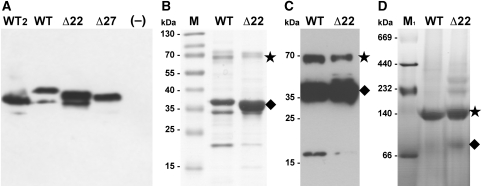
We asked whether helix 1 in NhaP1 is an essential part of the antiporter, or is present in the recombinant M. jannaschii protein merely because of incorrect processing by the E. coli membrane insertion machinery. To answer this question, we examined the pH-dependent transport activity of four NhaP1 constructs in everted vesicles. We generated two deletion constructs, namely, NhaP1Δ22 and NhaP1Δ27, which lacked the first 21 or 26 N-terminal residues of the NhaP1 sequence (Figure 3). These two constructs differed in the number of charged residues at the N-terminus, which we thought might affect expression levels or membrane insertion; this proved, however, not to be the case. Both deletion constructs carried a myc tag before a C-terminal hexa-histidine tag for purification. For comparison with earlier functional studies (Hellmer et al, 2002, 2003), we also generated a new WT construct with the same two affinity tags. As a fourth construct, we used the protein that yielded the 2D crystals and structural data in this and our previous study (Vinothkumar et al, 2005). This fourth construct had only the C-terminal histidine tag. All constructs were expressed separately in KNabc cells, an E. coli strain that is deficient in sodium–proton antiporters (Nozaki et al, 1996). Western blot analysis with an antibody against the C-terminus of NhaP1 indicated that the expression levels of all four constructs were comparable (Figure 6A). Proton efflux upon addition of NaCl was measured with everted vesicles, using the pH-sensitive fluorescence dye acridine orange.
Figure 6.
Biochemical characterization of NhaP1. (A) Western blot of everted vesicles used in the activity assay. 30 μg total protein was loaded per lane and detected with an antibody against the C-terminal helix of NhaP1 coupled to horseradish peroxidase. (B) Ni-NTA-affinity purified WT and Δ22 protein (5 μg) separated on a 12% gel and stained with Coomassie brilliant blue. (C) Western blot of Ni-NTA-affinity purified WT and Δ22 protein (5 μg) transferred to a PVDF membrane, immunostained with anti-myc antibody coupled to horseradish peroxidase. (D) Blue-native gel electrophoresis of WT (30 μg) and Δ22 (20 μg). Samples were run on 4–16% Bis/Tris native-PAGE and destained. The NhaP1 dimer is indicated with an asterisk, the monomer by a diamond. WT: NhaP1wt-Myc-His; WT2: NhaP1wt-His; Δ22: NhaP1Δ22-Myc-His; Δ27: NhaP1Δ27-Myc-His; –: untransformed KNabc cells; M: prestained protein marker (Fermentas); M1: HMW protein mix for native electrophoresis (GE Healthcare).
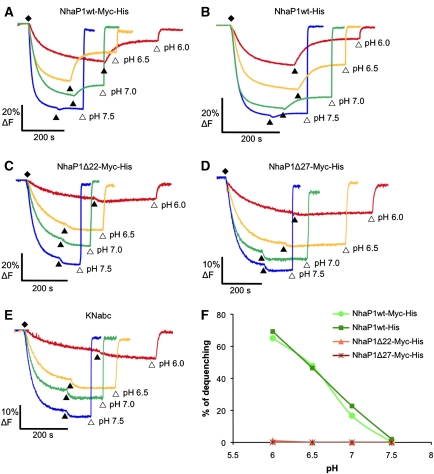
The activity profiles of both WT constructs were similar, indicating that the additional myc tag did not affect transport (Figure 7A and B). The his-tagged protein used for structural studies had already been shown to be highly active in reconstituted vesicles (Vinothkumar et al, 2005). Within the pH range tested, the two WT constructs were maximally active at pH 6. At increasing pH the activity decreased, and dropped to background level at pH ⩾7.5. Fluorescence traces for the deletion constructs Δ27 and Δ22 showed no detectable transport activity at any pH in the range tested (Figure 7C and D).
Figure 7.
Na+/H+ antiporter activity of NhaP1 constructs in everted vesicles. (A, B) Two WT constructs with one or two affinity tags had normal activity at pH 6, reduced activity at pH 6.5, and no detectable activity at pH ⩾7.5. Constructs Δ22 and Δ27 lacking helix 1 (C, D) or untransformed KNabc cells (E) showed no detectable activity in the pH range tested. (F) pH-dependent activity expressed as percentage of dequenching. Fluorescence measurements were performed with 40 μl vesicle suspension at 5 mg/ml final protein concentration in a thermostated (22°C), stirred cuvette containing 2 ml of 2 μM acridine orange in MTCM buffer (10 mM MES/Tris titrated to the pH indicated, 140 mM choline chloride, 5 mM MgCl2, 5 mM KCl). (♦) Vesicles preloaded with protons by addition of 2.5 mM Tris-DL-lactate titrated to the pH of the MTCM buffer; (▴) transport initiated with 25 mM NaCl; (▵) pH gradient dissipated with 25 mM NH4Cl. All concentrations are final. The excitation wavelength was 495 nm and emission was recorded at 530 nm.
To examine further whether the loss of activity was due to protein misfolding or an inability of the deletion constructs to dimerize in the membrane, we purified the WT and Δ22 proteins by Ni-NTA affinity chromatography. Both yielded stable membrane protein fractions that ran essentially as a single, symmetrical peak on a gel-filtration column in the presence of 0.03% DDM (not shown). CD spectra of the isolated proteins in detergent solution were virtually identical (Supplementary Figure S4), indicating the same high α-helix content in both forms. The 7-Å structure of NhaP1 shows that the WT consists almost entirely of membrane-spanning α-helices, and it is thus reasonable to assume that the Δ22 mutant is likewise fully folded into transmembrane helices. This suggests that the lack of activity of the truncated mutant is not due to misfolding.
Polyacrylamide gel electrophoresis (Figure 6B) and western blot analysis with anti-myc antibodies (Figure 6C) indicated that a significant fraction of both the WT and the Δ22 protein ran as a dimer, even in the presence of the harsh detergent SDS. The WT protein was subject to some proteolytic degradation, whereas the truncated form was largely resistant. Blue-native gel electrophoresis (Figure 6D) indicated that in the absence of SDS, virtually all the WT construct was dimeric. The dimer was the most prominent band also for the Δ22 mutant, while small amounts of monomer and two higher molecular weight bands, presumably unspecific oligomers, were also visible. The loss of transport activity in NhaP1 truncation mutants that lack helix 1 is therefore not due to proteolytic degradation or the inability of the protein to form dimers.
Finally, it is possible that helix 1 determines the orientation of the antiporter in the membrane, and that it is active in only one, its native, right-side out orientation. Given the characteristics of helix 1 as a signal sequence, it is not unlikely that a gene product lacking this sequence would be inserted into the membrane upside down. At least for the mitochondrial ADP-ATP carrier reconstituted into liposomes, it has been shown that only one of the two possible orientations is active (Kramer and Klingenberg, 1979). To eliminate this intriguing possibility for NhaP1, we conducted transport studies with the wt and Δ22 protein reconstituted into proteoliposomes in vitro. If the inactivity of the truncated form in everted vesicles is due to the upside-down orientation of the antiporter in the membrane, then the protein should be active in reconstituted vesicles, where both orientations are equally likely. We know from the p22121 symmetry of the 2D crystals that the wt protein inserts into reconstituted vesicles in both orientations.
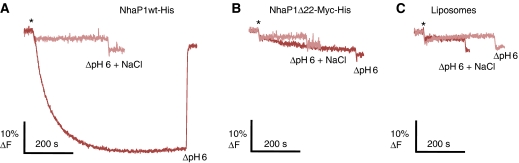
WT or Δ22 protein reconstituted into proteoliposomes were exposed to a sodium and pH gradient across the membrane (high pH and sodium concentration inside). When the protein is active, protons are pumped into the vesicles and acridine orange accumulates inside, resulting in dimerization and quenching of the fluorescence dye (Palmgren, 1991). In this assay, the reconstituted WT protein was highly active at pH 6. By contrast, Δ22 reconstituted into proteoliposomes in the same way did not show significant quenching (Figure 8A and B). Control experiments with empty liposomes or in which the sodium gradient was dissipated also showed no quenching (Figure 8). These results resemble those obtained with everted vesicles, and indicate that the inactivity of the Δ22 form is not due to an upside-down orientation of the protein in the membrane.
Figure 8.
Na+/H+ antiporter activity of NhaP1 constructs reconstituted into proteoliposomes. (A) WT construct shows normal activity at pH 6 and no detectable activity if the sodium gradient was dissipated. (B, C) Construct Δ22 lacking helix 1, or liposomes without protein showed no significant activity at any condition. Fluorescence measurements were performed by adding (*) 4 μl proteoliposomes in a thermostated (22°C), stirred cuvette containing to 2 ml of 2 μM acridine orange in MTCM buffer (10 mM MES/Tris pH 6, 140 mM choline chloride, 5 mM MgCl2, 5 mM KCl with or without 0.3 M NaCl). (▵) Proton gradient was dissipated by addition of NH4Cl to a final concentration of 25 mM. The excitation wavelength was 495 nm and emission was recorded at 530 nm.
In summary, we conclude that the absence of helix 1 in recombinant NhaP1 has no major effect on protein stability, α-helix content, or dimer formation. The only apparent difference between the two forms is that the WT is active, and the truncated mutants are not. There are several possible explanations: either helix 1 participates directly in ion transport. This seems unlikely since there is only minimal contact between it and the six-helix bundle, which most likely harbours the transport site (Figures 4 and 5). In the centre of this bundle, the transport site would be shielded from any direct contact to the helices on the dimer interface, including helix 1. Alternatively, it is possible that helix 1 is involved in the allosteric regulation of antiporter activity. Allosteric regulation of the substrate/product antiporter CaiT has recently been demonstrated (Schulze et al, 2010). The easiest way to investigate this would be by direct structural comparison of the WT and truncated protein, but so far the latter has resisted all our attempts to crystallize. In the absence of crystals, we are currently investigating the membrane topography of the truncated mutant, and whether or not the absent signal sequence does indeed cause the protein to insert into the membrane upside down.
Materials and methods
Cloning, expression, purification, and 2D crystallization
The NhaP1 construct described previously (Vinothkumar et al, 2005) was used for structural studies. The protein was expressed by auto-induction (Studier, 2005) in BL21(DE3)-pLysS cells. Cultures grown at 37° were harvested after 16–18 h. The protein was purified as before (Vinothkumar et al, 2005), with the following modifications. Protein for 2D crystallization was solubilized in 1.5% DDM. The detergent was changed to 1–1.5% OG while washing the protein on the Ni2+-NTA column. The eluted protein was dialysed for 2–3 h against 25 mM Na-acetate pH 4, 10% glycerol, 300 mM NaCl, 2 mM β-mercaptoethanol or 2–3 mM DTT, and 1% OG. 2D crystals were grown as described (Vinothkumar et al, 2005), except that the NaCl concentration in the dialysis buffer was reduced to 50 mM, and the E. coli polar lipids (Avanti Polar Lipids) added for 2D crystallization were solubilized in 1% DDM (Glycon).
For transport measurements in everted vesicles, a construct with an additional myc tag at the C-terminus, identical to one previously used (Hellmer et al, 2002, 2003), was recreated. Constructs were amplified from a template NhaP1 gene and cloned into the pTrcHis2-Topo plasmid via NcoI and EcorI restriction sites, with the primers NhaP1Δ22_NcoI forward: 5′-CCG CCG CCA TGG CCC TTA AAA TTG CTG AAA AGT TAA AAA TTC C-3′; NhaP1Δ27_NcoI forward: 5′-CCG CCG CCA TGG CCC TTT TAA AAA TTC CAG ATA TAC CGT TAT TG-3′; NhaP1_EcoRI reverse: 5′-CAA AGT ATA AAG AAG AAT CCC ACC ATA AGG GCG AAT TCG CCG CC-3′. For cloning of NhaP1wt into the pTrcHis2-Topo plasmid with only a C-terminal his-tag the following plasmids were used: NhaP1wtHis_NcoI forward: CCG CCG CCA TGG GCG AAC TTA TGA TGG CTA TTG G; NhaP1wtHis_EcorI reverse: CGG GAA TTC TCA GTG GTG GTG GTG GTG GTG CTC GAG GGA TTC.
Data collection and image processing
Specimens of tubular 2D crystals were prepared for data collection by the back-injection method (Wang and Kühlbrandt, 1991) in 4.5% trehalose (Vinothkumar et al, 2005). Usually, copper grids coated with 80–100 Å amorphous carbon were used for specimen preparation. Occasionally, copper grids coated with a conductive titanium–silicon metal glass (Rhinow and Kühlbrandt, 2008) were used for recording images at a 45° tilt. Images were recorded at liquid-helium temperature, with a JEOL 3000 SSF electron microscope at a specimen tilt of 0°, 20°, 30°, and 45°, an acceleration voltage of 300 kV, and a magnification of × 70 000 in spot-scan mode (Downing, 1991). For data collection at 0° or 20° tilt, a dose of ∼20 e/Å2 was used. At higher tilt angles, the dose was increased to compensate for the larger irradiated area. The defocus of all images was within a range of 0.2–1.6 μm. Crystal quality was evaluated by optical diffraction. Well-ordered areas of 4000 × 4000 or 6000 × 6000 pixels were digitized with a 7-μm step size on a Zeiss SCAI flat-bed scanner.
The 74 lattices included in the 3D reconstruction were processed with the MRC package (Henderson et al, 1986; Crowther et al, 1996) to correct for lattice distortions and the effect of the contrast transfer function (CTF). Initial estimates of tilt angles were calculated from lattice parameters using the EMTILT program (Roberts et al, 1981), and image data were merged to p22121 symmetry. Refinement of this first raw 3D data set was carried out in three rounds. In the first round, phase origins, tilt geometry, and CTF were refined including data up to 10 Å resolution. For the second and third round, these parameters were refined including data to 8 or 6 Å resolution, respectively. Each refinement step resulted in a significant reduction of background noise and improvement of vertical resolution. The point-spread function indicated a nominal maximum resolution of 6.5 Å in the membrane plane and 15 Å in the perpendicular direction. Image amplitudes were scaled using the program SCALIMAMP3D, applying an inverse temperature factor of B=−450 Å2 to compensate for the resolution-dependent degradation of image amplitudes. The 3D map was calculated with the CCP4 program suite (Collaborative Computational Project 4, 1994) and the molecular model of NhaP1 was built manually with COOT, using the X-ray structure of NhaA (pdb-id 1zcd) as a template. Superimpositions were carried out with the SSM Superposition routine (Krissinel and Henrick, 2004) within COOT (Emsley and Cowtan, 2004). Figures were generated using the programs PovScript+ (Fenn et al, 2003) and Povray (http://www.povray.org).
Activity in everted vesicles
Antiporter activity was measured as H+ efflux from everted vesicles. nhaA, nhaB, and nhaC deficient E. coli KNabc cells (Nozaki et al, 1996) transformed with the respective plasmids were grown overnight at 37°C in LBK medium (10 g/l tryptone, 5 g/l yeast extract, 87 mM KCl) to A600 nm=1. Protein expression was induced by addition of IPTG to 1 mM and cells were harvested after 3 h. Everted vesicles were prepared as described (Rosen, 1986), resuspended in TCDS buffer (10 mM Tris/HCl pH 7.5, 140 mM choline chloride, 250 mM sucrose, and 0.5 mM DTT), at a total protein concentration of 5 mg/ml and stored in liquid N2 for further use. The expressed gene products were identified and their concentrations in the everted vesicle fraction estimated by SDS–PAGE and western blot analysis with a polyclonal antibody against the C-terminus of NhaP1 (GenScript) coupled to horseradish peroxidase. Fluorescence measurements were performed with 40 μl everted vesicles in a stirred cuvette at 22°C containing 2 ml of MTCM buffer (10 mM MES/Tris, 140 mM choline chloride, 5 mM MgCl2, 5 mM KCL, titrated to the indicated pH) and 2 μM acridine orange. The vesicles were preloaded with protons by adding Tris-DL-lactate to 2.5 mM, titrated to the pH of the MTCM buffer. Once steady state was achieved, NaCl was added to a final concentration of 25 mM and fluorescence dequenching was monitored at 530 nm in a Hitachi fluorimeter, using an excitation wavelength of 495 nm. The transmembrane pH gradient was dissipated by the addition of NH4Cl to 25 mM.
Activity in proteoliposomes
E. coli polar lipids were dried and resuspended in MTCB (10 mM MES/Tris, 140 mM choline chloride pH 8, 5 mM KCl, 5 mM MgCl2) buffer with 0.3 M NaCl to a concentration of 20 mg lipid/ml. Liposomes were prepared by extrusion through a polycarbonate filter (pore size 0.4 μm), diluted four-fold, and destabilized by adding 20% Triton X-100. The optimal detergent concentration for protein insertion is reached when the absorption at 540 nm, a measure for the turbidity of the liposome suspension, begins to decline (Rigaud et al, 1995; Geertsma et al, 2008). Protein solubilized in 0.03% DDM was added at an LPR of 80:1 (w/w) and incubated for 30 min at room temperature. Detergent was removed by repeated addition of biobeads (Rigaud et al, 1998), first in two separate batches, each followed by shaking for 1 h at room temperature, then by adding twice the amount of biobeads followed by shaking overnight at 4°C, and finally by adding another batch and shaking for 45 min at 4°C. Proteoliposomes were collected and washed by centrifugation at 90.000 r.p.m. for 30 min, resuspended at a concentration of ∼60 mg/ml lipid, and stored in liquid N2 for further use.
Activity measurements were performed in a stirred cuvette at 22°C containing 4 μl of proteoliposomes or control liposomes, 2 ml MTCB pH 6 without or with 0.3 M NaCl and 2 μM of the fluorescence dye acridine orange. All measurements were monitored at 530 nm in a Hitachi fluorimeter, using an excitation wavelength of 495 nm. The transmembrane pH gradient was dissipated by the addition of 25 mM NH4Cl.
Supplementary Material
Acknowledgments
We thank Deryck Mills for assistance with electron microscopy, Karen Davies for advice on image processing, Remco Wouts for computer support, Carsten Zeilinger (University of Hannover, Germany) for a kind gift of the pTrcHis2-TOPO with NhaP1 plasmid, and Etana Padan for providing the E. coli KNabc strain. PG acknowledges financial support from the International Max Planck Research School on Biological Membranes, WK acknowledges support from the EU 7th Framework Programme FP7/2007-2013 under grant agreement n° HEALTH-F4-2007-201924, EDICT Consortium; from DFG SFB 628 and 807, and the DFG Cluster of Excellence Frankfurt “Macromolecular Complexes”. CP is supported by the fellowship SFRH/BD/62643/2009 from Fundação para a Ciência e a Tecnologia.
Footnotes
The authors declare that they have no conflict of interest.
References
- Appel M, Hizlan D, Vinothkumar KR, Ziegler C, Kuhlbrandt W (2009) Conformations of NhaA, the Na/H exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J Mol Biol 386: 351–365 [DOI] [PubMed] [Google Scholar]
- Brett CL, Donowitz M, Rao R (2005) Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol 288: C223–C239 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project 4C (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM (1996) MRC image processing programs. J Struct Biol 116: 9–16 [DOI] [PubMed] [Google Scholar]
- Downing KH (1991) Spot-scan imaging in transmission electron microscopy. Science 251: 53–59 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fenn TD, Ringe D, Petsko GA (2003) POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J Appl Crystallogr 36: 944–947 [Google Scholar]
- Geertsma ER, Nik Mahmood NA, Schuurman-Wolters GK, Poolman B (2008) Membrane reconstitution of ABC transporters and assays of translocator function. Nat Protoc 3: 256–266 [DOI] [PubMed] [Google Scholar]
- Hegde RS, Bernstein HD (2006) The surprising complexity of signal sequences. Trends Biochem Sci 31: 563–571 [DOI] [PubMed] [Google Scholar]
- Hellmer J, Patzold R, Zeilinger C (2002) Identification of a pH regulated Na(+)/H(+) antiporter of Methanococcus jannaschii. FEBS Lett 527: 245–249 [DOI] [PubMed] [Google Scholar]
- Hellmer J, Teubner A, Zeilinger C (2003) Conserved arginine and aspartate residues are critical for function of MjNhaP1, a Na+/H+ antiporter of M. jannaschii. FEBS Lett 542: 32–36 [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Downing KH, Lepault J, Zemlin F (1986) Structure of purple membrane from halobacterium-halobium - recording, measurement and evaluation of electron-micrographs at 3.5 a resolution. Ultramicroscopy 19: 147–178 [Google Scholar]
- Higy M, Junne T, Spiess M (2004) Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry 43: 12716–12722 [DOI] [PubMed] [Google Scholar]
- Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H (2005) Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435: 1197–1202 [DOI] [PubMed] [Google Scholar]
- Kedrov A, Wegmann S, Smits SHJ, Goswami P, Baumann H, Muller DJ (2007) Detecting molecular interactions that stabilize, activate and guide ligand-binding of the sodium/proton antiporter MjNhaP1 from Methanococcus jannaschii. J Struct Biol 159: 290–301 [DOI] [PubMed] [Google Scholar]
- Kramer R, Klingenberg M (1979) Reconstitution of adenine nucleotide transport from beef heart mitochondria. Biochemistry 18: 4209–4215 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Miyazaki E, Sakaguchi M, Wakabayashi S, Shigekawa M, Mihara K (2001) NHE6 protein possesses a signal peptide destined for endoplasmic reticulum membrane and localizes in secretory organelles of the cell. J Biol Chem 276: 49221–49227 [DOI] [PubMed] [Google Scholar]
- Nozaki K, Inaba K, Kuroda T, Tsuda M, Tsuchiya T (1996) Cloning and sequencing of the gene for Na+/H+ antiporter of Vibrio parahaemolyticus. Biochem Biophys Res Commun 222: 774–779 [DOI] [PubMed] [Google Scholar]
- Palmgren MG (1991) Acridine orange as a probe for measuring pH gradients across membranes: mechanism and limitations. Anal Biochem 192: 316–321 [DOI] [PubMed] [Google Scholar]
- Rhinow D, Kühlbrandt W (2008) Electron cryo-microscopy of biological specimens on conductive titanium-silicon metal glass films. Ultramicroscopy 108: 698–705 [DOI] [PubMed] [Google Scholar]
- Rigaud JL, Levy D, Mosser G, Lambert O (1998) Detergent removal by non-polar polystyrene beads—applications to membrane protein reconstitution and two-dimensional crystallization. Eur Biophys J Biophys 27: 305–319 [Google Scholar]
- Rigaud JL, Pitard B, Levy D (1995) Reconstitution of membrane proteins into liposomes: application to energy-transducing membrane proteins. Biochim Biophys Acta 1231: 223–246 [DOI] [PubMed] [Google Scholar]
- Roberts K, Shaw PJ, Hills GJ (1981) High-resolution electron microscopy of glycoproteins: the crystalline cell wall of Lobomonas. J Cell Sci 51: 295–313 [DOI] [PubMed] [Google Scholar]
- Rosen BP (1986) Ion extrusion systems in Escherichia coli. Methods Enzymol 125: 328–336 [DOI] [PubMed] [Google Scholar]
- Sato Y, Sakaguchi M (2005) Topogenic properties of transmembrane segments of Arabidopsis thaliana NHX1 reveal a common topology model of the Na+/H+ exchanger family. J Biochem 138: 425–431 [DOI] [PubMed] [Google Scholar]
- Schulze S, Koster S, Geldmacher U, Terwisscha van Scheltinga AC, Kuhlbrandt W (2010) Structural basis of Na(+)-independent and cooperative substrate/product antiport in CaiT. Nature 467: 233–236 [DOI] [PubMed] [Google Scholar]
- Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Taglicht D, Padan E, Schuldiner S (1991) Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem 266: 11289–11294 [PubMed] [Google Scholar]
- Vinothkumar KR, Smits SH, Kühlbrandt W (2005) pH-induced structural change in a sodium/proton antiporter from Methanococcus jannaschii. EMBO J 24: 2720–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi S, Pang T, Su X, Shigekawa M (2000) A novel topology model of the human Na(+)/H(+) exchanger isoform 1. J Biol Chem 275: 7942–7949 [DOI] [PubMed] [Google Scholar]
- Wang DN, Kühlbrandt W (1991) High-resolution electron crystallography of light-harvesting chlorophyll a/b-protein complex in three different media. J Mol Biol 217: 691–699 [DOI] [PubMed] [Google Scholar]
- Williams KA (2000) Three-dimensional structure of the ion-coupled transport protein NhaA. Nature 403: 112–115 [DOI] [PubMed] [Google Scholar]
- Williams KA, Geldmacher-Kaufer U, Padan E, Schuldiner S, Kühlbrandt W (1999) Projection structure of NhaA, a secondary transporter from Escherichia coli, at 4.0 A resolution. EMBO J 18: 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zizak M, Cavet ME, Bayle D, Tse CM, Hallen S, Sachs G, Donowitz M (2000) Na(+)/H(+) exchanger NHE3 has 11 membrane spanning domains and a cleaved signal peptide: topology analysis using in vitro transcription/translation. Biochemistry 39: 8102–8112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.