Atomic model of an infectious rotavirus particle
The structure of the entire rotavirus particle is revealed by high-resolution electron cryomicroscopy, providing insight into the structural rearrangements of the coat proteins involved in viral entry.
Keywords: cryoEM, non-enveloped virus entry, rotavirus
Abstract
Non-enveloped viruses of different types have evolved distinct mechanisms for penetrating a cellular membrane during infection. Rotavirus penetration appears to occur by a process resembling enveloped-virus fusion: membrane distortion linked to conformational changes in a viral protein. Evidence for such a mechanism comes from crystallographic analyses of fragments of VP4, the rotavirus-penetration protein, and infectivity analyses of structure-based VP4 mutants. We describe here the structure of an infectious rotavirus particle determined by electron cryomicroscopy (cryoEM) and single-particle analysis at about 4.3 Å resolution. The cryoEM image reconstruction permits a nearly complete trace of the VP4 polypeptide chain, including the positions of most side chains. It shows how the two subfragments of VP4 (VP8* and VP5*) retain their association after proteolytic cleavage, reveals multiple structural roles for the β-barrel domain of VP5*, and specifies interactions of VP4 with other capsid proteins. The virion model allows us to integrate structural and functional information into a coherent mechanism for rotavirus entry.
Introduction
To access the cytosol and initiate infection of a target cell, non-enveloped viruses—those that lack a lipid-bilayer membrane—must perforate or disrupt a membrane of the cell. Viruses of different structural classes have acquired distinct molecular machineries for carrying out this penetration step. In all cases, a triggering event that accompanies or follows receptor binding or endocytic engulfment exposes a previously buried, membrane-targeting structure, but for none of these mechanisms do we yet have a ‘molecular movie' comparable to the relatively unified picture of membrane fusion that has emerged from studies of enveloped viruses. Picornaviruses (e.g. poliovirus) and reoviruses release an N-terminally myristoylated peptide that oligomerizes into pores in the surrounding endosomal membrane (Danthi et al, 2003; Schiff et al, 2007). Parvoviruses contain a lipase, the N-terminal extension of a few of their capsid proteins, which emerges during penetration (Farr et al, 2005). Rotaviruses enter by a process that resembles the action of enveloped-virus fusion proteins: coupling of a large-scale conformational change in the penetration protein to a physical distortion of the cellular membrane that the virus will cross (Dormitzer et al, 2004).
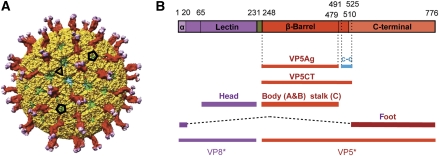
Rotaviruses are the principal agents of infectious dehydrating diarrhea of infants and the cause of nearly half a million childhood deaths per year (Parashar et al, 2006). They have a segmented, double-stranded RNA (dsRNA) genome, packaged within a multi-shelled virus particle (Estes and Kapikian, 2007). The outer protein layer of the virion—the molecular machinery for host-cell binding and penetration—contains two protein components, VP4 and VP7. These form, respectively, a set of spike-like projections and an extended shell (Figure 1A). During infectious entry, a sequence of molecular transformations in the outer-layer proteins strips them from the virion (the ‘triple-layered particle' or TLP) and delivers into the cytosol an inner capsid particle, known as the ‘double-layered particle' or DLP. The DLP does not uncoat further: when freed of the outer layer, it can synthesize, cap, and extrude mRNA transcribed from the packaged genomic segments (Estes and Kapikian, 2007). The structural components of the DLP shell are VP6 and VP2. Trimers of VP6 form a T=13 l icosahedral lattice, supported by a shell assembled from 120 copies of VP2. Associated with each dsRNA segment is one copy each of VP1 (the viral polymerase) and VP3 (the mRNA capping enzyme).
Figure 1.
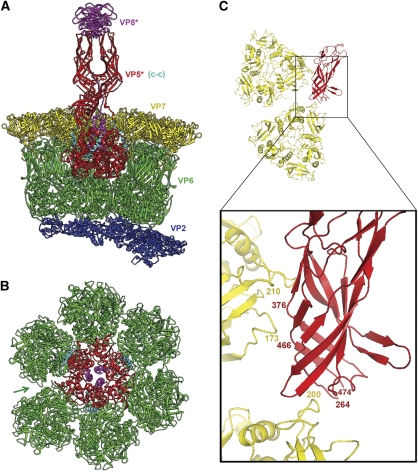
Structure of VP4 spike. (A) Overview of rotavirus TLP, in surface rendering from the cryoEM map and coordinates. VP8* is in magenta, VP5* is in red, and VP7 is in yellow. Selected icosahedral symmetry elements—two five-fold axes and a three-fold axis—are indicated by pentagons and a triangle, respectively. The VP7 trimers form a T=13 l icosahedral surface lattice, with six-coordinated positions intervening between the five-folds. There are two symmetrically distinct six-coordinated positions—VP4 spikes occupy those closer to the five-folds. Some of VP6 (green) and VP2 (blue) on the inner capsid particle can be seen through the gaps in the VP7 lattice at the six-coordinated positions not occupied by VP4 and at the five-folds. (B) Diagram of the organization of the VP4 polypeptide chain (Dormitzer et al, 2001, 2004). Tryptic cleavage generates VP8* (magenta in coloured bar) and VP5* (red), with excision of residues 232–247 (green). The segments and domains of VP8* and VP5* (α, lectin, β-barrel, c-c, C-terminal) are labelled; the β-barrel domain was called the ‘antigen domain' (VP5Ag) in some of our previous papers; the fragment generated from recombinant VP4 by successive cleavages with chymotrypsin and trypsin is called VP5CT. We have described the morphology of the spikes in the mature virion (after tryptic cleavage of VP4) as ‘head', ‘body', ‘stalk', and ‘foot'; the polypeptide-chain segments contributing to each of these structures are shown as labelled lines. The structure described in this paper reveals that the β-barrel domains of two subunits form the body and that of the third forms the stalk and that the foot includes both the segment at the N-terminus of VP8* labelled ‘α‘ and the C-terminal domain. The segment labelled ‘c-c' forms the three-chain coiled coil in the VP5CT trimer crystal structure, which probably shows the conformation of rearranged VP5* following membrane disruption (see Figure 7). Amino-acid residues at domain, segment, and fragment boundaries are designated by numbers above the coloured bar.
The VP7 shell partly covers the base of the VP4 spike and appears to lock VP4 onto the virion (Figure 1A). The lock depends on Ca2+, which stabilizes VP7 trimers by occupying two sites at each subunit interface (Aoki et al, 2009). Withdrawal of Ca2+ leads to dissociation of VP7 trimers, loss of VP7 from DLPs, and release of VP4 (Cohen et al, 1979; Dormitzer et al, 2000). From a crystal structure of VP7 (Aoki et al, 2009) and high-resolution electron cryomicroscopy (cryoEM) analysis of a VP7-recoated DLP (Chen et al, 2009), we know that each VP7 trimer clamps onto an underlying VP6 trimer, through contacts made almost entirely by the VP7 N-terminal arms (residues 51–70, approximately). A single-arm contact is apparently not sufficient to retain a VP7 on the DLP surface; only when three such contacts are correlated through stability of the VP7 trimer does the outer shell remain in place.
High-level infectivity requires that VP4 be cleaved by trypsin into two fragments, designated VP8* and VP5* (Figure 1B), with excision of residues 232–247 (Estes and Kapikian, 2007). Uncleaved VP4 is probably somewhat flexible, as the spikes only appear in cryoEM reconstructions of trypsin-treated particles (Crawford et al, 2001). The post-cleavage rearrangement that erects the VP4 spike yields a projection with obvious two-fold symmetry (Figure 1A), but the base of the spike is trimeric, and the two-fold axis of the projection is displaced by a diagonal ‘stalk' from the three-fold axis of the base (Yeager et al, 1994; Pesavento et al, 2005). The lectin-like, globular domain of VP8*, residues 65–224, accounts for each of the two heads at the tip of the trypsin-activated spike (Dormitzer et al, 2002). A β-barrel domain in VP5*, residues 264–474, accounts for each of the two halves of the ‘body' (Dormitzer et al, 2004). Molecular details of the diagonal stalk and the trimeric ‘foot' have hitherto been lacking.
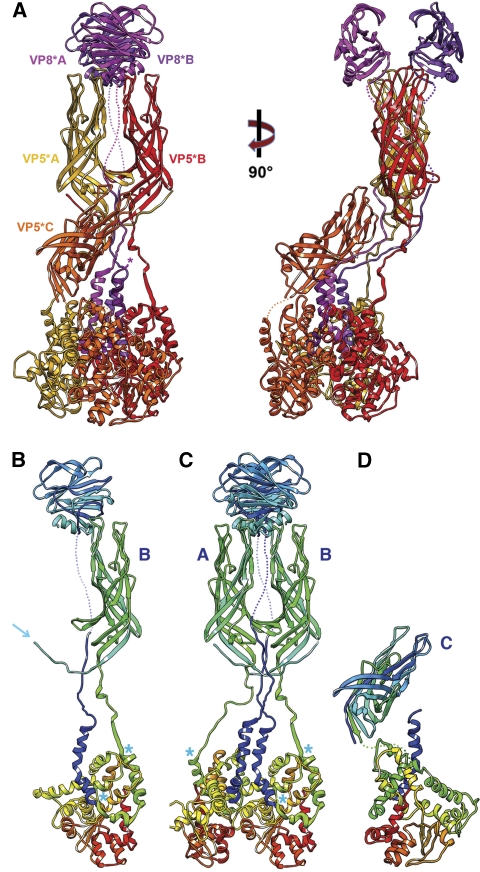
We describe here a near-atomic resolution cryoEM map of the virion. This map allows us to trace almost the complete VP4 polypeptide chain in each of the three subunits in a spike. The resulting picture of VP4 has two structurally unusual features (Figure 2). First, the initial 26 residues of VP8* are part of the base (‘foot'), with each of the three subunits contributing to a three-chain coiled coil along the local three-fold axis. Second, the β-barrel domain of the third VP5* subunit, previously assumed to be flexibly linked to the base, is actually the principal component of the stalk. Also contributing to the stalk are segments linking two of the VP8* N-terminal helices to the lectin domains at the far end of the spike and segments linking the two β-barrel domains of the body back to their C-terminal domains in the foot (Figure 2). These two unexpected features solve puzzles posed by previous structural work and lead to a more specific model for molecular events during penetration.
Figure 2.
(A) Ribbon representation of the VP4 (=VP8*+VP5*) spike. Two orthogonal views are shown. The VP8* fragments are in magenta (at the top of the molecule on the left, VP8*A is in the foreground and VP8*B is in the background); the VP5* fragments are in yellow (chain A), red (chain B), and orange (chain C). The models for the lectin- and β-barrel domains are based on the crystallographic coordinates (1KQR and 2B4H, respectively); those of the former were docked into the density, and those of the latter, docked and adjusted as described in Results. Connections evident from the map but not strong enough to represent in the deposited coordinates are shown as dotted lines. (B) The B chain (both VP8* and VP5*), coloured in a ‘rainbow' from blue (N-terminus) to red (C-terminus). The arrow shows the N-terminus of VP5*. Blue asterisks (both here and in C, D) designate ends of the segments that will form the three-chain coiled coil in the rearranged VP5* conformation. (C) The A and B chains, coloured as in (B). Note that the N-termini of VP5* A and B cross-over to interact with the β-barrel domain of the partner subunit. This exchange is also present in the crystal structure of a VP5Ag dimer (Yoder and Dormitzer, 2006); it may stabilize the asymmetric spike conformation after trypsin cleavage. (D) The C chain, coloured as in (B).
Results
Electron microscopy
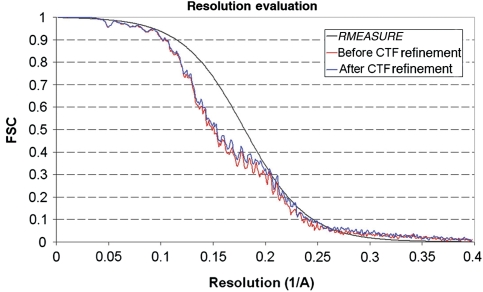
The procedures for obtaining images and determining and refining a three-dimensional reconstruction are described in Materials and methods. About 4000 particle images contributed to the final reconstruction. The molecular features in the DLP and VP7 regions were comparable in definition to those in the reconstruction of VP7-recoated DLP (7RP) (Chen et al, 2009), confirming the estimate of 4.3 Å resolution from Fourier shell correlation (FSC) (Figure 3). Density in the VP4 foot also was well defined. Moving outward in the spike, the features became less sharp. In the VP5* β-barrel domain, most of the β-strand backbone and some of the side chains were clear, and we can therefore describe the ‘effective resolution' in this domain (i.e. the resolution cutoff in X-ray crystallography that would give an equivalent density map from well-measured amplitudes and reasonably well-determined phases) as 4.5–5 Å. In the VP8* lectin domain, most of the β-strand backbone was not clearly resolved, but rod-like density for the one short helix was evident, and the effective resolution was probably about 7–8 Å.
Figure 3.
Resolution estimation. The red curve shows the FSC calculated for the icosahedral reconstruction using the CTF parameters measured by CTFTILT, which takes into account defocus variations across the image that arise from sample tilt (Mindell and Grigorieff, 2003). The FSC shown in blue was calculated for the reconstruction after refining the defocus of individual particles using restraints (Chen et al, 2009). This reconstruction was further analysed (black curve) using the computer program RMEASURE (Sousa and Grigorieff, 2007), which represents an alternative resolution estimate, confirming the estimate based on the FSC. The small discrepancy between the RMEASURE and FSC curves at lower resolution is due to the smoothed curve implemented in RMEASURE to represent its resolution estimate.
Atomic model of VP4
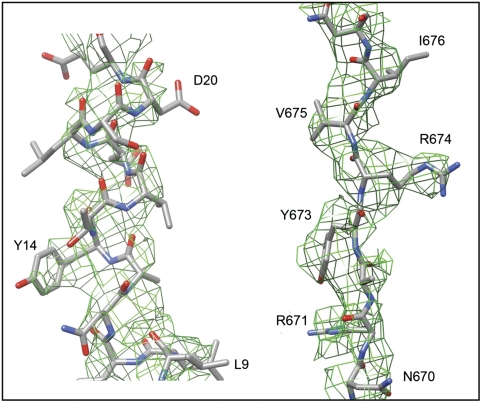
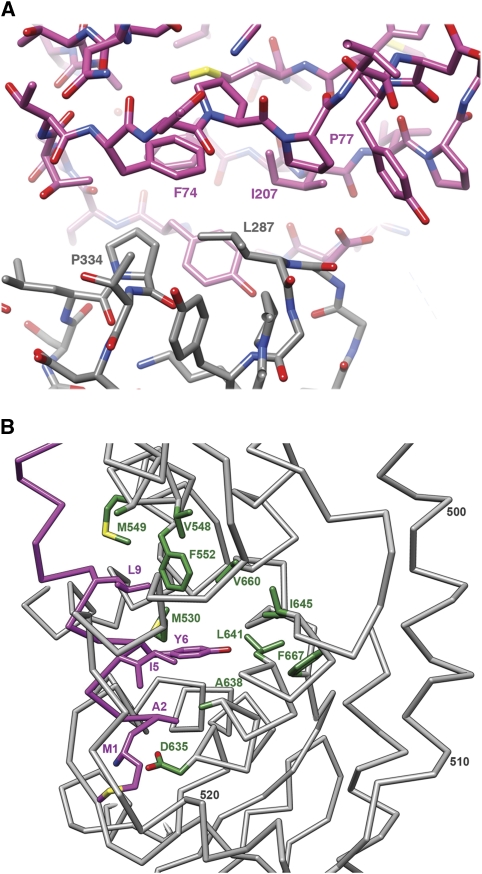
The structures of the DLP (McClain et al, 2010), VP6 (Mathieu et al, 2001), VP7 (Aoki et al, 2009), the lectin domain of VP8* (Dormitzer et al, 2002), and the β-barrel domain of VP5* (Dormitzer et al, 2004; Yoder and Dormitzer, 2006) have all been determined by X-ray crystallography; those of the DLP and the 7RP, by high-resolution cryoEM (Zhang et al, 2008; Chen et al, 2009). Interpreting the TLP image reconstruction in terms of an atomic model required de novo polypeptide-chain traces for the VP4 foot and segments linking domains, as well as insertion of known pieces into the overall structure. Most of the foot is three-fold symmetric. We could trace much of the polypeptide chain in each of the three subunits in the TLP map without local averaging. This partial model was then used to obtain the transformations needed to average the map with the program RAVE (Uppsala Software Factory). Regions that averaged well were rebuilt with the averaged map, and regions that were different (e.g. near the interface with VP6 or the linkers leading away from and back into the foot) were built only in the unaveraged map. Examples of the fit are shown in Figure 4; other examples are in Supplementary data. The de novo model ultimately included residues 1–49 and 474–776 for chains A and B and 1–29 and 500–776 for chain C. The connections for chains A and B between residues 49 and 65 (the first residue in the lectin domain) could be discerned but were not built. The averaged density for the VP4 foot was used to calculate structure factors for refinement of coordinates for the three-fold symmetric part of the model, residues 1–26 and 500–776, by masking the density carefully and moving it into a P1 unit cell (with RAVE and CCP4 programs). We carried out rigid body and minimization refinement in CNS, using the mlf target and data from 15 to 3.5 Å resolution. Figure-of-merit measures were appended to each reflection based on values from the FSC (Harauz and van Heel, 1986). R and Rfree values for the refined coordinates are shown in Table I. For the VP8* lectin domain and the VP5* β-barrel domain, the map was cut to include the entire spike, and structure factors were calculated in a P1 cell. These domains were steered as rigid bodies into the density, then modified smoothly to improve the fit with a round of rigid-body refinement (in CNS). Some further optimization of the VP5* β-barrel domains were then made as piece-wise rigid-body adjustments to improve the overall fit. The domain flexes slightly about its central β-sheet region with respect to the crystal structures, bringing the opposite ends of the barrel slightly closer together. In the final model, it became clear that key residues in the hydrophobic loops are buried in a hydrophobic surface cavity in VP8* (Figure 5A).
Figure 4.
Portions of the three-fold averaged density map of the VP4 foot, with the final model superposed. On the left is a part of the N-terminal helical region of VP8*, contoured at 1.5 σ; on the right is a strand in VP5* between residues 670 and 677, contoured at 1.0 σ.
Table 1. R and R-free, from refinement of VP5* foot coordinates, carried out as described in Materials and methods.
| Resolution cutoff (Å) | R-factor | R-free |
|---|---|---|
| 3.5 | 40.2 | 40.5 |
| 4.0 | 38.3 | 38.8 |
| 4.5 | 35.4 | 35.8 |
| 5.0 | 32.6 | 33.1 |
| 5.5 | 30.3 | 30.7 |
| 6.0 | 28.3 | 28.7 |
| 7.0 | 25.2 | 25.6 |
Figure 5.
(A) The contact between the hydrophobic loops of the VP5* β-barrel and the lectin domain of VP8*. (B) The N-terminal helix of VP8* (‘α‘ in Figure 1B) and its interactions with the C-terminal domain of VP5*. Selected residues are labelled with the single-letter code and the residue number in the VP4 polypeptide chain. The backbone and side chains of the N-terminal helix are in magenta; the side chains of selected residues in the foot (grey backbone) are in green.
The only region of VP4 not accounted for by density is the lectin domain of the third subunit. We suggest that it has been removed by a tryptic cleavage at Lys29 (in addition to the ‘normal' cleavage at 247), because the density stops abruptly at that position. The first clearly defined residue in the β-barrel density for the third subunit is Glu264; cleavage C-terminal to Lys258 or Lys263 may also have excised the segment 248–258/263. There is some evidence in the literature for cleavage at Lys258 (Crawford et al, 2001; Dormitzer et al, 2001). It is possible that, under some conditions of tryptic cleavage, the third VP8* remains flexibly tethered to its N-terminal segment through the linker between residues 29 and 65. Its presence would not change any of the conclusions below. We designate the two VP4 polypeptide chains that form the two-fold projection by the letters A and B, and the third chain, by the letter C.
VP4 conformations in the virion
The overall configuration of each VP4 subunit is that of a huge loop, with the N-terminal, helical segment anchored against the C-terminal domain. The unusual asymmetry of the assembly implies distinct configurations of the inter-domain connections (‘linkers') for each of the three polypeptide chains (Figure 2). The N-terminal segments of the A and B chains can be traced far enough towards the head that their connection to a particular VP8* lectin domain is unambiguous. Some density can be discerned for the bridge between residues 51 (at the C-terminal end of well-defined linker density) and 65 (the N-terminus of the globular lectin domain). A related argument can be made for the connection between the VP8* C-terminus and the VP5* N-terminus. These connectivities place each of the three VP8* N-termini in contact with the VP5* C-terminal foot domain of the same polypeptide chain, and each of the two VP8* lectin domains in contact with its corresponding VP5* β-barrel (Figure 2B). They further suggest that the N- and C-terminal regions of VP4 associate with each other even before the protein trimerizes, consistent with the observation that recombinant VP4 is a relatively compact monomer in solution (Dormitzer et al, 2001). Analysis of specific molecular interactions within the foot also supports this conclusion. VP8* residues 1–10, all α-helical, insert into the globular C-terminal domain of VP5*, residues 510–776. The side chains of Met1, Ala2, Ile5, Tyr6, and Leu9 are part of the hydrophobic core of the domain, and the carboxylate of Asp635 neutralizes the buried amino terminus (Figure 5B). The helices of the central coiled coil (residues 13–26) extend from the N-terminal helices, with a kink at residues 11–12 where the polypeptide chain emerges from the foot (Figure 2).
VP4 interactions with VP6
What holds the spike in place? The VP5* foot interacts both with VP6, in the DLP, and with VP7 (Figure 6A). The former interaction recruits cytosolic VP4 onto the DLP and presumably templates its trimerization. The lattice of VP6 trimers on the DLP surface has two sets of six-coordinated cavities—one closer to the icosahedral five-fold axis and the other closer to the icosahedral three-fold (Figure 1A). The VP4 spikes insert into the former set. The quasi-six-fold disposition of VP6 trimers around a VP4 spike allows each of the three VP4 foot domains to approach two VP6 trimers. Two of the C-terminal domains (subunits A and C) have essentially identical interactions with their VP6 neighbours; the third (subunit B) interacts with one of its neighbours just as do the two others, but the remaining VP6 neighbour (the sixth around the position of insertion) is out of contact range (Figure 6B). In other words, the position at which the spike inserts has approximate three-fold symmetry, relating two pairs of VP6 neighbours and one member of the third pair. The selective insertion of VP4 into one set of six-coordinated cavities depends on differences between the two locations in the DLP; the orientations of five-fold-proximal VP6 trimers shift when VP7 binds, diminishing those differences in the VP7-recoated DLP (Chen et al, 2009) and in the TLP analysed here.
Figure 6.
Interactions of VP4 in the TLP. (A) Cutaway view, with the VP4 spike in the orientation shown in Figure 2A. VP8* is in magenta and VP5* is in red. The VP5* segment that will form the coiled coil in the ‘post-entry' conformation (see Figure 7) is in cyan. In addition to the α-helix that runs axially at the periphery of the foot, this segment includes about 15 more C-terminal residues (mostly in one radially directed α-helix) at the interface between globular foot domains of the VP5* subunits (see also asterisks in Figure 2B and C). The foot is anchored at a six-coordinated position in the VP6 layer (green); a VP7 trimer (yellow) caps each trimer of VP6. Both form T=13 l icosahedral surface lattices. The VP6 lattice overlays the VP2 shell (dark blue), which surrounds the coiled RNA genome. (B) The VP5* foot and surrounding VP6 trimers, seen from outside the TLP as if the spike model in (A) were tipped towards the viewer. Thus, the two lower VP6 trimers in (B) are the two that are cut away in (A). Colours as in (A). Arrow marks a gap in VP6 packing, showing where the six-coordination deviates noticeably from local six-fold symmetry. The VP6 trimer immediately counter clockwise to the arrow is the only one of the six that does not contact the foot directly. Note the packing of the VP8* N-terminal helices (magenta) close to the three-fold axis of the foot. (C) Detail of the interaction between VP5*C and the two VP7 subunits to either side of the gap marked in (B). The small figure shows two VP7 trimers in the orientation and view that would (roughly) superpose them onto the two VP6 trimers flanking the arrow at the lower left of (B); the box shows the region illustrated in the detailed enlargement. VP7 is in yellow and VP5*C is in red. Numerals designate residues at or near the contact.
Interactions with VP7 and VP4 asymmetry
Addition of a VP7 trimeric cap onto each VP6 trimer locks VP4 into the virion. The contacts between VP7 and the VP4 foot are relatively tenuous, but VP7 creates a cover over the cavity into which the foot inserts, preventing dissociation (Figure 6A). In recoating experiments in vitro, VP4 must be added first, then VP7 (Trask and Dormitzer, 2006). A similar order of events presumably occurs during virus assembly in infected cells. The orientations of the six VP7 trimers surrounding a VP4 are such that the N-terminal arms of three VP7 subunits intrude into the foot cavity. Each contacts a VP4 subunit, at the point at which the VP5* polypeptide chain enters the foot (Figure 6). Density for the contacting segment of VP7 (which includes the glycan at Asn69) is much weaker than in other locations not adjacent to a VP4; thus, the contact itself probably makes at best a modest contribution to anchoring VP4. The order of assembly requires that the VP7 arm insert even with the VP4 foot in place. The conformation of the arm and the position of the VP4 foot are compatible with this requirement (Figure 6).
The spike adopts its unusual conformation only after tryptic cleavage (Crawford et al, 2001), which occurs following addition of VP7 (indeed, after exit from the cell in which the virus has assembled). Thus, VP7 contacts probably dictate how the spike projects. A determining feature is the gap between two of the VP7 trimers that surround the T=13 lattice position at which VP4 inserts (Figure 6B, arrow). The lower tip of the VP5*C β-barrel lies in this gap, where it has a number of interactions with the VP7 subunits to either side. In particular, the map shows that a VP7 loop containing residue 200 adjusts to contact VP5*C near residue 267; on the other side of the gap, VP7 residues in loops near 210 and 170 contact VP5* strands at 375 and 467 (Figure 6C). Collisions of these surfaces of VP5* with VP7 would prevent any of the other quasi-six-fold-related orientations, where the corresponding tips of the VP7 subunits are closer together. Thus, only the orientation actually adopted will allow the β-barrel of VP5*C to insert correctly and to form the diagonal stalk.
The contacts that determine the asymmetry of the spike also suggest how trypsin selects the observed cleavage points to generate the mature virion. The last residue of the VP5* β-barrel domain is at about position 475, and the linker (residues 475–490) into the C-terminal domain contains no trypsin-sensitive sites. The linker (residues 27–65) between the N-terminal α-helical segment of VP8* (residues 1–26) and the lectin domain also has no trypsin-sensitive sites, except at Lys29, which could readily be occluded on the immature particle by the surrounding β-barrel domains. Thus, the linker (residues 231–247) between VP8* and VP5* will be the only clearly exposed trypsin-sensitive VP4 segment. Following the known cleavages at residues 230 and 247, leading to excision of this segment, the lower tip of the C-subunit β-barrel (which contains its trypsin-generated N-terminus) will be free to insert into the VP7 gap described in the preceding paragraph, and the N-terminal residues of the VP5*A- and VP5*B-subunits will be free to form the interchanged strands seen in Figure 2B and C. The excised segment could bridge between the observed C-terminus of VP8* and the observed N-terminus of VP5*, so that the lectin and β-barrel domains could be docked against each other in uncleaved VP4 just as we see them in the trypsin-cleaved protein. When the asymmetric spike has fully assembled, Lys29 on VP8*C will have become relatively accessible to tryptic cleavage, leading to loss of the VP8*C lectin domain.
VP4 conformational transitions
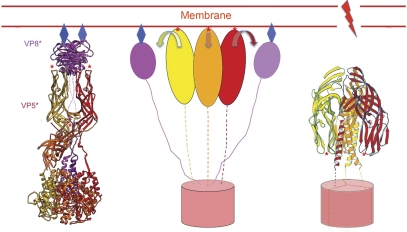
The asymmetric conformation of the VP4 spike on the virion is a trypsin-primed intermediate, awaiting release of the VP7 constraint to spring into a more stable state (Yoder et al, 2009). The crystal structure of a VP4 cleavage product, called VP5CT, shows the conformation of that stable end point (Dormitzer et al, 2004). Along with the VP4 structure described here and with a series of experiments on association of VP5* and VP5CT with lipid bilayers (Kim et al, 2010; Trask et al, 2010), they allow us to outline the conformational transitions leading to that end point and to describe the likely connection to membrane disruption and viral penetration. VP5CT is a truncated fragment of VP5* (prepared by chymotryptic and tryptic cleavage of VP4) that terminates at about residue 528 (Figure 1B). It forms a stable trimer, with the three β-barrel domains draped like the elements of a folded umbrella around a trimeric, α-helical coiled-coil stem (Figure 7). In the spike conformation, the hydrophobic apices of β-barrels A and B point away from the rest of the virion and hence away from the C-terminal foot; in VP5CT, the hydrophobic apices of all three β-barrels project towards the C-terminus of the coiled coil instead. Thus, a transition from the spike to the ‘umbrella' will include 180° rotation of β-barrel domains A and B with respect to the C-terminal part of the VP5* polypeptide chains (Figure 7). Preparation of a monoclonal antibody that recognizes the VP5CT trimer has allowed us to show that VP5* adopts the folded-back conformation when released from virions in vitro (by chelation of Ca2+) (Yoder et al, 2009). If released from virions in the presence of liposomes, VP5* associates with the lipid, but it cannot do so if liposomes are added later—presumably because the fold-back has already occurred (Trask et al, 2010). Moreover, if proteolysis of VP4 is carried out with liposomes present, the VP5CT fragment associates with the lipid, but not if liposomes are added after the cleavage is complete (Trask et al, 2010). These experiments show that an intermediate between the primed spike on the virion and the folded-back umbrella can bind lipid membranes.
Figure 7.
Model for the sequence of conformational changes in VP4 and how those changes link to interactions with a surrounding membrane. (Left) The spike (colours as in Figure 2A) binds receptor (blue diamonds) at the sialic acid-binding cleft on VP8* (magenta ribbons). The β-barrel domains (on which red asterisks designate hydrophobic loops) and C-terminal domains of VP5* are shown as ribbons in yellow, orange, and red for (A–C) subunits, respectively. (Centre) VP8* separates from the VP5* body but remains tethered to the foot through its N-terminal segment. The three VP5* β-barrel domains (yellow, red, and orange ovals) can then project outwards so that the hydrophobic loops at their apices (red asterisks) contact the membrane bilayer. We do not yet know whether the segment that will form the coiled coil in the folded-back conformation (right panel) pulls out from its packing at the trimer interface in the foot, or whether the foot (red cylinder) is still largely intact. (Right) Zipping up of the coiled coil (perhaps accompanied by withdrawal of some of its residues from the trimer interface in the foot) reconfigures the β-barrel domains (arrows in centre panel), so that their hydrophobic tips (red asterisks) now point towards the foot. Membrane disruption accompanies this conformational change.
The likely membrane-binding surfaces are the set of three hydrophobic loops at the apex of each VP5* β-barrel distal to the foot in the spike conformation (Dormitzer et al, 2004; Yoder and Dormitzer, 2006). Polar mutations in two of these loops reduce both viral infectivity and VP5* association with liposomes (Kim et al, 2010; Trask et al, 2010). Their conformational similarity to the fusion loops of class II and class III viral fusion proteins has been described (Dormitzer et al, 2004; Yoder and Dormitzer, 2006); one of them even has an amino-acid sequence related to the sequence of E1 fusion loop of the Semliki Forest virus (Mackow et al, 1988). The VP8* lectin domain covers two of these loops on chains A and B; the opposite ends of the A and B chain β-barrels cover the loops on chain C (Figures 2 and 5A). Thus, like the fusion loops and peptides of enveloped-virus entry proteins, these hydrophobic surfaces are hidden on the mature virion and exposed only during a conformational change triggered by events that accompany entry. A neutralizing monoclonal antibody that binds the one loop exposed on the A- and B-chain spikes and that efficiently blocks penetration but not attachment also interferes with liposome association (Mackow et al, 1988; Ruggeri and Greenberg, 1991; Tihova et al, 2001; Trask et al, 2010).
Discussion
The full structure of VP4 in the context of a complete virion and the accumulated experimental data just summarized yield a description of the molecular rearrangements that allow viral entry (Figure 7). VP8* mediates cell attachment—in the case of rhesus rotavirus (RRV), to terminal sialic acid moieties—through the lectin domains at the tip of the spike. The mechanism of endocytic uptake is not yet known, but there is good evidence that penetration is from an internal, membrane-bound compartment. The initial step in membrane disruption is likely to be interaction of the hydrophobic loops at the apices of the β-barrel domains with the target bilayer. To expose these loops, the lectin domains of VP8*A and VP8*B must dissociate from the β-barrels; barring further cleavage, the linker connecting them to their trimer-clustered N-termini will keep them tethered to the foot. Exposure of the loops at the tip of β-barrel domain C requires disruption of the contact between the top of the stalk and the bottom of the paired β-barrel domains that form the body. Each of the β-barrel domains—the two in the outer spike and the third in the stalk—has a 20-residue connector leading to the corresponding C-terminal domain; these connectors probably allow all three β-barrel domains to project independently towards the membrane, forming an extended intermediate (Figure 7, centre).
Rearrangements leading to an extended intermediate could in principle occur without loss of the VP7 layer, as neither the linker between the N-terminal segment (α) and the lectin domain of VP8* nor the connector between β-barrel and C-terminal domain is substantially hindered by unperturbed VP7. The altered form of VP4 produced by exposure of rotavirus particles to alkaline pH may represent this sort of intermediate (Pesavento et al, 2005). Moderate-resolution cryoEM reconstructions of high-pH inactivated virions shows that the spikes transform into three symmetrically positioned, projecting knobs. These knobs can be decorated with Fab fragments from mAb 2G4, an antibody that binds near the hydrophobic loops (Pesavento et al, 2005). Density for the Fab indicates that the VP5* β-barrels account for the knob density, with the hydrophobic loops facing outward. Moreover, as expected by the interactions of its anchored N-terminal segment, VP8* remains attached to the alkali-treated particle, and it can mediate sialic-acid-dependent cell binding after a return to neutral pH. Thus, loss of Ca2+, the event that leads to VP7 dissociation, need not occur until after the formation of the membrane interacting, extended intermediate. We and others have proposed that loss of Ca2+ and departure of VP7 might be a critical activating step (‘trigger') (Aoki et al, 2009), but the structure suggests the possibility of an alternative trigger for the initial VP4 conformational change. The molecular identity of that trigger remains to be determined. Strain on VP8* from binding to the membrane is one possibility: some monoclonal antibodies that recognize VP8* induce uncoating in vitro (Zhou et al, 1994).
Zipping up of the three-chain coiled coil probably drives the transition to a folded-back, trimeric VP5* ‘umbrella'. The polypeptide-chain segment that forms each chain of the coiled coil includes part of the linker between β-barrel and C-terminal domains (residues 491–510) and a further 15 residues sandwiched at the interface between two C-terminal domains (Figures 2B and C and 6). We can picture two alternative models for this step. In one, coiled-coil formation pulls these residues out of the interface, leaving the trimeric foot in place (potentially still retained by VP7). In the other, coiled-coil formation not only withdraws residues from the interface but in the process destabilizes the entire foot and releases it from its socket in the DLP. These alternatives have very different implications for the mechanism of membrane disruption. In the former, membrane distortion by the folding back of membrane-inserted β-barrel domains is coupled to the DLP, which provides a framework with respect to which the membrane is deformed and ultimately broken. In the latter, the inversion of the β-barrel umbrella must effect membrane perforation, even though the perforating protein has no fixed anchor except within the membrane itself. We represent the former model in Figure 7C.
Although closely related to rotaviruses in replication strategy and in many aspects of virion structure, the orthoreoviruses penetrate through a series of molecular events rather different from those just outlined. Reovirus attachment and penetration are functions of two distinct outer-layer proteins—σ1 and μ1, respectively (Schiff et al, 2007). The former projects (as trimers) at icosahedral five-fold positions. The latter, similar in structure to rotavirus VP6 but with an N-terminal myristoyl modification, forms, along with a ‘chaperone' protein, a T=13l icosahedral lattice (interrupted at the five-folds by a pentameric protein projecting from the core) (Dryden et al, 1993). Proteolytic activation of the virion removes the chaperone, σ3, creating a so-called ‘infectious subviral particle' (ISVP) and priming μ1 to undergo a series of conformational changes when triggered by endosomal events, not yet fully defined (Schiff et al, 2007). The μ1 conformational changes release a myristoylated N-terminal peptide (μ1N), which has separated from the rest of the subunit by an autolytic cleavage. Released μ1N oligomerizes into pores in the endosomal membrane (Agosto et al, 2006; Ivanovic et al, 2008; Zhang et al, 2009). A crystal structure of a (μ1)3(σ3)3 heterohexamer (Liemann et al, 2002), a ∼6.5 Å resolution cryoEM reconstruction of a virion (Zhang et al, 2005), and a recent 3.5 Å resolution reconstruction of an ISVP (Zhang et al, 2010b) have outlined some structural correlates of the rearrangements just described. Although the autolytic cleavage can occur without release of σ3 (Liemann et al, 2002), activation may potentiate it (Zhang et al, 2010b). The cleavage itself leads only to localized structural changes; release of μ1N requires extensive unfolding of the μ1 subunit and dissociation of the μ1 trimer and hence some additional stimulus (Zhang et al, 2006).
Release of a pore-forming, N-myristoylated peptide, and conformational rearrangement of a fusion-protein-like structure are two distinct modes of membrane disruption, adopted by otherwise rather closely related viruses. A reovirus penetration-protein homologue (VP6) is present in the rotavirus particle, but it lacks the N- and C-terminal elaborations in μ1 that surround μ1N, and it appears to function mainly as a structural adaptor between rotavirus core and outer layer. Orbiviruses, a third major group of dsRNA viruses, may have a penetration machinery similar to the rotavirus outer layer in the way it disrupts a membrane, but different in its detailed molecular structure (Zhang et al, 2010a).
The structure described here is a substantial step forward towards understanding the molecular mechanisms of early events in rotavirus cell entry—and non-enveloped virus entry more generally. When correlated with results from assembly and disassembly experiments in vitro and from infectivity experiments with recoated particles, the structure raises the following new questions. What are the characteristics of the endocytic event and the properties of the resulting membrane-bound vesicle? What triggers each of the steps in Figure 7 (or is the second step spontaneous and stochastic)? Do loss of Ca2+ and dissociation of VP7 precede VP4 rearrangement, or are these processes instead the final steps in uncoating to generate a DLP? Can we determine the lifetime and structure of the postulated extended intermediate? How does the transition from extended intermediate to folded-back umbrella disrupt the membrane into which the hydrophobic loops have inserted? The structure leads directly to the design of constrained mutants (e.g. with disulphide links) that we and others can deploy to answer these questions and thus to arrive at a yet more complete mechanism for one mode of non-enveloped viral entry.
Materials and methods
Virus preparations
RRV, serotype G3, P5B[3], was grown in MA104 cells and purified as described previously (Trask and Dormitzer, 2006; Trask et al, 2010).
Electron microscopy and image processing
Data collection and processing followed previously published procedures (Zhang et al, 2008; Chen et al, 2009). Briefly, RRV particles (TLPs) were embedded in vitreous ice on C-flat holey-carbon grids by plunge freezing to ∼90°K. Single-particle image data were collected at × 59 000 nominal magnification and 300 keV on an FEI Tecnai F30 electron microscope equipped with a field-emission source; 110 micrographs were recorded on Kodak SO163 film, at a dose of ∼20 e−/Å2 and at an underfocus between 1.2 and 3.0 μm. To minimize magnification changes over the defocus variation, we performed coma-free alignment of the microscope before the data acquisition. Moreover, we kept the sample height constant throughout the session and compensated for any deviation (e.g. originating from grid tilt or local bending) by adjusting the eucentric height of the stage. We used cut-plate film holders to reduce image background from electrons back-scattered from the metal film plate. The micrographs were digitized on a Zeiss SCAI scanner at 7 μm step-size, a scanning resolution of 1.233 Å/pixel after magnification calibration. From these micrographs, 4187 particles were selected by using the program SIGNATURE (Chen and Grigorieff, 2007). The defocus parameters of each particle were evaluated by the program CTFTILT (Mindell and Grigorieff, 2003) and individually refined in the final phase of the model reconstruction (Chen et al, 2009). These parameters were used for CTF correction in the program FREALIGN (Grigorieff, 2007) to achieve precise particle alignment and density map reconstruction.
In the initial particle alignment, TLP images were 2 × -binned and aligned to a previously reconstructed EM density map of 7RP (Chen et al, 2009) (a virus-like particle containing VP2, VP6 and VP7) through a systematic search at a 1° angular interval (FREALIGN mode 3). Subsequently, images at the full resolution were used to refine alignment parameters, as described previously (Chen et al, 2009). In the refinement, a shell mask (Rin=220 Å, Rout=400 Å) was used to retain the best-ordered density corresponding to the three protein layers (VP2, VP6, and VP7) and to exclude density corresponding to the genome and the VP4 spikes. Masking of the VP4 region during refinement reduced the possibility of model bias in this part of the reconstruction, in analogy to crystallographic omit maps. To improve the resolution, particle images with alignment phase residual in the bottom 10th percentile were not included in the final reconstruction. At the end of the model refinement, the averaged phase residual was 70.1°. The resolution of the icosahedral reconstruction was 6.5 Å according to the FSC criterion (Harauz and van Heel, 1986) at FSC=0.5, and 4.3 Å at FSC=0.143 (Figure 3). In addition to icosahedral symmetry, the rotavirus capsid has a T=13 quasi-equivalent arrangement of subunits in the VP6 and VP7 layers. Following the procedure implemented in the previous work on the EM reconstruction of the 7RP (Chen et al, 2009), local averaging improved the resolution of the VP6–VP7 region to at least 4 Å. As different parts of the VP4 spikes have different local symmetries (three-fold at the base, two-fold in the outer parts of the projections, no symmetry in the transitions between these regions), several different maps (including the ‘raw' icosahedrally averaged map) were used for subsequent model building and structure analysis.
Supplementary Material
Acknowledgments
We thank Scott Aoki, Marina Babyonyshev, and Chen Xu for advice and assistance. The work was supported by NIH Grants P01 GM-62580 (to SCH and NG), CA-13202 (to SCH), AI-53174 (to PRD), and by an Ellison Medical Foundation New Scholars in Global Infectious Diseases Award (to PRD). NG and SCH are investigators in the Howard Hughes Medical Institute. Coordinates and maps have been deposited with PDB ID codes 3N09 and 3IYU and map code EMD-5199. ECS and JZC designed and carried out the experiments and interpreted the data; PRD designed the experiments; NG and SCH designed the experiments and interpreted the data. All authors participated in writing the paper.
Footnotes
PRD and ECS are employees and shareholders of Novartis Vaccines and Diagnostics, Inc.
References
- Agosto MA, Ivanovic T, Nibert ML (2006) Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc Natl Acad Sci USA 103: 16496–16501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki ST, Settembre EC, Trask SD, Greenberg HB, Harrison SC, Dormitzer PR (2009) Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 324: 1444–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JZ, Grigorieff N (2007) SIGNATURE: a single-particle selection system for molecular electron microscopy. J Struct Biol 157: 168–173 [DOI] [PubMed] [Google Scholar]
- Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N (2009) Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci USA 106: 10644–10648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J, Laporte J, Charpilienne A, Scherrer R (1979) Activation of rotavirus RNA polymerase by calcium chelation. Arch Virol 60: 177–186 [DOI] [PubMed] [Google Scholar]
- Crawford SE, Mukherjee SK, Estes MK, Lawton JA, Shaw AL, Ramig RF, Prasad BV (2001) Trypsin cleavage stabilizes the rotavirus VP4 spike. J Virol 75: 6052–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danthi P, Tosteson M, Li QH, Chow M (2003) Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J Virol 77: 5266–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer PR, Greenberg HB, Harrison SC (2000) Purified recombinant rotavirus VP7 forms soluble, calcium-dependent trimers. Virol 277: 420–428 [DOI] [PubMed] [Google Scholar]
- Dormitzer PR, Greenberg HB, Harrison SC (2001) Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core. J Virol 75: 7339–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer PR, Nason EB, Prasad BV, Harrison SC (2004) Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430: 1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer PR, Sun ZY, Wagner G, Harrison SC (2002) The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J 21: 885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS (1993) Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol 122: 1023–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes MK, Kapikian AZ (2007) Rotaviruses. In Fields Virology, Knipe DM, Howley PM (eds) 5th edn, pp 1918–1974. Philadelphia: Lippincott, Williams & Wilkins [Google Scholar]
- Farr GA, Zhang LG, Tattersall P (2005) Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci USA 102: 17148–17153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N (2007) FREALIGN: high-resolution refinement of single particle structures. J Struct Biol 157: 117–125 [DOI] [PubMed] [Google Scholar]
- Harauz G, van Heel M (1986) Exact filters for general geometry three dimensional reconstruction. Optik 73: 146–156 [Google Scholar]
- Ivanovic T, Agosto MA, Zhang L, Chandran K, Harrison SC, Nibert ML (2008) Peptides released from reovirus outer capsid form membrane pores that recruit virus particles. EMBO J 27: 1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Trask SD, Babyonyshev M, Dormitzer PR, Harrison SC (2010) Effect of mutations in VP5* hydrophobic loops on rotavirus cell entry. J Virol 84: 6200–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC (2002) Structure of the reovirus membrane-penetration protein, μ1, in a complex with is protector protein, σ3. Cell 108: 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackow ER, Shaw RD, Matsui SM, Vo PT, Dang MN, Greenberg HB (1988) The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci USA 85: 645–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Petitpas I, Navaza J, Lepault J, Kohli E, Pothier P, Prasad BV, Cohen J, Rey FA (2001) Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J 20: 1485–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain B, Settembre E, Temple BR, Bellamy AR, Harrison SC (2010) X-ray crystal structure of the rotavirus inner capsid particle at 3.8 Å resolution. J Mol Biol 397: 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Parashar UD, Gibson CJ, Bresse JS, Glass RI (2006) Rotavirus and severe childhood diarrhea. Emerg Infect Dis 12: 304–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JB, Crawford SE, Roberts E, Estes MK, Prasad BV (2005) pH-induced conformational change of the rotavirus VP4 spike: implications for cell entry and antibody neutralization. J Virol 79: 8572–8580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri FM, Greenberg HB (1991) Antibodies to the trypsin cleavage peptide VP8* neutralize rotavirus by inhibiting binding of virions to target cells in culture. J Virol 65: 2211–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff LA, Nibert ML, Tyler KL (2007) Orthoreoviruses and their replication. In Fields' Virology, Knipe DM, Howley PM (eds) 5th edn, pp 1853–1915. Philadelphia: Lippincott Williams & Wilkins [Google Scholar]
- Sousa D, Grigorieff N (2007) Ab initio resolution measurement for single particle structures. J Struct Biol 157: 201–210 [DOI] [PubMed] [Google Scholar]
- Tihova M, Dryden KA, Bellamy AR, Greenberg HB, Yeager M (2001) Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: implications for cell entry. J Mol Biol 314: 985–992 [DOI] [PubMed] [Google Scholar]
- Trask SD, Dormitzer PR (2006) Assembly of highly infectious rotavirus particles recoated with recombinant outer capsid proteins. J Virol 80: 11293–11304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask SD, Kim IS, Harrison SC, Dormitzer PR (2010) A rotavirus spike protein conformational intermediate binds lipid bilayers. J Virol 84: 1764–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Berriman JA, Baker TS, Bellamy AR (1994) Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J 13: 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JD, Dormitzer PR (2006) Alternative intermolecular contacts underlie the rotavirus VP5* two- to three-fold rearrangement. EMBO J 25: 1559–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JD, Trask SD, Vo TP, Binka M, Feng N, Harrison SC, Greenberg HB, Dormitzer PR (2009) VP5* rearranges when rotavirus uncoats. J Virol 83: 11372–11377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Agosto MA, Ivanovic T, King DS, Nibert ML, Harrison SC (2009) Requirements for the formation of membrane pores by the reovirus myristoylated μ1N peptide. J Virol 83: 7004–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chandran K, Nibert ML, Harrison SC (2006) Reovirus μ1 structural rearrangements that mediate membrane penetration. J Virol 80: 12367–12376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Boyce M, Bhattacharya B, Schein S, Roy P, Zhou ZH (2010a) Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins. Proc Natl Acad Sci USA 107: 6292–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ji Y, Zhang L, Harrison SC, Marinescu DC, Nibert ML, Baker TS (2005) Features of reovirus outer capsid protein μ1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution. Structure 13: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH (2010b) 3.3 Å cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141: 472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N (2008) Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA 105: 1867–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YJ, Burns JW, Morita Y, Tanaka T, Estes MK (1994) Localization of rotavirus VP4 neutralization epitopes involved in antibody-induced conformational changes of virus structure. J Virol 68: 3955–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.