Caspase-8 and caspase-7 sequentially mediate proteolytic activation of acid sphingomyelinase in TNF-R1 receptosomes
Tumour necrosis factor (TNF)-induced cell death involves the generation of ceramide by endosomal acid sphingomyelinase (A-SMase). This study reveals that in TNF-receptosomes, A-SMase is activated by caspase-7-mediated proteolytic cleavage downstream of caspase-8 activation.
Keywords: acid sphingomyelinase, caspase-7, caspase-8, ceramide, TNF receptosomes
Abstract
We previously demonstrated that tumour necrosis factor (TNF)-induced ceramide production by endosomal acid sphingomyelinase (A-SMase) couples to apoptosis signalling via activation of cathepsin D and cleavage of Bid, resulting in caspase-9 and caspase-3 activation. The mechanism of TNF-mediated A-SMase activation within the endolysosomal compartment is poorly defined. Here, we show that TNF-induced A-SMase activation depends on functional caspase-8 and caspase-7 expression. The active forms of all three enzymes, caspase-8, caspase-7 and A-SMase, but not caspase-3, colocalize in internalized TNF receptosomes. While caspase-8 and caspase-3 are unable to induce activation of purified pro-A-SMase, we found that caspase-7 mediates A-SMase activation by direct interaction resulting in proteolytic cleavage of the 72-kDa pro-A-SMase zymogen at the non-canonical cleavage site after aspartate 253, generating an active 57 kDa A-SMase molecule. Caspase-7 down modulation revealed the functional link between caspase-7 and A-SMase, confirming proteolytic cleavage as one further mode of A-SMase activation. Our data suggest a signalling cascade within TNF receptosomes involving sequential activation of caspase-8 and caspase-7 for induction of A-SMase activation by proteolytic cleavage of pro-A-SMase.
Introduction
Tumour necrosis factor α (TNF-α) belongs to the TNF superfamily of cytokines. It is involved in a variety of cellular processes such as inflammation, differentiation, control of cell proliferation and initiation of apoptosis. TNF is known to bind to two receptors of the TNF-receptor superfamily: TNF-receptor 1 (TNF-R1) and TNF-receptor 2 (TNF-R2). TNF-R1 is a member of the death receptor subgroup within this superfamily (Guicciardi and Gores, 2009). The death receptors share a common ‘death domain' (DD) in their cytoplasmic tail, which is necessary for the activation of apoptosis. TNF-R1 is the only receptor that mediates apoptosis in response to TNF-α stimulation.
Binding of TNF-α to TNF-R1 can induce non-apoptotic or apoptotic signalling pathways. First, the adaptor protein TNFR-associated DD (TRADD) is recruited to the DD of the TNF-R1 and serves as platform for binding of further proteins. By recruitment of RIP1, TRAF2 and c-IAP1 into an assembly termed ‘complex I' (Micheau and Tschopp, 2003), the cellular level of FLIPL is positively regulated (Micheau et al, 2001). The resulting block of apoptosis leads to cell survival. Conflicting data exist regarding the complex formation that induces apoptosis after TNF stimulation. In the model of Micheau and Tschopp, most ‘complex I' proteins become modified by ubiquitination and subsequently dissociate from TNF-R1. Thereby, the DD of TRADD is liberated and allows FADD binding that enables the recruitment of caspase-8 and -10 through their death effector domains, leading to the induction of apoptosis (complex II). In contrast to this model, we previously reported that after recruitment of TRADD, RIP and TRAF2 to TNF-R1 at the cell surface, the receptor is rapidly internalized and FADD and pro-caspase-8 are recruited, forming the death-inducing signalling complex (DISC) still associated with the TNF receptor at endosomal vesicles (TNF receptosomes) (Schneider-Brachert et al, 2004, 2006). Subsequently, pro-caspase-8 is cleaved and activated. These observations fit to the emerging concept that receptor endocytosis—previously regarded merely as a mechanism to switch off receptor signalling—may be necessary for full activation of certain signalling events (reviewed by McPherson et al, 2001; Sorkin and von Zastrow, 2002; Scita and Di Fiore, 2010). In line with this concept deletion of a region termed TNF-R1 internalization domain (TRID), was found to block endocytosis of TNF-R1 and to prevent the recruitment of FADD and caspase-8. These results indicated that TNF-R1 internalization is necessary for efficient induction of apoptosis (for review see Schütze et al, 2008; Guicciardi and Gores, 2009).
It is well established that TNF-receptor triggering activates the endolysosomal enzyme SMPD1 (Lansmann et al, 2003), also known as acid sphingomyelinase (A-SMase) (Schütze et al, 1992; Wiegmann et al, 1994, 1999; Kolesnick and Krönke, 1998; Schwandner et al, 1998). This enzyme generates the potent proapoptotic lipid second messenger ceramide from sphingomyelin (reviewed by Lin et al, 2000b; Morales et al, 2007; Hannun and Obeid, 2008; Jenkins et al, 2009). A role of A-SMase in transmitting apoptotic signals of death receptors has been reported not only for TNF-R1 (Monney et al, 1998; García-Ruiz et al, 2003; Heinrich et al, 2004), but also for CD95 (Cifone et al, 1994; Kolesnick et al, 1994; Martin et al, 1995; Herr et al, 1997; Brenner et al, 1998; De Maria et al, 1998; Lin et al, 2000a; Lin et al, 2000b) and TRAIL receptors (Dumitru and Gulbins, 2006; Thon et al, 2006).
Apparently, A-SMase can be activated in several ways. We reported that diacyl glycerol, generated by hydrolysis of phosphatidylcholine by phosphatidylcholine-specific phospholipase C activates protein kinase C (PKC) as well as A-SMase (Schütze et al, 1992). Zeidan and Hannun (2007) showed that phosphorylation at Ser 508 by PKCδ leads to activation and translocation of A-SMase. Other studies reported that reactive oxygen species (ROS) are involved in the activation of A-SMase (Charruyer et al, 2005; Lang et al, 2007; review Dumitru et al, 2007). On the other hand, it was reported that ROS production occurs downstream of A-SMase activation as well (Reinehr et al, 2006).
TNF-induced A-SMase activation was found to be mediated through the TNF-R1 adaptor proteins TRADD and FADD (Schwandner et al, 1998) and is prevented by deletion of the DD of TNF-R1 (Schneider-Brachert et al, 2004). As mentioned above, the recruitment of TRADD and FADD to TNF-R1 is also blocked in cells expressing the TNF-R1 ΔTRID deletion mutant, implying that internalization of the TNF-R1 is necessary for A-SMase activation as well. Ceramide generated within the endolysosomal compartment by A-SMase activates the aspartic acid-protease cathepsin D (CTSD) by inducing the autoproteolytic cleavage of the CTSD zymogen (Heinrich et al, 1999, 2000, 2004; Schneider-Brachert et al, 2004). Ceramide-mediated activation of CTSD leads to translocation of the enzyme through the endosomal membrane into the cytosol, where it cleaves Bid to generate the pro-apoptotic fragment tBid. Activated tBid then initiates the intrinsic apoptotic signalling pathway by releasing cytochrome C from mitochondria, formation of the apoptosome and activation of caspase-9 and -3. Using a selective caspase-8 inhibitor, the study of Heinrich et al (2004) indicated that caspase-8 might be involved in the TNF-induced activation of A-SMase. The molecular pathway of TNF-R1 apoptosis signalling via internalized TNF receptosomes was reviewed recently (Schütze et al, 2008).
The complete molecular link between TNF-R1 and A-SMase activation in internalized TNF receptosomes is still elusive and subject of the present report.
We show here for the first time by confocal microscopy and by analysing immunomagnetically isolated TNF receptosomes that both active caspase-8 and caspase-7 partially colocalize with A-SMase as well as with TNF-R1. We demonstrate that both caspase-8 and caspase-7 are indispensible for TNF-induced A-SMase activation, which is characterized by the appearance of a 57-kDa proteolytic cleavage fragment of pro-A-SMase. In contrast to caspase-8, only caspase-7 is capable of directly cleaving and activating immunoprecipitated endogenous as well as recombinant pro-A-SMase. A sequential activation of all three enzymes, caspase-8, caspase-7, and pro-A-SMase was observed within the same time frame in magnetically isolated TNF receptosomes.
Our data suggest that TNF-mediated stimulation of A-SMase requires sequential activation of caspase-8 and -7 resulting in proteolytic cleavage of pro-A-SMase within TNF receptosomes.
Results
Caspase-8 deficiency prevents TNF activation of A-SMase and cathepsin D
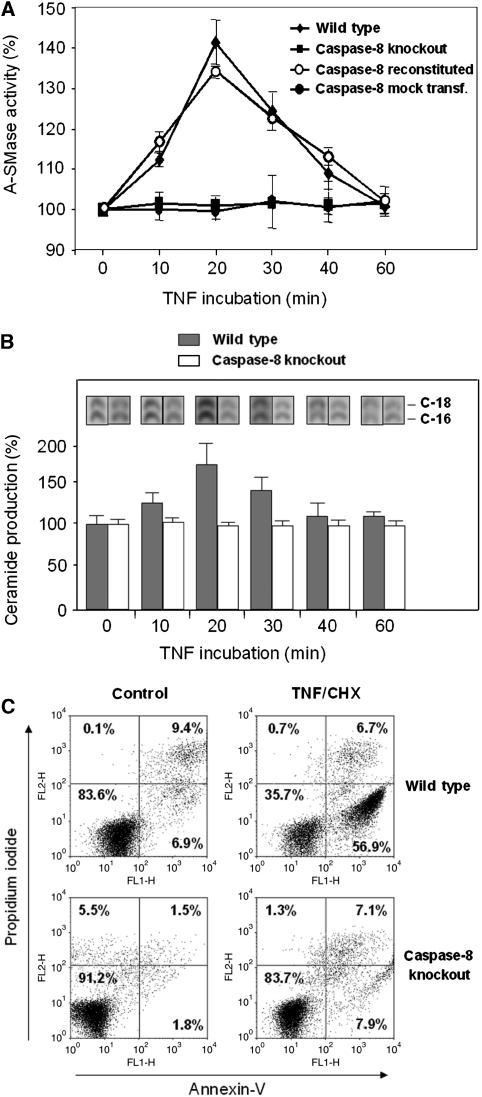
A strict dependence of TNF-induced A-SMase activation on functional caspase-8 was observed by comparing caspase-8-deficient Jurkat cells with wild-type cells. As shown in Figure 1A, TNF treatment leads to a transient increase in A-SMase activity in lysates from wild-type Jurkat cells, while there is no change in A-SMase activity in caspase-8-deficient cells. Retransfection of caspase-8-deficient Jurkat cells with an expression plasmid for caspase-8 restores the ability of the transfected cells to respond to TNF stimulation by increased A-SMase activity. Furthermore, A-SMase can be activated in vitro by the addition of exogenous caspase-8 to lysates from caspase-8-deficient Jurkat cells (Supplementary Figure S1). Also, the production of C-16/C-18 ceramide is not increased upon TNF treatment in caspase-8-deficient Jurkat cells, while wild-type Jurkat cells display a clear transient increase in C-16/C-18 ceramide levels after TNF stimulation (Figure 1B). Caspase-8-deficient Jurkat cells were almost completely resistant to TNF/CHX treatment, demonstrating the critical role of caspase-8 in TNF-induced apoptosis (Figure 1C).
Figure 1.
Impaired A-SMase activation and apoptosis after TNF stimulation in caspase-8-deficient Jurkat cells. (A) Time course of A-SMase activity determined in Jurkat cell lysates after TNF treatment. Wild-type Jurkat cells are compared with caspase-8-deficient (caspase-8 knockout) Jurkat cells as well as to caspase-8-deficient Jurkat re-transfected with caspase-8 expression plasmid (caspase-8 reconstituted) and caspase-8-deficient Jurkat mock-transfected with an empty plasmid (caspase-8 mock transf.). (B) Time course of ceramide production determined in Jurkat cell lysates after TNF treatment. Wild-type Jurkat cells are compared with caspase-8-deficient Jurkat cells. Data of two experiments are shown, each performed in triplicates (±s.e.m.). (C) Induction of apoptosis in Jurkat wild-type and caspase-8-deficient cells after treatment with 0.5 μg/ml CHX and 100 ng/ml TNF for 4 h, analysed by annexin-V/propidium iodide staining.
Active caspase-8 colocalizes with internalized TNF-R1 receptosomes
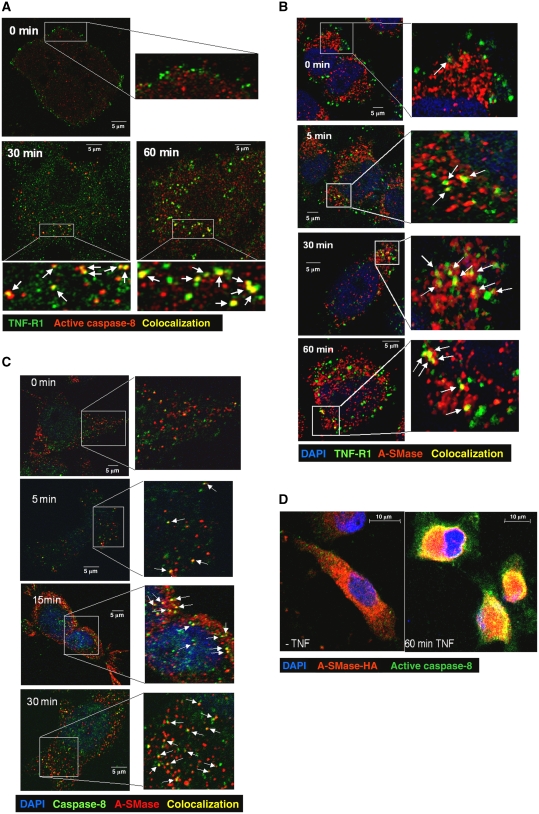
We next asked, whether the molecular components of a potential signalling cascade from TNF-R1 to A-SMase via caspase-8 actually localize in the same subcellular compartment. To this, we performed synchronized internalization experiments using biotinylated TNF coupled to streptavidin-FITC for labelling of TNF/TNF-receptor complexes. Simultaneous immunofluorescence detection of ligand-bound TNF receptors and cleaved caspase-8, respectively, revealed a time-dependent appearance of endocytic vesicles that are positive for both molecules in HeLa cells. As shown in Figure 2A, at 0 min, before internalization is started, fluorescently labelled TNF receptors can be found almost exclusively at the plasma membrane while a punctate staining of low intensity in the cell interior is observed for cleaved caspase-8. After 30 min, a fraction of small endocytic vesicles containing labelled TNF receptors is also positively stained for cleaved caspase-8. At later time points (45 and 60 min), the number and volume of double-positive endocytic vesicles is increased. These observations demonstrate that a significant amount of activated caspase-8 is still bound to the TNF receptor during endocytosis, which is in line with previous observations obtained after immunomagnetic isolation of TNF receptosomes (Schneider-Brachert et al, 2004, 2006).
Figure 2.
Partial colocalization of caspase-8 and A-SMase with TNF receptosomes. (A) Merged confocal microscopic images of HeLa cells labelled with biotin-TNF/FITC-avidin complexes (green) and anti-cleaved capase-8 monoclonal antibody (red) at indicated times of TNF-receptor internalization. Colocalization of TNF receptor and cleaved caspase-8 is indicated in yellow (see arrows). (B) Partial colocalization (yellow, see arrows) of endogenous A-SMase (red) and TNF receptosomes (green), and (C) partial colocalization (yellow, see arrows) of endogenous A-SMase (red) and caspase-8 (green) as well as (D) recombinant HA-tagged A-SMase (red) and cleaved caspase-8 (green) at several time points after TNF-receptor internalization.
Endogenous A-SMase colocalizes with internalized TNF-R1 receptosomes
Investigation of the intracellular distribution of endogenous A-SMase by staining with an antibody generated against a synthetic A-SMase peptide (Perrotta et al, 2007; Bianco et al, 2009) also revealed partial colocalization of A-SMase with biotinylated TNF/streptavidin-FITC-labelled internalized TNF receptosomes, detectable already after 5 min of incubation with biotinylated TNF at 37°C (Figure 2B).
Active caspase-8 and A-SMase colocalize in the same compartment
Simultaneous staining of HeLa cells for endogenous A-SMase and active caspase-8 revealed partial colocalization of both proteins, detectable also after only 5 min of TNF treatment (Figure 2C). A pronounced colocalization of active caspase-8 and A-SMase was also observed in cells expressing pro-A-SMase-HA (Figure 2D). Together, these observations indicate a possible interaction between caspase-8 and pro-A-SMase in the same subcellular compartment.
Activation of A-SMase by TNF correlates with the proteolytic generation of a 57-kDa fragment
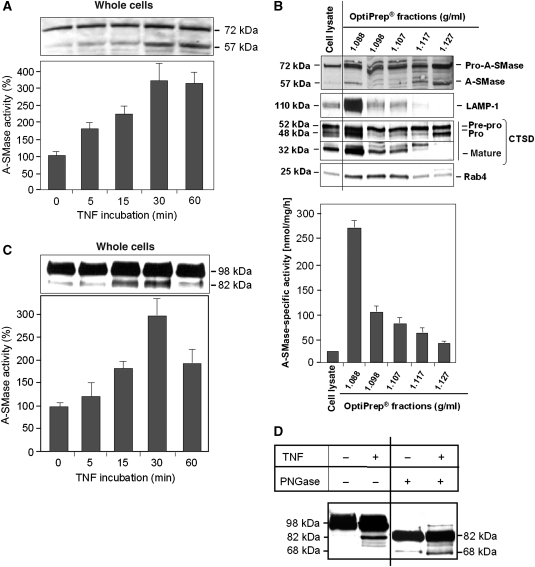
Stimulation of HeLa cells with TNF results in enhanced enzymatic A-SMase activity paralleled by the appearance of a 57-kDa protein in addition to a 72-kDa protein, both detected by the anti-human A-SMase antibody in the cell lysates prepared from the same samples that were used for the enzymatic assay (Figure 3A). The 72-kDa band corresponds to the A-SMase precursor and the 57-kDa band to the maturated A-SMase enzyme described by Ferlinz et al (1994). Notably, the 57-kDa A-SMase cosediments with organelle fractions containing the lysosomal marker proteins LAMP-1 and mature cathepsin D after separation of cell lysates on OptiPrep-gradients (at a density of 1.088 g/ml in Figure 3B). This fraction contained the highest A-SMase activity. Theses observations suggest that the 57-kDa protein is identical to the mature lysosomal A-SMase. Both pre-pro- and pro-A-SMase proteins of 75 and 72 kDa, respectively, were detected predominantly in non-lysosomal fractions at densities of 1.117 and 1.127 g/ml, which also contain 57 kDa proteins but exhibited only minor enzymatic A-SMase activity. The 57-kDa bands in these fractions may represent unglycosylated pro-A-SMase molecules, which possess reduced enzymatic activity (see below).
Figure 3.
A-SMase cleavage and activation after TNF stimulation. (A) HeLa cells were stimulated with TNF for indicated times and cell lysates were analysed for endogenous A-SMase by western blotting using anti-A-SMase antibodies that detect the 72-kDa pro-A-SMase and a 57-kDa cleavage product. A-SMase processing is accompanied by an increase of A-SMase activity. Data are from four experiments (+s.e.m.), each performed in triplicates. (B) Cell lysate and OptiPrep fractions from unstimulated HeLa cells were analysed by western blotting using the indicated antibodies and A-SMase assay. A-SMase activity peaked in the lysomal fraction 2. (C) HeLa cells expressing recombinant A-SMase-GFP were stimulated with TNF for indicated times. Western blots were performed using anti-GFP antibodies, detecting pro-A-SMase-GFP at 98 kDa and the cleavage product of A-SMase-EGFP at 82 kDa, which is also accompanied by an increase of A-SMase activity. Data are shown for three experiments (±s.e.m.), each performed in triplicates. (D) Immunoprecipitated pro-A-SMase-EGFP of untreated or TNF-treated cells was deglycosylated by PNGase F treatment and detected by anti-GFP antibody in western blot. Glycosylated pro-A-SMase-EGFP migrates at 98 kDa. The glycosylated cleavage fragment as well as the deglycosylated pro-A-SMase-EGFP migrate at 82 kDa. The deglycosylated cleavage fragment of A-SMase-EGFP migrates at 68 kDa (figure assembled from cropped lanes of the same gel (see primary scans in Supplementary data)).
In order to test, whether the 57-kDa A-SMase is derived from proteolytic cleavage of the 72-kDa A-SMase precursor molecule, we transfected HeLa cells with a fusion protein of pro-A-SMase (72 kDa) and EGFP (26 kDa), together displaying a molecular weight of 98 kDa in western blots. After stimulation of stably transfected HeLa cells with TNF, a time-dependent appearance of an 82-kDa cleavage product of pro-A-SMase-EGFP was observed after staining with anti-GFP antibody, correlating with an increase in A-SMase enzymatic activity (Figure 3C). This cleaved protein corresponds to a 57-kDa mature A-SMase molecule after subtraction of the 26-kDa EGFP.
Characterization of immunoprecipitated fusion protein for posttranslational modifications revealed that the pro-A-SMase-EGFP and its 82 kDa cleavage fragment are glycosylated. After deglycosylation by PNGase F, the pro-form of A-SMase-EGFP (98 kDa) displays a molecular weight of 82 kDa similar to the observed cleavage fragment. However, also the TNF-induced 82 kDa fragment is glycosylated and shows up as a 68-kDa band after deglycosylation (Figure 3D). This experiment suggests that the 82-kDa band is probably a mixture of unglycosylated pro-A-SMase-EGFP and the cleavage fragment, and explains the weak constitutive 82 kDa band in untreated cells.
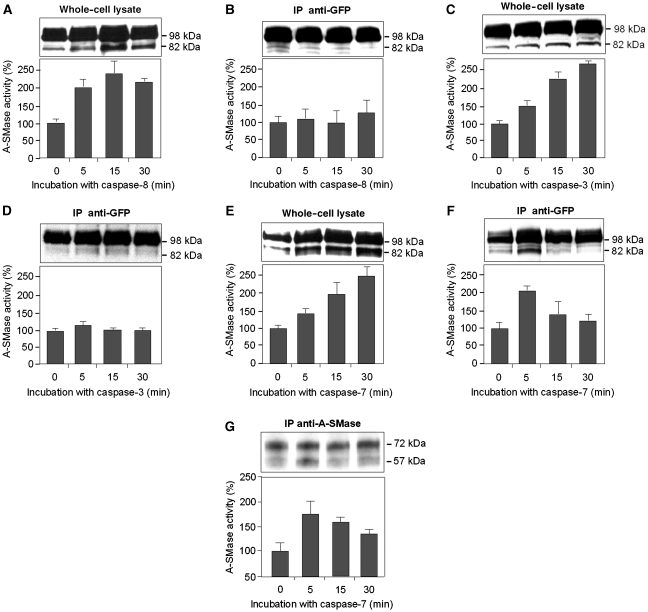
To test the involvement of caspase-8 in A-SMase activation, cell lysates and purified pro-A-SMase-EGFP were treated with recombinant active caspase-8 in vitro (Figure 4A and B). The 82-kDa proteolytic cleavage fragment was observed in cell lysates, when these were treated in vitro with recombinant active caspase-8 (Figure 4A). Again the A-SMase activity profile paralleled the appearance of the proteolytic fragment. However, when the immunoprecipitated pro-A-SMase-EGFP protein was treated in vitro with active caspase-8, enhanced cleavage was not observed and also A-SMase activity remained unaltered (Figure 4B). This observation implies that caspase-8 is not able to directly cleave and activate pro-A-SMase. We subsequently tested known caspase-8 substrates in vitro to see, whether one of these could be the intermediate protease that directly cleaves and activates pro-A-SMase. As shown in Figure 4C, incubation of cell lysates with exogenous caspase-3 showed a similar increase in A-SMase enzymatic activity and enhanced pro-A-SMase cleavage as observed before for exogenous caspase-8 treatment. Incubation of immunoprecipitated pro-A-SMase with caspase-3, however, also failed to significantly enhance A-SMase activity and A-SMase processing, suggesting that pro-A-SMase is not a direct substrate for caspase-3. In addition, confocal laser-scan analysis revealed that colocalization of active caspase-3 and TNF-R1 could not be detected after 30 min of TNF internalization (Supplementary Figure S2) or at any other time points investigated between 0 and 60 min (data not shown).
Figure 4.
Cleavage and activation of pro-A-SMase in cell lysates and immunoprecipitated material after caspase incubation. Western blot analysis with an anti-GFP antibody and A-SMase activity assay was performed on (A) whole-cell lysate of A-SMase-EGFP-transfected cells incubated with caspase-8, (B) immunoprecipitated pro-A-SMase-EGFP incubated with active caspase-8, (C) whole-cell lysate of pro-A-SMase-EGFP-transfected cells incubated with active caspase-3, (D) immunoprecipitated pro-A-SMase-EGFP incubated with caspase-3, (E) whole-cell lysate of pro-A-SMase-EGFP-transfected cells incubated with caspase-7, and (F) immunoprecipitated pro-A-SMase-EGFP incubated with active caspase-7 for the indicated times. In experiments with whole-cell lysates, all caspases induce cleavage and activation of A-SMase-EGFP (A, C, E), while cleavage and activation are not observed with immunoprecipitated pro-A-SMase-EGFP treated with caspase-8 (B) or very weak with caspase-3 (D). Only immunoprecipitated pro-A-SMase-EGFP treated with caspase-7 clearly displays cleavage and activation of A-SMase-EGFP, which is maximal after 5 min (F). In this case, both the kinetics of cleavage and the activation of A-SMase-EGFP run in parallel. (G) Western blot analysis of immunoprecipitated endogenous A-SMase from wild-type HeLa cells incubated with active caspase-7 and A-SMase activity assay. Both the kinetics of cleavage and the activation of A-SMase run in parallel. A-SMase activity data (±s.e.m.) are from four (A) or three (B–G) experiments each conducted as triplicate measurements, respectively.
We next investigated the effects of caspase-7 treatment on pro-A-SMase and found that caspase-7 in fact can activate pro-A-SMase not only when added to cell lysates (Figure 4E), but also when added to pro-A-SMase-EGFP purified by immunoprecipitation (Figure 4F). Again, after addition of active caspase-7, the intensity of the 82-kDa cleavage fragment derived from the purified pro-A-SMase-EGFP protein coincides with enhanced A-SMase activity. In order to prove, that also endogenous pro-A-SMase can be cleaved and activated by caspase-7, we treated immunoprecipitated endogenous pro-A-SMase with exogenous caspase-7, which also resulted in generation of the 57-kDa A-SMase protein and enhanced A-SMase activity (Figure 4G), suggesting that also endogenous pro-A-SMase is a direct substrate for caspase-7. Together, these results demonstrate that caspase-7 is involved in the activation of both the endogenous and the ectopically expressed form of A-SMase.
Colocalization of caspase-7 with TNF-R1-receptosomes and pro-A-SMase
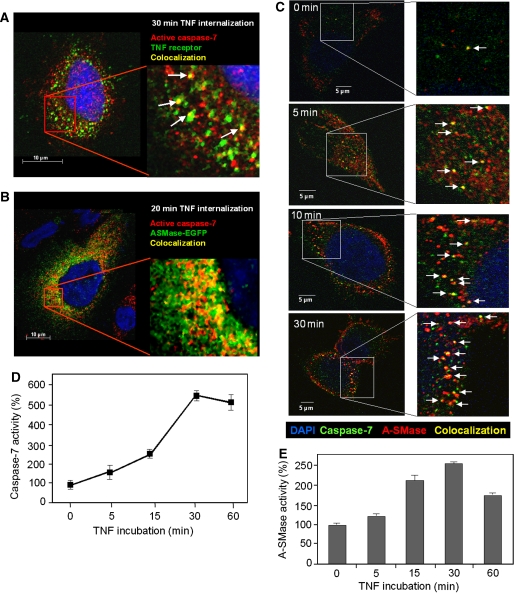
Simultaneous immunofluorescence detection of TNF receptors and cleaved caspase-7 revealed endocytic vesicles positive for both proteins (Figure 5A). As shown in Figure 5B, partial colocalization was also visible for active caspase-7 and ectopically expressed pro-A-SMase-EGFP as well as for caspase-7 and endogenous A-SMase, detected by the anti-A-SMase antibody (Figure 5C).
Figure 5.
Partial colocalization of active caspase-7 with TNF receptosomes and pro-A-SMase-EGFP as well as with endogenous A-SMase. Merged confocal microscopic images of (A) HeLa cells fluorescence labelled with biotin-TNF/avidin-FITC complexes (green) and anti-cleaved capase-7 monoclonal antibody (Cell signaling) (red) after 30 min of TNF-receptor internalization. (B) HeLa cells labelled with anti-cleaved capase-7 monoclonal antibody (red) and EGFP-tagged A-SMase (green) 20 min after TNF-receptor internalization, (C) HeLa cells labelled with anti-capase-7 antibody (Abcam) (green) and antibodies against endogenous A-SMase at various time points after TNF-receptor internalization. Colocalization of the respective fluorescently labelled molecules is indicated by yellow colour (marked with arrows). (D, E) TNF induction of caspase-7 and A-SMase run in parallel. Pro-A-SMase-EGFP-transfected cells were incubated with TNF for the indicated times. Cell lysates were used for caspase-7 activity assays (D) and A-SMase activity assays (E). Both time courses show a time-dependent increase of activity with the maximum after 30 min of TNF incubation. Data from three experiments (±s.e.m.) are shown.
Notably, the kinetics of TNF induction of caspase-7 and A-SMase also run in parallel (Figure 5D and E), giving further support to a possible functional link of both enzymes in situ.
Caspase-8, caspase-7 and pro-A-SMase are sequentially activated in magnetically isolated TNF-R1 receptosomes
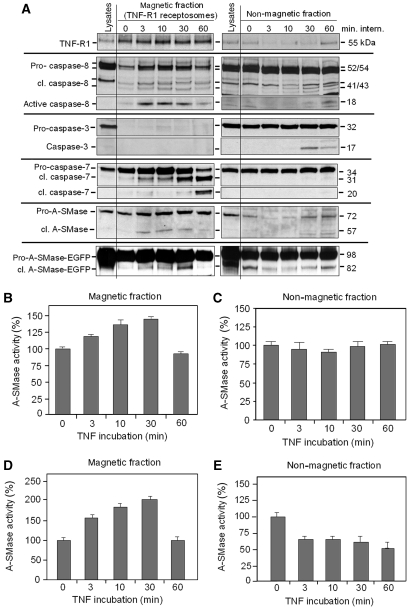
In order to confirm a direct colocalization of caspase-8, caspase-7 and pro-A-SMase in TNF-R1 containing membrane compartments with an independent method, we isolated TNF-R1 receptosomes from wild-type HeLa cells as well as from pro-A-SMase-EGFP-transfected HeLa cells using our immunomagnetic separation system (Schneider-Brachert et al, 2004, 2006; Liao et al, 2008; Tchikov and Schütze, 2008; Yazdanpanah et al, 2009). Purified TNF receptosomes (Figure 6A, left panel) and the remaining non-magnetic lysate fractions (Figure 6, right panel) were analysed by western blotting. The pro-forms of caspase-8 and -7 as well as the 72-kDa form of endogenous pro-A-SMase and the 98-kDa form of pro-A-SMase-EGFP were associated with magnetically isolated TNF-R1 receptosomes already at 0 min and were transiently enriched further on, indicating recruitment of these enzymes during internalization of TNF-R1. Already after 3 min pro-caspase-8 is activated within TNF receptosomes as evident from the appearance of the 41/43-kDa and the 18-kDa mature caspase-8 molecules. In non-magnetic fractions, active 18 kDa caspase-8 was observed at later time points (30–60 min) only after prolonged exposure of the blots.
Figure 6.
Cleavage and activation of caspase-8, caspase-7 and pro-A-SMase in magnetically isolated TNF receptosomes and non-magnetic fractions. (A) Time course of intracellular TNF receptosome trafficking (left panel) and corresponding non-magnetic fractions (right panel) in HeLa wild-type and pro-A-SMase-EGFP-transfected HeLa cells. Total cell lysate, magnetic and non-magnetic fractions derived after indicated times of TNF-receptor internalization were analysed for cleavage of pro-caspase-8 (54/52 kDa) to the active form of 18 kDa, cleavage of pro-caspase-3 to the active 17 kDa form (Cell signaling), cleavage of procaspase-7 (35 kDa) to the active forms of 31 and 20 kDa, cleavage of endogenous pro-A-SMase (72 kDa) to the more active 57 kDa form, and cleavage of recombinant pro-A-SMase-EGFP (98 kDa) to the more active 82 kDa form. All blots performed with the same antibody were assembled from the same gels with exception of the blot for active caspase-8 in the non-magnetic fractions, which was taken from a longer exposure of the same gel, which was blotted for pro- and cleaved caspase-8 (see primary scans in Supplementary data). Significant differences between the magnetic TNF receptosome fractions and the non-magnetic fractions become apparent (see text for details). Magnetic and non-magnetic fractions were used to measure endogenous A-SMase activation in HeLa wild-type cells (B, C) and to measure activation of ectopically expressed A-SMase-EGFP in stably transfected HeLa cells (D, E) after indicated times of TNF-receptor internalization. Results from two experiments (±s.e.m.) are shown.
Neither caspase-3 nor cleaved caspase-3 are present in receptosomes. In contrast, caspase-3 and also cleaved caspase-3 (after 30 min) were detected in the non-magnetic fractions, indicating that a caspase-3 activation occurs after TNF stimulation at later time points and is not associated with TNF receptosomes, as already indicated by the lack of colocalization (see above and Supplementary Figure S2).
The generation of the 18-kDa active caspase-8 fragment in TNF receptosomes appears slightly in advance of cleaved caspase-7 in the magnetic fractions, primarily the 31-kDa fragment. This 31 kDa caspase-7 molecule represents the processed active caspase-7 form lacking the N-terminal peptide of the pro-enzyme (Denault and Salvesen, 2003). The TNF-induced caspase-7 activation seems to take place exclusively within TNF receptosomes, since the corresponding non-magnetic fractions did not display any caspase-7 activation (Figure 6A, right panel). The appearance of the enzymatically active 31 kDa form of caspase-7 in TNF receptosomes is paralleled by the generation of the active 57 kDa fragment of endogenous A-SMase in wild-type HeLa cells and also by the generation of the 82-kDa A-SMase-EGFP fragment in the A-SMase-EGFP-transfected cells. In both cases, this processing of pro-A-SMases is paralleled by a concomittant increase in the respective A-SMase activities within the isolated TNF receptosomes (Figure 6B and D). The TNF-induced A-SMase processing and activation again is restricted to TNF receptosomes, since these events were not observed in the non-magnetic fractions (Figure 6A, right panel; Figure 6C and E).
The substantial amount of pro-A-SMase(-EGFP) detected in the receptosomes at 0 min may be explained by the fact that these vesicles are formed from resealed plasma membrane patches, which engulf all proteins within or close to the plasma membrane at the time of their formation. We have observed A-SMase localization at or close to the plasma membrane in confocal microscopic images (see Supplementary Figure S4), which may have caused the unexpected occurrence of A-SMase in the TNF receptosomes at the 0-min time point.
Direct binding of caspase-8 to TNF-R1 on internalized receptosomes has previously been demonstrated by coimmunoprecipitation experiments (Schneider-Brachert et al, 2004). To address a possible association of caspase-7 with TNF-R1, we here also performed coimmunoprecipitation of TNF-R1 and caspase-7 from isolated receptosomes and found no direct association of these two proteins (data not shown). Therefore we can conclude, that in contrast to caspase-8, caspase-7 does not bind directly to TNF-R1 but rather is associated with the receptosomal membrane in close proximity to TNF-R1.
The very early recruitment of caspases and pro-A-SMases prompted us to investigate the maturation of internalized TNF receptosomes: as can be taken from Supplementary Figure S3A, TNF receptosomes rapidly recruit Rab5B, a marker protein of endosomal trafficking, and the SNARE-protein Vti1b, located on trans-Golgi vesicles. Early fusion of Vti1b-positive vesicles and internalized TNF receptosomes was also detected by confocal laser-scan microscopy (Supplementary Figure S3B), indicating a very early (within 3–5 min) fusion of TNF receptosomes with trans-Golgi compartments, which contain the zymogen of A-SMase. At these early time points, the pH within the multivesicular compartments should not be very low. We measured the activity of caspase-7 in vitro at various pH values and found ∼50% activity at pH 6.5 compared with 100% at pH 7.5 (Supplementary Figure S5).
Caspase-7 binds directly to pro-A-SMase
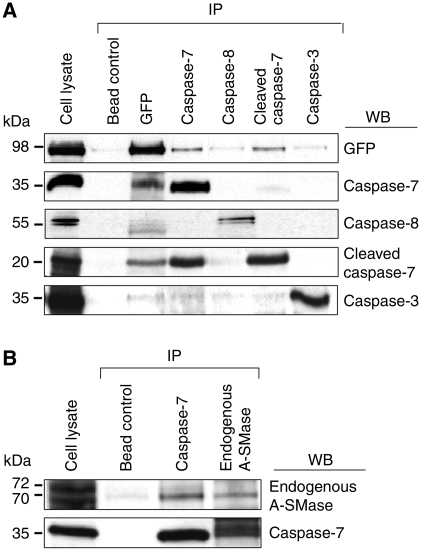
To analyse a possible interaction between caspase-7 and A-SMase, coimmunoprecipitation experiments were performed using an anti-GFP antibody for purification of recombinant pro-A-SMase-EGFP (Figure 7A) or the anti-A-SMase antibody for immunoprecipitation of endogenous A-SMase (Figure 7B). Copurification of A-SMase and caspase-7 occurred irrespective of the partner used for immunoprecipitation, confirming a direct association of caspase-7 and A-SMase within TNF receptosomes. In contrast, caspase-8 did not display firm association neither with caspase-7 nor with A-SMase. In experiments examining the coimmunoprecipitation of caspase-3 with A-SMase, no association of both molecules could be observed.
Figure 7.
Coimmunoprecipitation of A-SMase with caspase-8, caspase-7, cleaved caspase-7 and caspase-3. (A) HeLa cells stably transfected with GFP-tagged A-SMase were lysed and anti-GFP (Invitrogen, Molecular Probes), anti-caspase-8 (Santa Cruz), anti-caspase-7 (Abcam) and anti-caspase-3 (Cell signaling) antibodies were used for precipitation. Western blots performed with anti-GFP (Invitrogen, Molecular Probes), anti-caspase-8 (Santa Cruz), anti-caspase-7 (Abcam), anti-cleaved-caspase-7 (Cell signaling) and anti-caspase-3 (Cell signaling) antibodies demonstrate that caspase-7 coprecipitates with pro-A-SMase-GFP and vice versa in contrast to caspase-8 and caspase-3, which are neither precipitated with anti-GFP nor with anti-caspase-7 antibodies (in the case of caspase-8). (B) Immunoprecipitation of endogenous A-SMase with the anti-A-SMase antibody and of caspase-7 with anti-caspase-7 antibodies (Abcam) from HeLa wild-type cell lysates. Western blots show that caspase-7 coimmunoprecipitates with A-SMase and vice versa, in line with the results shown in (A). Figure assembled from cropped lanes of the same blots of the respective antibody (see primary scans in Supplementary data).
Deletion of caspase-7 inhibits TNF-induced cleavage and activation of A-SMase
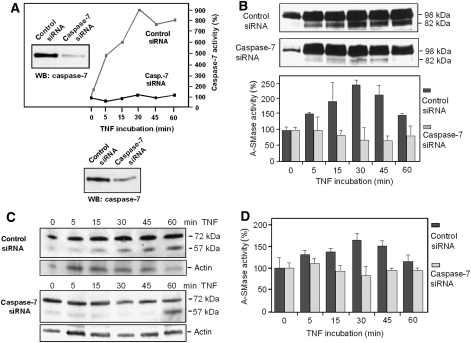
Finally, we used caspase-7 knockdown by siRNA to confirm its functional role in TNF signalling leading to A-SMase activation. Transfection of HeLa cells with caspase-7-specific siRNA led to a marked decrease in caspase-7 protein and a complete inhibition of TNF-induced caspase-7 enzymatic activity (Figure 8A; Supplementary Table S2). In cells expressing pro-A-SMase-EGFP transfected with control siRNA, an increase in the amount of the 82-kDa fragment could be observed after TNF stimulation, which was accompanied by an increase in A-SMase activity (Figure 8B). In contrast, generation of this fragment was almost invisible in caspase-7 knockdown cells and also the A-SMase activity did not increase during TNF treatment. These findings were also confirmed for endogenous A-SMase, since pro-A-SMase cleavage and generation of the 57-kDa fragment detected by the anti-A-SMase antibody still occurred in control siRNA transfected HeLa cells, whereas caspase-7 knockdown again displayed a significantly reduced pro-A-SMase processing (Figure 8C). In addition, caspase-7 siRNA down modulation also resulted in inhibition of the activation of endogenous A-SMase (Figure 8D).
Figure 8.
TNF-induced A-SMase processing and activation, in caspase-7 knockdown cells. (A) Down modulation of caspase-7 protein expression by siRNA analysed by western blotting and lack of TNF-induced caspase-7 activation in caspase-7 knockdown cells. (B) HeLa cells stably transfected with EGFP-tagged pro-A-SMase, or HeLa wild-type cells (C, D) were transfected with caspase-7 siRNA or control siRNA. Cells incubated with TNF for the indicated times were analysed by western blot with anti-GFP (B) or anti-A-SMase antibodies (C), respectively, and A-SMase activity assays (B, D). The knockdown of caspase-7 blocked the cleavage and activation of recombinant as well as endogenous pro-A-SMase, while they were unaltered in cells transfected with control siRNA.
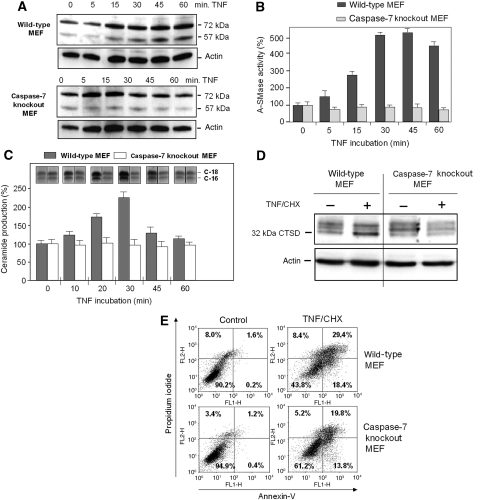
In line with these observations, TNF-treated mouse embryonic fibroblasts (MEFs) deficient for caspase-7 also did not respond with pro-A-SMase cleavage and enzymatic activation (Figure 9A and B). Caspase-7 deficiency in these cells also resulted in a lack of TNF-induced ceramide production (Figure 9C) and subsequent downstream cathepsin D activation (Figure 9D), indicated in the wild-type sample by the increased appearance of the mature 32 kDa isoform (Heinrich et al, 1999, 2000).
Figure 9.
TNF-induced A-SMase processing and activation, ceramide production, cathepsin D activation and apoptosis in caspase-7 knockout MEFs. (A, B) Wild-type MEFs or caspase-7 knockout MEFs were incubated with TNF for the indicated times and analysed by western blot and A-SMase activity assays. While in wild-type MEFs cleavage of endogenous pro-A-SMase and activation of A-SMase is observed, both events are completely blocked in caspase-7 knockout MEFs. (C) Wild-type MEFs or caspase-7 knockout MEFs were treated with TNF for indicated times and the production of ceramide analysed by TLC. TNF-induced ceramide production in caspase-7 knockout MEFs is completely prevented in contrast to wild-type MEFs. (D) Wild-type MEFs or caspase-7 knockout MEFs were treated with TNF for 12 h and analysed for expression of active 32 kDa cathepsin D protein. Caspase-7 deficiency in caspase-7 knockout MEFs prevents TNF induction of cathepsin D processing, which can be detected in wild-type MEFs. (E) Wild-type MEFs or caspase-7 knockout MEFs treated with CHX and TNF for 24 h were analysed for apoptosis by annexin-V/propidium iodide staining.
As a consequence, the blockade of the pro-apoptotic caspase-7/A-SMase/cathepsin D cascades in caspase-7-deficient MEF resulted in a reduced apoptotic response following TNF/CHX treatment (from 18.4 to 13.8% in early apoptosis and from 29.4 to 19.8% in late apoptosis in wild-type cells and caspase-7-deficient MEFs, respectively (see Figure 9E)). The residual apoptosis rates in the caspase-7-deficient MEFs are most likely due to unimpeded caspase-8/caspase-3 activities after TNF treatment.
Identification of the caspase-7 cleavage site in pro-A-SMase
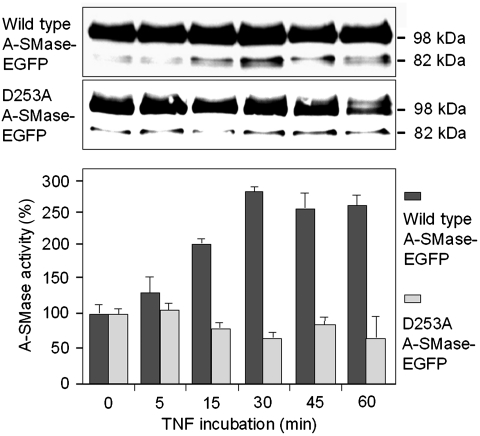
We next aimed to determine the exact cleavage site for caspase-7 in pro-A-SMase-EGFP. According to the observed molecular weight of the pro-A-SMase-EGFP cleavage fragment (82 kDa) four possible cleavage sites were taken into consideration (Supplementary Figure S6; Supplementary Table 1). Mutation of aspartate 225 (D225A) and a simultaneous triple mutation of aspartates 220, 222 and 225 (3D220A), respectively, did not result in a change of TNF-induced A-SMase cleavage and activation (data not shown). In contrast, mutation of aspartate 253 (D253A) resulted in a loss of TNF-induced A-SMase activation and an almost completely suppressed generation of the 82-kDa cleavage fragment (Figure 10). This result indicates that caspase-7 most probably cleaves pro-A-SMase at a non-canonical cleavage site after aspartate 253.
Figure 10.
Identification of a putative caspase-7 cleavage site in pro-A-SMase. The pro-A-SMase-EGFP mutants (see Supplementary Figure S7) were treated with TNF and cell lysates were analysed by western blot with an anti-GFP antibody and A-SMase activity assays. Compared with wild-type pro-A-SMase-EGFP only the mutant D253A showed an inhibition of A-SMase activation in combination with reduced occurrence of the cleavage fragment. Activity data (±s.e.m.) are from three experiments.
The 82-kDa band of unmutated A-SMase-EGFP in Figure 10 is actually resolved as a double band, which represents a mixture of a cleavage product of glycosylated A-SMase produced by caspase-7 and the unglycosylated form of uncleaved pro-A-SMase. In the western blot of the D253A-mutant samples in Figure 10, there is only a single band at 82 kDa, which represents in this case only the unglycosylated form of uncleaved pro-A-SMase, which is constitutively present and largely unchanged by TNF stimulation. In contrast, the double band at 82 kDa of wild-type A-SMase-EGFP shows a clear peak in intensity 30 min after TNF stimulation, which we attribute to an increased generation of the cleavage fragment by caspase-7.
Discussion
The molecular events that lead to the activation of A-SMase after ligand binding to death receptors and particularly to TNF-R1 are still poorly defined. The importance of caspase-8 in A-SMase activation has previously been shown for CD95 in a variety of cells (Grullich et al, 2000; Fanzo et al, 2003; Grassme et al, 2003; Rotolo et al, 2005; Reinehr et al, 2008) and for TNF in one earlier report by our group (Heinrich et al, 2004). Our present observation in caspase-8-deficient Jurkat cells supports this functional link and points to a general signalling pathway that couples death receptors via caspase-8 to A-SMase activation.
The partial colocalization of TNF and active caspase-8 in the same endosomal compartment, which we have observed in HeLa cells, suggests a possible interaction of active caspase-8 and A-SMase at this intracellular site. Using a polyclonal anti-A-SMase antibody, generated against a synthetic peptide of human A-SMase (Perrotta et al, 2007; Bianco et al, 2009) as well as by expression of recombinant A-SMase-EGFP fusion proteins we could indeed detect TNF-dependent colocalization of A-SMase and active caspase-8 in HeLa cells. The colocalization was most prominent in an intermediate space between the trans-Golgi-network (TGN) and the plasma membrane, indicating fusion of pro-A-SMase containing TGN vesicles with TNF-R1 containing endosomes. Such a fusion event has already been documented in our previous report (Schneider-Brachert et al, 2004) by electron microscopy and by biochemical analysis of isolated TNF receptosomes and is further substantiated by our present data showing a very early fusion within 5 min after TNF-receptor internalization.
The observation of a protein band of 57 kDa detected by the anti-A-SMase antibody in HeLa cells and of a 82-kDa EGFP-tagged protein detected in pro-A-SMase-EGFP expressing HeLa cells that both correlated with the induction of A-SMase activity after TNF stimulation (see Figure 3A and C), suggested that this band represents a truncated A-SMase-protein, which has been proteolytically cleaved at its N-terminus and activated thereby. Ferlinz et al (1994) already reported the existence of two molecular forms of A-SMase, a 70/72-kDa precursor protein and a mature 57 kDa protein, which correlate well with the 72-kDa pro-form and the 57-kDa cleaved form of the endogenous A-SMase as well as with the 98-kDa pro-form of A-SMase-EGFP and the corresponding 82 kDa cleavage fragment observed by us. From our cell fractionation experiments (see Figure 3B), we can conclude that the cellular 57 kDa protein indeed corresponds to the mature lysosomal A-SMase, which is reported to exhibit a higher activity than the 75/72-kDa pre-pro- and pro-A-SMase enzymes, respectively. This is also in line with our findings, that the appearance of the 57-kDa A-SMase molecule is associated with an increase of enzymatic activity and that a blocked generation of the 57-kDa species is associated with suppressed A-SMase activation (see Figures 3A and C; 4A, C and E–G; 6A, B and D; 8B–D; 9A and B and 10). The enzymatic activity of the cleavage fragments was calculated to be ∼5- to 14-fold higher than that of the respective pro-forms. Under all conditions tested, there is only a certain fraction of the pro-A-SMase pool (up to 46% of endogenous and up to 42% of recombinant enzyme) that becomes converted to the activated cleavage fragment. This restriction of the amount of total A-SMase cleaved and activated may be caused by a restricted accessibility of the pro-A-SMase to cleavage by caspase-7 in whole cells. TNF receptosomes fuse only to a certain portion of pro-A-SMase containing compartments, as can be seen in the confocal micrographs in Figure 2B and also when comparing the amounts of pro-A-SMase recruited to TNF receptosomes with the portion that remains in the non-magnetic fractions, shown in Figure 6.
On the other hand, the cleavage and activation of pro-A-SMase might be regulated by posttranslational modifications such as phosphorylation or glycosylation of the enzyme (reviewed by Jenkins et al, 2009), which may restrict the percentage of cleavable substrate for caspase-7. As an example for the regulatory function of substrate phosphorylation, the requirement of phosphorylation of the reticulon protein Nogo-B at Ser 16 within its non-canonical caspase-7 cleavage site was described recently (Schweigreiter et al, 2007). The identification of aspartate 253 as the putative cleavage site in A-SMase supports this explanation for the observed partial cleavage, since serine 250 could well function as a regulatory phosphorylation site within this non-canonical target sequence. Alternatively, the low abundance of cleaved A-SMase is caused by a reduced stability. Proteolytic degradation of the enzyme is one mechanism to terminate its enzymatic activity (Kölzer et al, 2004). This scenario may explain the transient appearance of the cleaved A-SMase forms (Figures 3C; 4A, F and G; 6A; 8A).
The glycosylation status of A-SMase has a crucial role for enzymatic activity and transport (Newrzella and Stoffel, 1996). The observed mobility shift after treatment with PNGase F for the pro-form as well as for the cleavage fragment of pro-A-SMase-EGFP (Figure 3D) identified both as glycoproteins. Intriguingly, the apparent molecular weight of the deglcosylated pro-form of 82 kDa is almost indistinguishable from the mobility of the glycosylated cleavage fragment. Ideally, like in Figure 10 (upper panel; wild-type A-SMase-EGFP), a double band at about 82 kDa can be discriminated. This finding also explains the constitutive presence of a weak 82 kDa band, which most likely represents the unglycosylated pro-A-SMase-EGFP protein and the constitutive presence of a weak 57 kDa band detected by the anti-A-SMase antibody in wild-type cells.
A proteolytic activation of A-SMase in response to receptor stimulation has not yet been described. Rather, in the context of death receptor signalling, A-SMase activation has so far been attributed to PKCδ-dependent phosphorylation (Zeidan and Hannun, 2007), acidification of endosomal compartments (Reinehr et al, 2008) or a ROS-dependent activation involving oxidation of the C-terminal cystein residue of A-SMase (Qiu et al, 2003). Whether these proposed activation mechanisms of A-SMase operate in parallel or sequentially, whether they have a general relevance for death receptor signalling or are restricted to certain cell types or types of death receptors, has not yet been established.
How A-SMase and caspase-7 become associated with TNF receptosomes is currently unknown. At least the association of active caspase-7 with membrane vesicles of the microsomal fraction has been observed previously after CD95 stimulation (Chandler et al, 1998; Zhivotovsky et al, 1999). Since we were unable to demonstrate a physical interaction of caspase-7 with the TNF-R1, it may rather be associated with the cytosolic face of the of TNF receptosomal membrane (as depicted in our scheme in Figure 11). For caspase-3 plasma membrane, localization in lipid rafts and a Fas ligand-induced recruitment to the Fas receptor has been reported in Jurkat cells (Aouad et al, 2004). In this case, caspase-3 was in close proximity to caspase-8 and became part of the DISC after Fas triggering. An analogous mechanism might be possible in the case of caspase-7 recruitment to TNF receptosomes. This topology allows for contact between TNF-R1-associated caspase-8 and caspase-7, resulting in caspase-8-mediated caspase-7 activation as well as providing contact to pro-A-SMase after fusion of TNF receptosomes with trans-Golgi vesicles.
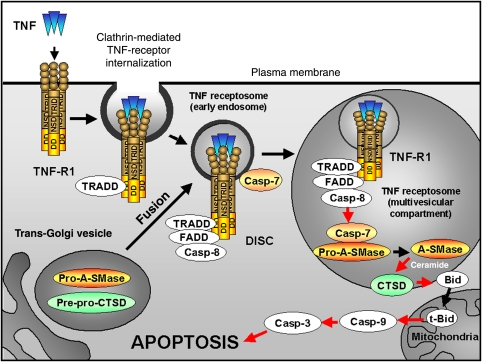
Figure 11.
Compartmentalization of TNF-R1 signalling and A-SMase activation. Binding of TNF ligand to TNF-R1 initializes the clathrin-dependent endocytosis and the recruitment of adaptor proteins TRADD, FADD and caspase-8. Within the receptosome-bound death-inducing signalling complex (DISC), caspase-8 is activated. Along the endocytotic pathway, TNF receptosomes fuse with trans-Golgi vesicles containing pro-acid sphingomyelinase (pro-A-SMase) and pre-pro-cathepsin D (pre-pro-CTSD) to form multivesicular organelles. There, activation of caspase-7 by active caspase-8 leads to the cleavage and activation of pro-A-SMase. Activated A-SMase generates ceramide, which activates CTSD and mediates its translocation from the late endosome to the cytosol. In this compartment, the proapoptotic protein Bid is cleaved by CTSD to tBid resulting in release of cytochrome C from mitochondria and activation of caspase-9 and caspase-3 leading to apoptotic cell death.
The stable association of pro-caspase-7 with A-SMase-EGFP as well as with endogenous A-SMase, which we observed in HeLa cells (Figure 7), is in fact remarkable, since the enzymatic interaction during cleavage should only be transient. Since predominantly the proform of caspase-7 is associated with A-SMase, this may indicate the involvement of the N-terminal propeptide of caspase-7 in the interaction with A-SMase.
It is presently unknown how A-SMase and pro-caspase-7 can come into direct contact inside the cell, since A-SMase in the lumen of the endolysosomal compartment and caspase-7 at the cytosolic surface of the TNF receptosomes are supposed to be separated by a lipid membrane. Whether an intraluminal membrane breakdown after vesicular fusion or a translocation of proteins through the lipid bilayer within the multivesicular compartment will lead to contact with each other is an open question. The same topology problem is unresolved for the proposed regulation of A-SMase secretion and activation by PKCδ (Zeidan and Hannun, 2007; Jenkins et al, 2010) as well as for the observed association of caspase-3 and A-SMase upon NO exposure (Castillo et al, 2007). So far only acid ceramidase has been found to associate with secreted A-SMase (He et al, 2003). We are currently trying to solve this important question by immunoelectron microscopic analysis.
Based on our confocal microscopy analysis, ∼5–20% of cellular endogenous A-SMase was detected colocalized with TNF receptosomes. Only 1–2% of the total cellular A-SMase and 0.4–0.6% of the total overexpressed A-SMase was calculated to be recovered in isolated TNF receptosomes, indicating that part of the receptosomes was lost during the isolation process (which takes several hours at 4°C), or the enzyme was partly degraded. However, the high amount of A-SMase cleaved and activated by TNF in whole cells compared with the ratio of TNF receptosome/A-SMase fusion events and A-SMase activation observed in isolated TNF receptosomes may also reflect the fact that additional mechanisms of activation exist for A-SMase.
As caspase-3 is considered to be functionally redundant with caspase-7 (for review see Luthi and Martin, 2007; Timmer and Salvesen, 2007), the role of caspase-3 in TNF-induced A-SMase activation had to be clarified. From experiments addressing the ability of caspase-3 to directly cleave and activate pro-A-SMase, the localization of caspase-3 in receptosomes, its possible colocalization with TNF-R1, and its possible direct interaction with A-SMase, we found no indications for an involvement of caspase-3 in TNF-R1-mediated activation of A-SMase (see Figures 4D, 6A, 7A; Supplementary Figure S2). Rather, our data suggest that by its membrane localization and its interaction with A-SMase, caspase-7 has a particular specificity for A-SMase as its substrate. This has to be distinguished from the mere catalytic specificity for peptide substrates in vitro, which largely overlaps with that of caspase-3. Of note, a difference in the physiological substrate specificity between caspases-3 and -7 has been observed (Walsh et al, 2008; Demon et al, 2009).
The aspartate residue 253 of pro-A-SMase, which we identified by mutation analysis as the most probable cleavage site for caspase-7 does not match the canonical consensus sequence Asp-X-X-Asp. Still it may be cleaved by caspase-7 (and also caspase-3) as predicted by an algorithm determining differentially preferred cleavage sites for caspase-3 and caspase-7 (Demon et al, 2009). It is known for some time that caspases are able to cleave also at non-canonical sites (Kipp et al, 2000), in particular caspase-7 (Agniswamy et al, 2007; Schweigreiter et al, 2007).
In summary, our present findings are closing an open gap in the pro-apoptotic TNF-R1 signalling cascade: the sequential activation of caspase-8, followed by caspase-7, and finally A-SMase within TNF receptosomes (depicted in Figure 11). The resulting ceramide production in these organelles ultimately leads—via activation of cathepsin D—to induction of the mitochondrial amplification loop contributing to apoptosis.
Materials and methods
Antibodies used in this study, cell culture, preparation of cell lysates and the detailed description of molecular cloning, transfection of cell lines, and caspase-7 knockdown, as well as the protocol for confocal microscopy can be found in Supplementary data.
Immunoprecipitation
For immunoprecipitation, 3 × 107 cells were trypsinized, washed with PBS and resuspended in 40 mM HEPES pH 7.4, 150 mM KCl, 0.2% NP-40, and protease inhibitor cocktail (Roche Complete, Roche Diagnostics). After freezing and thawing, the cell suspension was centrifuged for 10 min at 20 000 g. The cell lysate was pre-incubated with protein G-Sepharose (GE-Healthcare) at a ratio of 15 μl per 1 mg total protein for 45 min at 4°C with continuous rotation followed by centrifugation at 4°C for 1 min at 14 000 g. A measure of 1–2 μg monoclonal or 2–4 μg polyclonal antibody was added and incubated for 1.5 h at 4°C with continuous rotation. Subsequently, protein G sepharose (30 μl per 1 mg total protein) was added and further incubated for 1.5 h at 4°C with continuous rotation. The mixture was centrifuged at 4°C for 5 min at 14 000 g and the sepharose pellet was washed three times in 20 mM HEPES pH 7.4, 10 mM MgCl2, 5 mM DTT, and 0.2% NP-40 including protease inhibitors 0.1 mM Na3VO4, 0.1 mM Na2MoO4, 1 mM PMSF, 10 μM Leupeptin and 10 μM pepstatin and 750 μM ATP.
Immunoprecipitation of endogenous A-SMase
HeLa cells were lysed in buffer containing 150 mM NaCl, 50 mM Tris–HCl pH 8.0, 1% Triton X-100 and the protease inhibitor cocktail Complete (Roche). After addition of 50 μl protein G magnetic microbeads (Miltenyi) and 15 μl anti-A-SMase antibody (Areta International s.r.l. Gerenzano (Va), Italy), the cell lysate was incubated for 30 min on ice followed by separation of the complexes on a custom-built free-flow magnetic chamber (Tchikov and Schütze, 2008). After washing with lysis buffer for 1 h, the microbeads were eluated from the column and centrifuged for 10 min at 20 000 g, resuspended in buffer containing 500 mM NaCl, 50 mM Tris–HCl pH 8.0, 0,1% NP-40 and sonicated for 5 min. The samples were incubated with caspase-7 as described above and A-SMase cleavage and enzymatic activity was analysed by western blotting and A-SMase activity assays.
Deglycosylation of recombinant A-SMase
Immunoprecipitated A-SMase-EGFP was deglycosylated by incubation with PNGase F (New England Biolabs, Ipswich) according to the manufacturer's instructions. The reaction mixture was boiled with Laemmli buffer for 5 min and samples were analysed by western blotting.
Caspase cleavage assay
Cell lysates or IP material were homogenized in 20 mM HEPES pH 7.4, 10 mM MgCl2, 5 mM DTT and 0.2% NP-40 including protease inhibitors 0.1 mM Na3VO4, 0.1 mM Na2MoO4, 1 mM PMSF, 10 μM Leupeptin, 10 μM pepstatin and 750 μM ATP. In all, 30 units of active caspase-8 (Becton Dickinson) or 50 units of active caspase-7 (Biomol) were added to 100 μg of total protein and incubated at 37°C for distinct time points (0–30 min).
A-SMase activity assay
Activity of A-SMase in cell lysates or IP material was measured using N-methyl-[14C]-sphingomyelin (0.5 μCi/ml, 0.55 Ci/mmol, Hartman Inc.) as substrate in 250 mM Na-acetate pH 5.0, 1 mM EDTA, 0.1% Triton X-100. A measure of 2–10 μg of protein was incubated with the substrate for 2 h at 37°C in a total volume of 150 μl. The reaction mixture was extracted using 0.750 ml chloroform/methanol (2:1). Radioactivity of enzymatically liberated radioactive phosphorylcholine in a 250-μl aliquot of the aqueous phase was measured in a β-counter.
Evaluation of ceramide generation
Lipids were extracted from cells and the amount of ceramide was determined by thin-layer chromatography and charring densitometry following standard procedures (Schütze et al, 1992). For quantification, standard ceramide type III (Sigma; mostly composed of N-stearoylsphingosine) was used.
Subcellular fractionation by iodixanol density gradient centrifugation
HeLa cells were homogenized mechanically in 0.25 M sucrose, 10 mM triethanolamine-10 mM acetic acid buffer, pH 7.8. A top-loaded multistep iodixanol discontinuous density gradient (Graham et al, 1994) of 12, 14, 16, 18, 20% (w/v) was prepared following the recommendations of the OptiPrep manufacturer manual instructions (Axis-Shield PoC AS, Oslo, Norway). Osmolarity was adjusted to 290 mOsm, with 2.3 M sucrose according to the recommendation of the Lysosome Isolation Kit (Sigma). Fractionation was performed by ultracentrifugation in 4 ml Ultra-Clear centrifuge tubes in a Beckman SW60Ti rotor, 150 000 g for 5 h at 4°C in a Beckman Coulter Optima XL-80 Ultracentrifuge according to Schmidt et al (2009). After centrifugation, the fractions were collected from the top, placed into fresh centrifugation tubes, loaded with the homogenization medium up to 4 ml of the total volume and the material was sedimented by centrifugation at 150 000 g for 20 min, resuspended in homogenization buffer and stored as above.
Western blot analysis
Cells were lysed as described above and proteins were separated on 10–15% SDS–PAGE and immunoblotting was performed using one of the primary antibodies indicated above and secondary antibody horseradish peroxidase conjugates (Dianova). Blots were developed using the ECL detection reagent (Amersham-Pharmacia).
Preparation and analysis of magnetic TNF-R1 containing membrane fractions
Cells were incubated in a total volume of 250 μl cold PBS with 100 μl (400 ng) of biotinylated TNF (Fluorkine-Kit; R&D Systems, Wiesbaden, Germany) for 1 h on ice. Afterwards, 200 μl of 50 nm MACS Streptavidin microbeads solution (Miltenyi Biotech, Bergisch Gladbach, Germany) were added and cells were incubated for 1 h on ice. Magnetically labelled cells were washed with ice-cold PBS and pelleted by centrifugation at 100 g for 10 min at 4°C. Formation of magnetically labelled TNF receptosomes was achieved by incubation of cells at 37°C for various times as indicated in the figure legends. Subsequently, cells were mechanically homogenized using glass beads in a 0.25-M sucrose buffer, supplemented with 15 mM HEPES pH 7.4, 0.5 mM MgCl2 and the Complete® protease inhibitor set (Roche Diagnostics, Mannheim, Germany) at 4°C. A postnuclear supernatant was submitted to magnetic separation of TNF receptosomes in a high-gradient magnetic field generated in a custom-built free-flow magnetic chamber (Tchikov and Schütze, 2008). Magnetic fractions were separated by SDS–PAGE and analysed by western blotting, or were used directly for A-SMase activity assays.
Caspase activity assays
Caspase-7 activity was determined using the Apo-ONE® Homogenous caspase assay (Promega) and caspase-8 activity was measured using caspase-Glo® 8 assay (Promega) according to the manufacturer's instructions.
Apoptosis assay
Cell death was monitored by seeding 5 × 105 cells in six-well plates and stimulation with 100 ng/ml TNF and 0.5 μg/ml CHX for distinct time points (4 or 24 h). The cells were harvested and washed once with PBS and centrifuged 5 min at 200 g. The cell pellet was resuspended in Annexin-V-FLOUS labelling solution containing annexin-V and propidium iodide (dilutions according to the manufacturer Roche instructions) and incubated for 10–15 min at room temperature. Stained cells were analysed using the FACS-Calibur (BD) and the Cell-Quest 3.3 software.
Supplementary Material
Acknowledgments
We thank Andrea Hethke, Fereshteh Ebrahim and Casimir Malanda for excellent technical assistance as well as Heiner Oberg and Sandra Ussat for their help with FACS sorting of stably transfected cells. We are grateful to Peter Vandenabeele and Tom Vanden Berghe (Flanders Institute for Biotechnology, Ghent, Belgium) for analysis of the A-SMase sequence with regard to non-canonical caspase cleavage sites. We are indebted to John Blenis (Harvard Medical School, Boston) for caspase-8-deficient Jurkat cells and Richard Flavell (Yale School of Medicine, New Haven) for providing caspase-7 knockout MEFs as well as to Hamid Kashkar (University of Cologne, Germany) for the A-SMase-EGFP plasmid. This work was supported by grants from the Germany Research Foundation (DFG) Collaborative Research Center SFB 415, the DFG Sphingolipid Priority Program SPP 1267 and by the Schleswig-Holstein Cluster of Excellence ‘Inflammation at Interfaces' given to SS.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agniswamy J, Fang B, Weber IT (2007) Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis. FEBS J 274: 4752–4765 [DOI] [PubMed] [Google Scholar]
- Aouad SM, Cohen LY, Sharif-Askari E, Haddad EK, Alam A, Sekaly RP (2004) Caspase-3 is a component of Fas death-inducing signalling complex in lipid rafts and its activity is required for complete caspase-8 activation during Fas-mediated cell death. J Immunol 172: 2316–2323 [DOI] [PubMed] [Google Scholar]
- Bianco F, Perrotta C, Novellino L, Francolini M, Riganti L, Menna E, Saglietti L, Schuchman EH, Furlan R, Clementi E, Matteoli M, Verderio C (2009) Acid sphingomyelinase actively triggers microparticle release from glia cells. EMBO J 28: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner B, Ferlinz K, Grassmé H, Weller M, Koppenhoefer U, Dichgans J, Sandhoff K, Lang F, Gulbins E (1998) Fas/CD95/Apo-I activates the acidic sphingomyelinase via caspases. Cell Death Differ 5: 29–37 [DOI] [PubMed] [Google Scholar]
- Castillo SS, Levy M, Wang C, Thaikoottathil JV, Khan E, Goldkorn T (2007) Nitric oxide-enhanced caspase-3 and acidic sphingomyelinase interaction: a novel mechanism by which airway epithelial cells escape ceramide-induced apoptosis. Exp Cell Res 313: 816–823 [DOI] [PubMed] [Google Scholar]
- Chandler JM, Cohen GM, MacFarlane M (1998) Different subcellular distribution of caspase-3 and caspase-7 following Fas-induced apoptosis in mouse liver. J Biol Chem 273: 10815–10818 [DOI] [PubMed] [Google Scholar]
- Charruyer A, Grazide S, Bezombes C, Müller S, Laurent G, Jaffrézou JP (2005) UV-C light induces raft-associated acid sphingomyelinase and JNK activation and translocation independently on a nuclear signal. J Biol Chem 280: 19196–19204 [DOI] [PubMed] [Google Scholar]
- Cifone MG, De Maria R, Roncaioli P, Rippo MR, Azuma M, Lanier LL, Santoni A, Testi R (1994) Apoptotic signaling through CD95 (Fas/Apo-1) activates an acidic sphingomyelinase. J Exp Med 180: 1547–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maria R, Rippo MR, Schuchman EH, Testi R (1998) Acidic sphingomyelinase (ASM) is necessary for fas-induced GD3 ganglioside accumulation and efficient apoptosis of lymphoid cells. J Exp Med 187: 897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demon D, van Damme P, Vanden Berghe T, Deceuninck A, Van Durme J, Verspurten J, Helsens K, Impens F, Wejda M, Schymkowitz J, Rousseau F, Madder A, Vandekerckhove J, Declercq W, Gevaert K, Vandenabeele P (2009) Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol Cell Proteomics 8: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denault JB, Salvesen GS (2003) Human caspase-7 activity and regulation by its N-terminal peptide. J Biol Chem 278: 34042–34050 [DOI] [PubMed] [Google Scholar]
- Dumitru CA, Gulbins E (2006) TRAIL activates acid sphingomyelinase via a redox mechanism and releases ceramide to trigger apoptosis. Oncogene 25: 5612–5625 [DOI] [PubMed] [Google Scholar]
- Dumitru CA, Zhang Y, Li X, Gulbins E (2007) Ceramide: a novel player in reactive oxygen species-induced signalling? Antioxid Redox Signal 9: 1535–1540 [DOI] [PubMed] [Google Scholar]
- Fanzo JC, Lynch MP, Phee H, Hyer M, Cremesti A, Grassme H, Norris JS, Coggeshall KM, Rueda BR, Pernis AB, Kolesnick R, Gulbins E (2003) CD95 rapidly clusters in cells of diverse origins. Cancer Biol Ther 2: 392–395 [DOI] [PubMed] [Google Scholar]
- Ferlinz K, Hurwitz R, Vielhaber G, Suzuki K, Sandhoff K (1994) Occurrence of two molecular forms of human acid sphingomyelinase. Biochem J 301: 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ruiz C, Colell A, Marí M, Morales A, Calvo M, Enrich C, Fernández-Checa JC (2003) Defective TNF-alpha-mediated hepatocellular apoptosis and liver damage in acidic sphingomyelinase knockout mice. J Clin Invest 111: 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J, Ford T, Rickwood D (1994) The preparation of subcellular organelles from mouse liver in self-generated gradients of iodixanol. Anal Biochem 220: 367–373 [DOI] [PubMed] [Google Scholar]
- Grassme H, Cremesti A, Kolesnick R, Gulbins E (2003) Ceramide-mediated clustering is required for CD95-DISC formation. Oncogene 22: 5457–5470 [DOI] [PubMed] [Google Scholar]
- Grullich C, Sullards MC, Fuks Z, Merrill AH Jr, Kolesnick R (2000) CD95(Fas/APO-1) signals ceramide generation independent of the effector stage of apoptosis. J Biol Chem 275: 8650–8656 [DOI] [PubMed] [Google Scholar]
- Guicciardi ME, Gores GJ (2009) Life and death by death receptors. FASEB J 23: 1625–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid L (2008) Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 9: 139–150 [DOI] [PubMed] [Google Scholar]
- He X, Okino N, Dhami R, Dagan A, Gatt S, Schulze H, Sandhoff K, Schuchman EH (2003) Purification and characterization of recombinant, human acid ceramidase. Catalytic reactions and interactions with acid sphingomyelinase. J Biol Chem 278: 32978–32986 [DOI] [PubMed] [Google Scholar]
- Heinrich M, Neumeyer J, Jakob M, Hallas C, Tchikov V, Winoto-Morbach S, Wickel M, Schneider-Brachert W, Trauzold A, Hethke A, Schütze S (2004) Cathepsin D links TNF-induced acid sphingomyelinase to Bid-mediated caspase-9 and-3 activation. Cell Death Differ 11: 550–563 [DOI] [PubMed] [Google Scholar]
- Heinrich M, Wickel M, Schneider-Brachert W, Sandberg C, Gahr J, Schwandner R, Weber T, Saftig P, Peters C, Brunner J, Krönke M, Schütze S (1999) Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J 18: 5252–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich M, Wickel M, Winoto-Morbach S, Schneider-Brachert W, Weber T, Brunner J, Saftig P, Peters Ch, Krönke M, Schütze S (2000) Ceramide as an activator lipid of cathepsin D. Adv Exp Med Biol 477: 305–315 [DOI] [PubMed] [Google Scholar]
- Herr I, Wilhelm D, Böhler T, Angel P, Debatin KM (1997) Activation of CD95 (APO-1/Fas) signaling by ceramide mediates cancer therapy-induced apoptosis. EMBO J 16: 6200–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins RW, Canals D, Hannun YA (2009) Roles and regulation of secretory and lysosomal acid sphingomyelinase. Cell Signal 21: 836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins RW, Canals D, Idkowiak-Baldys J, Simbari F, Roddy P, Perry DM, Kitatani K, Luberto C, Hannun YA (2010) Regulated secretion of acid sphingomyelinase: implications for selectivity of ceramide formation. J Biol Chem 285: 35706–35718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipp M, Schwab BL, Przybylski M, Nicotera P, Fackelmayer FO (2000) Apoptotic cleavage of scaffold attachment factor A (SAF-A) by caspase-3 occurs at a noncanonical cleavage site. J Biol Chem 275: 5031–5036 [DOI] [PubMed] [Google Scholar]
- Kolesnick RN, Haimovitz-Friedman A, Fuks Z (1994) The sphingomyelin signal transduction pathway mediates apoptosis for tumor necrosis factor, Fas, and ionizing radiation. Biochem Cell Biol 72: 471–474 [DOI] [PubMed] [Google Scholar]
- Kolesnick RN, Krönke M (1998) Regulation of ceramide production and apoptosis. Annu Rev Physiol 60: 643–665 [DOI] [PubMed] [Google Scholar]
- Kölzer M, Werth N, Sandhoff K (2004) Interactions of acid sphingomyelinase and lipid bilayers in the presence of the tricyclic antidepressant desipramine. FEBS Lett 559: 96–98 [DOI] [PubMed] [Google Scholar]
- Lang PA, Schenck M, Nicolay JP, Becker JU, Kempe DS, Lupescu A, Koka S, Eisele K, Klarl BA, Rübben H, Schmid KW, Mann K, Hildenbrand S, Hefter H, Huber SM, Wieder T, Erhardt A, Häussinger D, Gulbins E, Lang F (2007) Liver cell death and anemia in Wilson disease involve acid sphingomyelinase and ceramide. Nat Med 13: 164–170 [DOI] [PubMed] [Google Scholar]
- Lansmann S, Schuette CG, Bartelson O, Hoernschemeyer J, Linke T, Weisgraber J, Sandhoff K (2003) Human acid sphingomyelinase. Eur J Biochem 270: 1076–1088 [DOI] [PubMed] [Google Scholar]
- Liao W, Xiao Q, Liao Z, Wincovitch S, Garfield S, Yang W, Tchikov V, El-Deiry W, Schütze S, Srinivasula SM (2008) CARP-2 is an endosomal ubiquitin protein ligase for RIP and regulates TNF-induced NF-B activation. Curr Biol 18: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Genestier L, Pinkoski MJ, Castro A, Nicholas S, Mogil R, Paris F, Fuks Z, Schuchmann EH, Kolesnick RN, Green DR (2000a) Role of acidic sphingomyelinase in Fas/CD95-mediated cell death. J Biol Chem 275: 8657–8663 [DOI] [PubMed] [Google Scholar]
- Lin X, Fuks Z, Kolesnick R (2000b) Ceramide mediates radiation-induced death of endothelium. Crit Care Med 28: N87–N93 [DOI] [PubMed] [Google Scholar]
- Luthi AU, Martin SJ (2007) The CASBAH: a searchable database of caspase substrates. Cell Death Differ 14: 641–650 [DOI] [PubMed] [Google Scholar]
- Martin SJ, Newmeyer DD, Mathias S, Farschon DM, Wang HG, Reed JC, Kolesnick RN, Green DR (1995) Cell-free reconstitution of Fas-, UV radiation- and ceramide-induced apoptosis. EMBO J 14: 5191–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson PS, Kay BK, Hussain NK (2001) Signaling on the endocytic pathway. Traffic 2: 375–384 [DOI] [PubMed] [Google Scholar]
- Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J (2001) NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol 21: 5299–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Tschopp J (2003) Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114: 181–190 [DOI] [PubMed] [Google Scholar]
- Monney L, Olivier R, Otter I, Jansen B, Poirier GG, Borner C (1998) Role of an acidic compartment in tumor-necrosis-factor-alpha-induced production of ceramide, activation of caspase-3 and apoptosis. Eur J Biochem 251: 295–303 [DOI] [PubMed] [Google Scholar]
- Morales A, Lee H, Goñi FM, Kolesnick R, Fernandez-Checa JC (2007) Sphingolipids and cell death. Apoptosis 12: 923–939 [DOI] [PubMed] [Google Scholar]
- Newrzella D, Stoffel W (1996) Functional analysis of the glycosylation of murine acid sphingomyelinase. J Biol Chem 271: 32089–32095 [DOI] [PubMed] [Google Scholar]
- Perrotta C, Bizzozero L, Falcon S, Rovere-Querini P, Prinetti A, Schuchman EH, Sonnino S, Manfredi AA, Clementi E (2007) Nitic oxide boosts chemoimmunotherapy via inhibition of acid sphingomyelinase in a mouse model of melanoma. Cancer Res 67: 7559–7564 [DOI] [PubMed] [Google Scholar]
- Qiu H, Edmunds T, Baker-Malcolm J, Karey KP, Estes S, Schwarz C, Hughes H, Van Patten SM (2003) Activation of human acid sphingomyelinase through modification or deletion of C-terminal cysteine. J Biol Chem 278: 32744–32752 [DOI] [PubMed] [Google Scholar]
- Reinehr R, Becker S, Braun J, Ebeler A, Grether-Beck S, Häussinger D (2006) Endosomal acidification and activation of NADPH oxidase isoforms are upstream events in hyperosmolarity-induced hepatocyte apoptosis. J Biol Chem 281: 23150–23166 [DOI] [PubMed] [Google Scholar]
- Reinehr R, Sommerfeld A, Keitel V, Grether-Beck S, Haussinger D (2008) Amplification of CD95 activation by caspase 8-induced endosomal acidification in rat hepatocytes. J Biol Chem 283: 2211–2222 [DOI] [PubMed] [Google Scholar]
- Rotolo JA, Zhang J, Donepudi M, Lee H, Fuks Z, Kolesnick R (2005) Caspase-dependent and -independent activation of acid sphingomyelinase signaling. J Biol Chem 280: 26425–26434 [DOI] [PubMed] [Google Scholar]
- Schmidt H, Gelhaus C, Lucius R, Nebendahl M, Leippe M, Janssen O (2009) Enrichment and analysis lysosomes from lymphocyte populations. BMC Immunol 10: 41, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Brachert W, Tchikov V, Kruse ML, Lehn A, Jakob M, Hallas C, Hildt E, Held-Feindt J, Kabelitz D, Krönke M, Schütze S (2006) Inhibition of TNF receptor 1 internalization by adenovirus E3-14.7K as a novel immune escape mechanism. J Clin Invest 116: 2901–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J, Heinrich M, Merkel O, Ehrenschwander M, Adam D, Mentlein R, Kabelitz D, Schütze S (2004) Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity 21: 415–428 [DOI] [PubMed] [Google Scholar]
- Schütze S, Potthoff K, Machleidt T, Berkovic D, Wiegmann K, Krönke M (1992) TNF activates NF-kappa-B by phosphatidylcholine-specific phospholipase-C-induced acidic sphingomyelin breakdown. Cell 71: 765–776 [DOI] [PubMed] [Google Scholar]
- Schütze S, Tchikov V, Schneider-Brachert W (2008) Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat Rev Mol Cell Biol 9: 655–665 [DOI] [PubMed] [Google Scholar]
- Schwandner R, Wiegmann K, Bernardo K, Kreder D, Krönke M (1998) TNF receptor death domain-associated proteins TRADD and FADD signal activation of acid sphingomyelinase. J Biol Chem 273: 5916–5922 [DOI] [PubMed] [Google Scholar]
- Schweigreiter R, Stasyk T, Contarini I, Frauscher S, Oertle T, Klimaschewski L, Huber LA, Bandtlow CE (2007) Phosphorylation-regulated cleavage of the reticulon protein Nogo-B by caspase-7 at a noncanonical recognition site. Proteomics 7: 4457–4467 [DOI] [PubMed] [Google Scholar]
- Scita G, Di Fiore PP (2010) The endocytic matrix. Nature 463: 464–473 [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M (2002) Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol 3: 600–614 [DOI] [PubMed] [Google Scholar]
- Tchikov V, Schütze S (2008) Immunomagnetic isolation of tumor necrosis factor receptosomes. Methods Enzymol 442: 101–123 [DOI] [PubMed] [Google Scholar]
- Timmer JC, Salvesen GS (2007) Caspase substrates. Cell Death Differ 14: 66–72 [DOI] [PubMed] [Google Scholar]
- Thon L, Mathieu S, Kabelitz D, Adam D (2006) The murine TRAIL receptor signals caspase-independent cell death through ceramide. Exp Cell Res 312: 3808–3821 [DOI] [PubMed] [Google Scholar]
- Walsh JG, Cullen SP, Sheridan C, Lüthi AU, Gerner C, Martin SJ (2008) Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci USA 105: 12815–12879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmann K, Schütze S, Machleidt T, Witte D, Krönke M (1994) Functional dichotomy of neutral and acidic sphingomyelinases in tumor necrosis factor signaling. Cell 78: 1005–1015 [DOI] [PubMed] [Google Scholar]
- Wiegmann K, Schwandner R, Krut O, Yeh WC, Mak TW, Krönke M (1999) Requirement of FADD for tumor necrosis factor-induced activation of acid sphingomyelinase. J Biol Chem 274: 5267–5270 [DOI] [PubMed] [Google Scholar]
- Yazdanpanah B, Wiegmann K, Tchikov V, Krut O, Pongratz C, Schramm M, Kleinridders A, Wunderlich T, Kashkar H, Utermöhlen O, Brüning JC, Schütze S, Krönke M (2009) Riboflavin kinase couples TNF receptor 1 to NADPH oxidase. Nature 460: 1159–1163 [DOI] [PubMed] [Google Scholar]
- Zeidan YH, Hannun YA (2007) Activation of acid sphingomyelinase by protein kinase Cdelta-mediated phosphorylation. J Biol Chem 282: 11549–11561 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Samali A, Gahm A, Orrenius S (1999) Caspases: their intracellular localization and translocation during apoptosis. Cell Death Differ 6: 644–651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.