Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore
Here, centromeric histone marks on a human artificial chromosome are found to resemble the chromatin landscape in transcribed genes, and selective manipulation shows them to govern the incorporation of the centromere-specifying CENP-A histone variant.
Keywords: CENP-A, centromere, chromatin, kinetochore, non-coding RNA
Abstract
Kinetochores assemble on distinct ‘centrochromatin' containing the histone H3 variant CENP-A and interspersed nucleosomes dimethylated on H3K4 (H3K4me2). Little is known about how the chromatin environment at active centromeres governs centromeric structure and function. Here, we report that centrochromatin resembles K4–K36 domains found in the body of some actively transcribed housekeeping genes. By tethering the lysine-specific demethylase 1 (LSD1), we specifically depleted H3K4me2, a modification thought to have a role in transcriptional memory, from the kinetochore of a synthetic human artificial chromosome (HAC). H3K4me2 depletion caused kinetochores to suffer a rapid loss of transcription of the underlying α-satellite DNA and to no longer efficiently recruit HJURP, the CENP-A chaperone. Kinetochores depleted of H3K4me2 remained functional in the short term, but were defective in incorporation of CENP-A, and were gradually inactivated. Our data provide a functional link between the centromeric chromatin, α-satellite transcription, maintenance of CENP-A levels and kinetochore stability.
Introduction
The inner kinetochore is a specialized domain of centromeric chromatin defined by the presence of the centromere-specific histone H3 variant CENP-A. As cells enter mitosis, interphase pre-kinetochores undergo a dramatic structural change, assembling on CENP-A chromatin a superstructure consisting of dozens of proteins and protein complexes to which microtubules attach (reviewed in Maiato et al, 2004; Carroll and Straight, 2006; Cheeseman and Desai, 2008). In higher eukaryotes, centromeres typically consist of repeated DNA arrays—for example α-satellite (alphoid DNA) in primates (Musich et al, 1980; Masumoto et al, 1989; Mitchell, 1996). However, centromere function is defined epigenetically and is independent of the underlying DNA sequence. This is best illustrated by stable dicentric chromosomes in which only one of two α-satellite regions on a single chromosome supports kinetochore assembly (Earnshaw and Migeon, 1985; Earnshaw et al, 1989; Sullivan and Schwartz, 1995; Sugata et al, 2000) and by rare neocentromeres that assemble kinetochores at loci lacking alphoid DNA (Warburton et al, 1997; Saffery et al, 1999; Alonso et al, 2007). The key to this epigenetic determination of kinetochore assembly is thought to lie in the pattern of histones and their modifications present at centromeric loci.
Examination of extended kinetochore chromatin fibers reveals that CENP-A nucleosomes occupy discrete domains interspersed with chromatin containing canonical histone H3 (Zinkowski et al, 1991; Blower et al, 2002; Sullivan and Karpen, 2004). The H3-rich chromatin in extended fibers from both human and Drosophila centromeres contains histone H3 dimethylated on lysine 4 (H3K4me2) (Sullivan and Karpen, 2004). Other marks characteristic of open euchromatin, including H3K4me3 and acetylated forms of H3 and H4 were not detected, nor was H3K9me3, a modification generally regarded as a mark of inactive chromatin. More recently, a study using superresolution microscopy found H3K9me3 within the CENP-A domain in chicken chromosomes (Ribeiro et al, 2010). This specialized chromatin landscape was referred to as ‘centrochromatin' (Sullivan and Karpen, 2004). Our studies aim to determine whether and/or how this specific chromatin landscape influences the structure, function and identity of human centromeres.
We recently described a synthetic human artificial chromosome (HAC) designed to specifically probe and manipulate centromere and kinetochore structure in vivo (Nakano et al, 2008). The HAC centromere is assembled on a synthetic higher-order alphoidtetO array that contains tet operator (tetO) sequences and CENP-B boxes in alternating alphoid monomers (Figure 1A). This allows sequence-specific discrimination of this centromere from other endogenous centromeres, as well as its specific targeting in vivo by tet repressor (tetR) fusions. The alphoidtetO centromere is fully functional, and tethering of tetR-EYFP on its own does not interfere with the local chromatin state or HAC kinetochore function. However, targeting of either a heterochromatin-nucleating transcriptional repressor or a transcriptional activator, interferes with HAC centromere structure and function (Nakano et al, 2008; Cardinale et al, 2009).
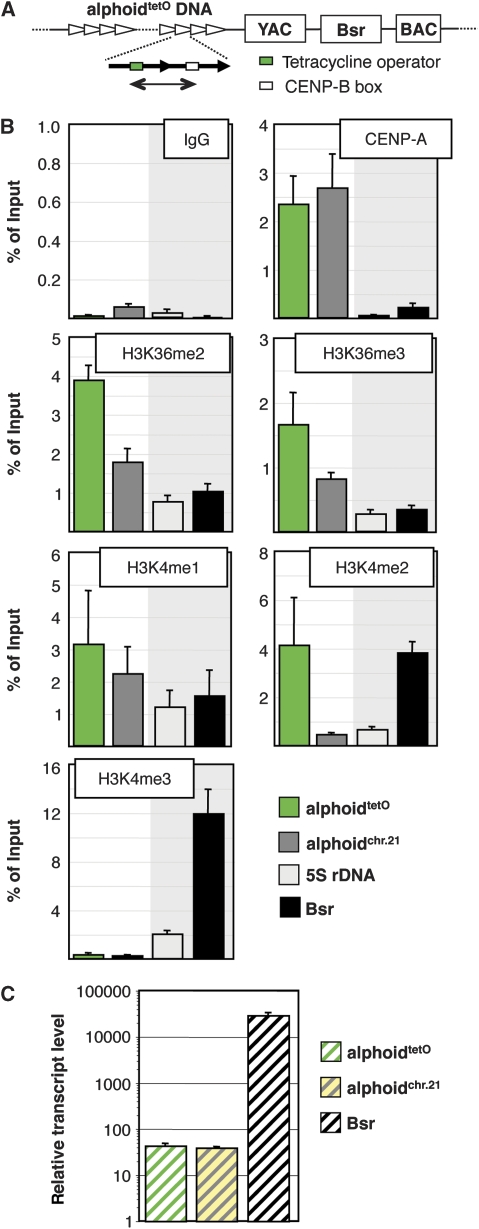
Figure 1.
Active centromere chromatin displays the signature of elongating RNA polymerase. (A) Schematic of the HAC, derived from Nakano et al (2008), indicating the synthetic alphoidtetO array (green: tetO; white: CENP-B box) and the HAC vector with YAC and BAC cassettes and the blasticidin (Bsr) resistance marker. The region of the alphoidtetO array analysed by ChIP is indicated by green line. (B) ChIP analysis in AB2.2.18.21 cells using antibodies of the indicated reactivity. The synthetic (alphoidtetO) centromere, endogenous chromosome 21 α21-I satellite DNA (alphoidchr.21), the 5S rDNA loci and the Bsr gene on the HAC vector were assessed. Data represent the mean and s.d. of three independent ChIP experiments. Note the different scaling of individual panels reflecting different efficiencies of individual antibodies. (C) Real-time RT–PCR analysis of synthetic HAC centromere (alphoidtetO), actively transcribed Bsr marker and endogenous chromosome 21 satellite (alphoidchr.21). Expression data are normalized to the copy number of the genomic regions and β-actin mRNA levels (see Materials and methods) and displayed as arbitrary numbers. Data represent the mean and s.e.m. of three independent experiments. Note the log scale.
In the present study, we show that centrochromatin at the HAC centromere comprises CENP-A in a chromatin environment resembling that in the downstream body of transcribed housekeeping genes. We then directly manipulate this ‘epigenetic' chromatin landscape by targeted removal of H3K4me2 specifically from the alphoidtetO HAC centromere, leaving all other centromeres untouched. Our results reveal that H3K4me2 is required for kinetochore stability, possibly by influencing the levels of local non-coding transcription and targeting of the CENP-A chaperone HJURP (Dunleavy et al, 2009; Foltz et al, 2009).
Results
Centromere chromatin displays the signature of RNA polymerase elongation activity
In order to characterize the chromatin environment of a single active centromere, we performed chromatin immunoprecipitation (ChIP) experiments followed by real-time PCR analysis of the alphoidtetO HAC (Nakano et al, 2008) using a panel of well-characterized monoclonal antibodies against histone modifications (Kimura et al, 2008). As expected, the synthetic HAC centromere and the 11-mer alphoid repeat of native chromosome 21, but not the endogenous 5S rDNA locus, were enriched for the kinetochore histone CENP-A (Figure 1B).
The HAC centromere also displayed a pattern of histone H3 modifications characteristic of the downstream open reading frames of actively transcribed genes and non-coding RNAs (Schneider et al, 2004; Vakoc et al, 2006; Barski et al, 2007; Guttman et al, 2009; Filion et al, 2010). The centromere was highly enriched for histone H3 mono-methylated on lysine 4 (H3K4me1), as well as H3K4me2, H3K36me2 and H3K36me3 (Figure 1B). The HAC centromere chromatin had low levels of histone H3 acetylated on K9 and K27 (Supplementary Figure S1A) and of H3K4me3, which is typically found near the transcription start site of active genes (Figure 1B). Similar levels of these histone modifications were also seen at the centromere of chromosome 21, with the exception of H3K4me2, which was present at lower levels (Figure 1B). H3K4me2 was previously found to be interspersed with CENP-A at active centromeres (Sullivan and Karpen, 2004), so the difference between the HAC and chromosome 21 data could conceivably reflect the presence of heterochromatic monomeric alphoid 21-II sequences flanking the kinetochore in the endogenous array or be due to other properties of the chromosome 21 α-satellite array (Ikeno et al, 1994; Masumoto et al, 1998).
AlphoidtetO HAC centromere chromatin also displayed significant levels of H3K9me3, a modification often attributed to pericentromeric heterochromatin (Supplementary Figure S1A). However, H3K9me3 has also been described in transcribed chromatin (Vakoc et al, 2005, 2006), so it may be simplistic to equate this modification solely with repressed chromatin. Levels of unmethylated H3K4, H3K36me1, H3K27me1, H3K27me2 and H3K27me3 were all higher on the endogenous chromosome 21 alphoid DNA, but were also detected at the synthetic alphoidtetO centromere at levels above background (Supplementary Figure S1A).
In keeping with the similarity of the chromatin profile of centromeres to the body of transcribed genes, real-time reverse transcription (RT)–PCR analysis revealed low levels of transcripts derived from the alphoidtetO and chromosome 21 centromeres (Figure 1C). Their detection was sensitive to low doses of actinomycin D (Supplementary Figure S1B). Strikingly, normalization of the transcript copy number to the copy number of the corresponding genomic locus revealed virtually identical transcript-to-template ratios for the alphoidtetO and chromosome 21 centromeres, consistent with similar levels of RNA polymerase activity at these loci. RNA corresponding to the alphoidtetO HAC and endogenous chromosome 21 was also detected by RNA immunoprecipitation analysis of formaldehyde-crosslinked cells using an antibody against histone H3K36me2, demonstrating a close connection of centromeric transcripts with this elongation-associated histone modification (Supplementary Figure S1C).
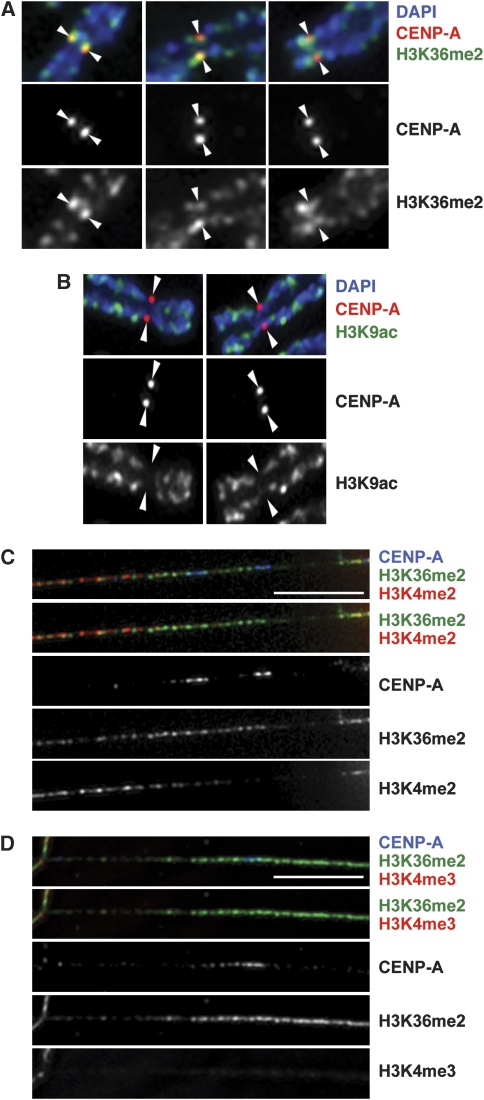
This is the first description of high levels of methylated H3K36 at human centromeres, so we independently assessed the centromeric localization of this modification on human chromosomes by immunofluorescence analysis. On mitotic chromosomes, staining for H3K36me2 overlapped the CENP-A staining (Figure 2A). In contrast, acetylation of H3K9 was not detected at centromeres of metaphase chromosomes (Figure 2B).
Figure 2.
H3K36me2 forms part of the CENP-A chromatin domain. (A–D) Immunofluorescence (IF) analysis of unfixed HT1080 chromosomes (A, B) and kinetochore fibres (C, D) using antibodies against CENP-A and the indicated histone modifications. Scale bars: 5 μm.
H3K36me2 nucleosomes were interspersed with and extended beyond the width of the centromeric CENP-A nucleosome domain on chromatin fibres stretched from endogenous centromeres (Figure 2C and D). The centromeric H3K36me2 domain was partially interspersed with H3K4me2 nucleosomes. However, in contrast to H3K36me2, H3K4me2 nucleosome density appeared to decrease asymmetrically towards the CENP-A domain (Figure 2C). H3K4me3 nucleosomes were virtually undetectable on these fibre domains (Figure 2D).
Together, these results establish that human centromeres are transcriptionally active chromatin compartments with a chromatin modification landscape resembling the body of actively transcribed genes. This landscape includes H3K36me2, a previously unrecognized component of CENP-A ‘centrochromatin'.
Centromeric H3K4me2 is not required for kinetochore function
To test the hypothesis that H3K4me2 has a direct role in maintaining centromere identity (Sullivan and Karpen, 2004), we generated an expression construct encoding tetR-EYFP fused to full-length Lysine-specific demethylase 1 (LSD1) (tetR-EYFP-LSD1; Figure 3A). LSD1 (also known as hKDM1) is a H3K4me2-specific histone demethylase (Shi et al, 2004). TetR-EYFP-LSD1 would be expected to remove H3K4me2 from the alphoidtetO HAC kinetochore, leaving chromatin at other kinetochores untouched.
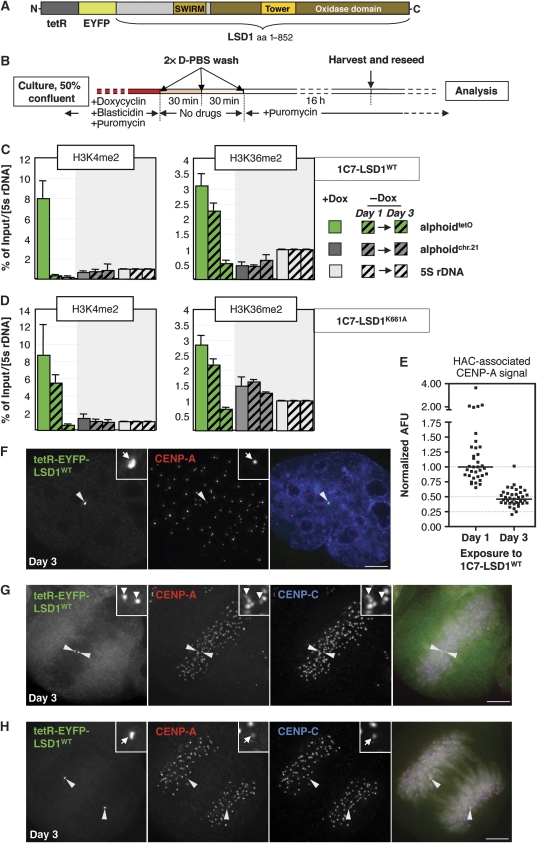
Figure 3.
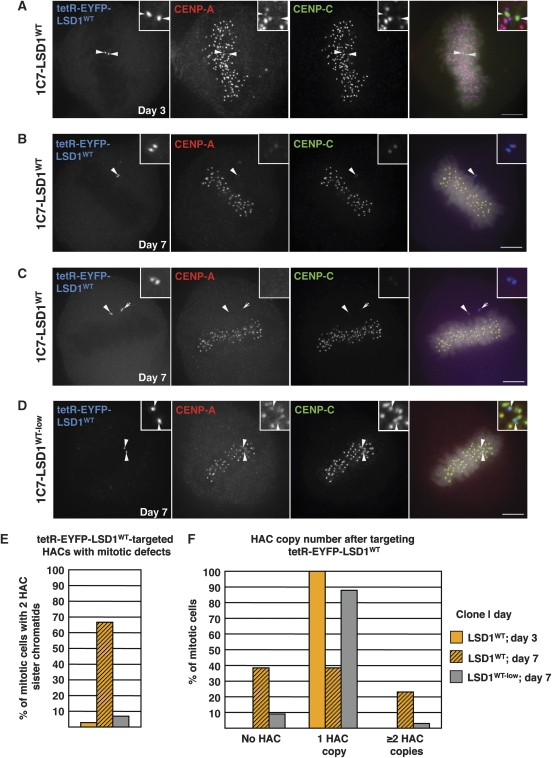
Centromeric H3K4me2 is not directly required for immediate kinetochore structure or function. (A) Schematic drawing of the tetR-EYFP-LSD1 fusion construct used in transient transfections and for the generation of stable 1C7-LSD1 cell lines. (B) Schematic diagram of the protocol used to wash out doxycycline to allow binding of the tetR fusion construct to the alphoidtetO array. (C, D) ChIP analysis of 1C7 cells stably expressing the wild-type LSD1 (1C7-LSD1WT, C) or the catalytically inactive LSD1K661A (D) tetR fusion constructs prior to, 1 and 3 days after washing out of doxycycline (Dox) to allow binding to the HAC centromere. Targeting of LSD1K661A reduces H3K4me2 levels at the HAC centromere only insignificantly (P=0.16, t-test) within 24 h. Both constructs cause a gradual decrease in centromeric H3K36me2 levels consistent with their ability to repress local transcription (see main text and Figure 6). Two to four independent ChIP experiments where carried out. Percentage of input values for each time point were normalized to the 5S rDNA locus. Error bars represent s.e.m. (E) Quantification of HAC-associated CENP-A staining in 1C7-LSD1WT interphase cells at the indicated time points after washing out of doxycycline. A full time course of this experiment is reproduced in Supplementary Figure S3. Values were normalized to the median value of ‘day 1'. Solid bars indicate the median. (F–H) IF analysis of 1C7-LSD1WT cells 3 days after washing out Dox. Interphase CENP-A staining (F) associated with the targeted HAC centromere (arrowheads) remains compacted. At this time point, the HAC sister kinetochores remain structurally and functionally intact as judged by staining for CENP-A and -C in metaphase (G) and anaphase (H). Scale bars: 5 μm.
We expressed this construct in HeLa 1C7 cells that maintain a single copy of the HAC, display conservation of the HAC centromere-associated chromatin signature (including the interspersed pattern of H3K4me2 and H3K36me2 nucleosomes on kinetochore fibres) and show a similar HAC mitotic stability to the original HT1080 cells (Cardinale et al (2009) and data not shown). TetR-EYFP-LSD1 caused a loss of detectable H3K4me2 staining from targeted interphase and mitotic HAC centromeres at 2 days after transfection (Supplementary Figure S2A and B). The global pattern and intensity of H3K4me2 staining appeared essentially unaffected in these cells, indicating specificity of the chimeric construct for the site of tetR:tetO tethering on the HAC. In controls, distinct H3K4me2 staining overlapped by CENP-A was observed at both interphase and mitotic HAC centromeres targeted with tetR-EYFP (Supplementary Figure S2A and B). Thus, tetR-EYFP-LSD1 is catalytically active and can be used to specifically deplete H3K4me2 from the HAC centromere in vivo.
We next generated 1C7 cells stably expressing tetR-EYFP-LSD1 (1C7-LSD1WT). Clones were selected and grown in the presence of doxycycline, which prevents binding of tetR to the HAC. The protocol for washing out the drug and inducing targeting to the HAC is shown in Figure 3B. Cellular levels of the fusion construct remain unchanged before and after washout, thereby ensuring that effects measured at the HAC are the specific result of direct targeting by the tethered tetR-EYFP-LSD1.
ChIP analysis of 1C7-LSD1WT cells showed that within 24 h of doxycycline washout, H3K4me2 levels fell dramatically at the HAC centromere, becoming essentially undetectable in the presence of the fusion construct at 3 days after washout (Figure 3C). Importantly, this removal of centromeric H3K4me2 did not result in a significant increase in centromeric heterochromatin marked by H3K9me3 or hypermethylated H3K27 (Supplementary Figure S3A).
The distribution of CENP-A at the HAC kinetochore appeared normal at both interphase and mitotic HACs at day 3 after doxycycline washout in 1C7-LSD1WT cells (Figure 3F–H), although the levels of CENP-A appeared to have decreased by ∼50% (Figure 3E). CENP-C staining was still present on the HAC although its levels also fell in parallel with those of CENP-A (Figure 3G and H and data not shown). Importantly, HAC kinetochores with tethered LSD1WT remained functional at this time point, with HAC sister chromatids aligning under tension on the metaphase plate (Figure 3G) and segregating normally in anaphase (Figure 3H). Together, these data demonstrate that kinetochores remain functional despite a near-complete lack of H3K4me2 and a >50% decrease in the levels of associated CENP-A and CENP-C. Parenthetically, this suggests that centromeres contain more CENP-A molecules than are directly required for the formation of a functional kinetochore.
Centromere tethering of LSD1 interferes with the long-term maintenance of kinetochore structure
When tetR-EYFP-LSD1WT was expressed in 1C7 cells following transient transfection, we observed a statistically significant decrease of HAC-associated CENP-C immunofluorescence 4 days after transfection compared with controls targeted with tetR-EYFP in cells expressing comparable levels of the fusion constructs (Supplementary Figure S4A and B). Indeed, tethering of tetR-EYFP does not interfere with HAC centromere structure or kinetochore function either after transient expression or during stable targeting into the HAC kinetochore for up to 30 days (Nakano et al, 2008; Cardinale et al, 2009; JHB and WCE, unpublished data).
LSD1 exerts a double-barrelled effect on gene activity. In the short term, it rapidly demethylates H3K4, thereby interfering with transcriptional memory (Li et al, 2007a; Muramoto et al, 2010). In addition, LSD1 is part of the histone-deacetylase-containing BHC complex involved in repression of neuronal promoters (Humphrey et al, 2001; Hakimi et al, 2002; Shi et al, 2003; Lee et al, 2005). As the catalytically inactive point mutation LSD1K661A (Stavropoulos et al, 2006) retains full association with the BHC complex (Lee et al, 2005), we used this mutant to distinguish between the effects of H3K4 demethylation and H3K4me-independent effects of tethering the LSD1 fusion construct.
In contrast to the rapid loss of H3K4me2 seen at HAC centromeres in cells stably expressing tetR-EYFP-LSD1WT, cells stably expressing the LSD1K661A mutant fusion displayed only a mild, statistically insignificant reduction of alphoidtetO-associated H3K4me2 within the initial 24 h (Figure 3D), despite expressing the construct at two-fold higher levels compared with 1C7-LSD1WT cells (Supplementary Figure S5). Consistently, immunofluorescence analysis failed to show the strong reduction of H3K4me2 staining at interphase or mitotic HACs observed with the LSD1WT fusion (Supplementary Figure S2A and B). However, LSD1K661A tethering eventually resulted in reduced centromeric H3K4me2 after 3 days (Figure 3D). This is consistent with a loss of H3K4 methylation secondary to repression of transcription (see below).
Although HACs targeted with tetR-EYFP-LSD1K661A showed no significant difference in their CENP-C staining compared with tetR-EYFP after transient transfection (Supplementary Figure S4A and B), in longer-term experiments with the 1C7-LSDK661A stable cell line, tethering of the LSD1 mutant did cause an eventual loss of kinetochore structure. Reproducibly, CENP-A loss from the HAC kinetochore was slower and less complete with tethered LSD1K661A than it was with tethered LSD1WT (Figure 4A and B). By day 5 after doxycycline washout, most HACs in 1C7-LSD1WT cells had lost ⩾80% of their CENP-A staining relative to day 1 (for complete time course see Supplementary Figure S3B). In contrast, the median staining in 1C7-LSDK661A cells remained about 50% of the initial value, and even at 7 days after targeting the fusion protein to kinetochores, most HACs retained >25% of their CENP-A (Figure 4B). Apparently, active demethylation of centromeric H3K4me2 significantly augments the loss of CENP-A induced by tethered LSD1 constructs.
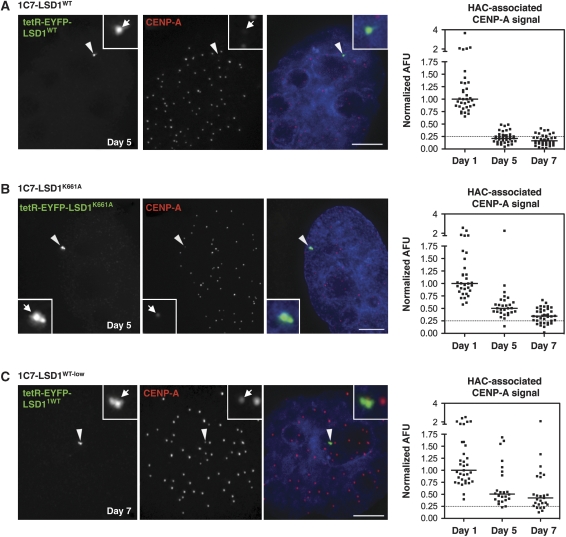
Figure 4.
LSD1 tethering interferes with the maintenance of CENP-A levels at the HAC. (A–C) CENP-A IF analysis of interphase 1C7 cells expressing (A) tetR-EYFP-LSD1WT, (B) a catalytically inactive mutant fusion, LSD1K661A or (C) a seven-fold lower level of tetR-EYFP-LSD1WT 5 days after targeting the respective construct to the HAC centromere. Displayed cells represent the median arbitrary fluorescence unit (AFU) of the quantifications shown to the right. Arrowheads depict the HAC. For (C) a ‘day 7' median cell is shown. Scale bars: 5 μm. (Right panels) CENP-A IF signal quantification at HACs targeted by the indicated fusion constructs at the given time points after washing out of Dox. AFU values were normalized to the median value of the ‘day 1' time point. The dotted line indicates the 25% signal mark for orientation. Solid vertical lines represent the median. For 1C7-LSD1WT cells, the corresponding full time course (days 1, 3, 5, 7) quantifications are shown in Supplementary Figure S3B, and day 1 and day 3 values are reproduced in Figure 3E.
We isolated an additional 1C7 clone (1C7-LSD1WT-low) expressing seven-fold lower levels of tetR-EYFP-LSD1WT than 1C7-LSD1WT cells. 1C7-LSD1WT-low cells display a less pronounced loss of CENP-A from the HAC centromere than that seen in the original 1C7-LSD1WT cells (Figure 4C). In 1C7-LSD1WT-low cells, most HACs retained CENP-A signals well above 25% of the values measured 1 day after doxycycline washout.
For this and all other 1C7 clones, expression levels detected by flow cytometry (Supplementary Figure S5A) correlated well with the amount of fusion construct bound to interphase HAC centromeres as determined by direct fluorescence signal quantification of microscopic images (Supplementary Figure S5B and data not shown). Thus, these levels of expressed proteins do not saturate the binding sites on the HAC.
Removal of H3K4me2 results in loss of kinetochore maintenance
Although kinetochores remained functional at 3 days after LSD1 targeting and H3K4me2 depletion, by 7 days HAC sister chromatids in 1C7-LSD1WT cells frequently failed to align on the metaphase plate and underwent missegregation in anaphase resulting in aberrant HAC copy numbers (Figure 5A–C and F). As HACs showed no mitotic defects after 3 days in the presence of the tetR-EYFP-LSD1WT fusion construct (Figures 3G and H and 5A and E), this suggested that gradual loss of CENP-A from the HAC centromere ultimately resulted in inactivation of the kinetochore. In contrast, few mitotic abnormalities were observed in 1C7-LSD1WT-low cells at the later time point, with the majority of HACs aligning on the metaphase plate (Figure 5D), and only a few cells showing gain or loss of HAC copies (Figure 5F). This dose response is consistent with the centromere inactivation arising as a result of the direct activity of LSD1 on the HAC centromere. Subsequent experiments investigated the mechanism by which LSD1 interfered with kinetochore maintenance.
Figure 5.
LSD1 activity at the HAC centromere correlates with loss of kinetochore function. (A–C) IF analysis of metaphase 1C7-LSD1WT cells expressing the fusion construct at high levels at 3 (A) and 7 days (B, C) after Dox washout. Cells were stained for CENP-A (red) and -C (green). Arrowheads indicate the HAC, also shown in the insets. Arrowheads in (A) indicate HAC sister kinetochores under tension. Arrowheads in (B, C) point to unaligned HACs. Arrow in (C) indicates extra HAC. Scale bars: 5 μm. (D) IF analysis as in (A) shows a metaphase 1C7-LSD1WT-low cell 7 days after Dox washout. (E) Analysis of the indicated 1C7-LSD1 clonal lines at the given time points. Late prometa-, meta- and anaphase cells with two detectable HAC sister chromatids (n=36, n=15 and n=29 for clones LSD1WT/day 3, LSD1WT/day 7 and LSD1WT-low/day 7, respectively) were scored. HAC-specific mitotic defects where considered as apparently unaligned HACs in late prometa- and metaphases (see e.g. B, C), as well as mis-segregating HAC sister chromatids in anaphase. (F) Quantification of HAC copy numbers as determined by the EYFP spot pairs in 1C7-LSD1WT clonal cells (n=36, n=39 and n=33 for clones LSD1WT/day 3, LSD1WT/day 7 and LSD1WY-low/day 7, respectively).
Demethylation of centromeric H3K4me2 abrogates local transcription
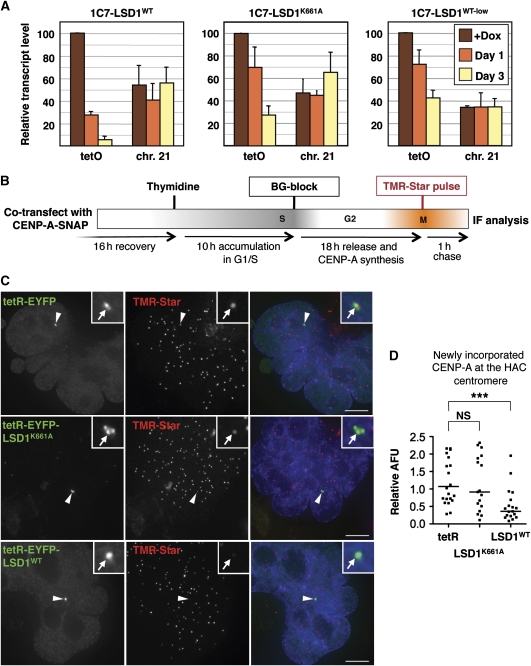
Real-time RT–PCR analysis in 1C7-LSD1WT as well as 1C7-LSD1K661A cells revealed that the tetR-EYFP-LSD1WT fusion construct is an efficient repressor of transcription, reducing alphoidtetO transcript copy numbers by >70% within the first 24 h after doxycycline washout (Figure 6A). By day 3, transcripts derived from the HAC centromere were barely detectable. Indeed, the low levels of RNA polymerase II reproducibly detected at the alphoidtetO array became undetectable 24 h after doxycycline washout (Supplementary Figure S6).
Figure 6.
Tethering of LSD1 reduces centromeric transcription and impairs the incorporation of newly synthesized CENP-A into the HAC centromere. (A) Real-time RT–PCR analysis of HAC (tetO) and chromosome 21 (chr.21) centromere transcripts in 1C7 cells stably expressing the indicated fusion constructs (WT versus K661A) and the LSD1WT fusion at different levels (high versus low) before, 1 and 3 days after washing out Dox. Differences in the repressive activity of wild-type (high) and mutant LSD1 fusions is statistically significant at both time points (P=0.015 and P<0.001 for day 1 and day 3, respectively; t-test). Data represent the means and s.e.m. of at least three independent experiments. (B) Schematic workflow to determine incorporation of newly synthesized CENP-A into centromeres. (C) 1C7 cells co-transfected with tetR-EYFP, tetR-EYFP-LSD1WT or tetR-EYFP-LSD1K661A and a construct expressing SNAP-tagged CENP-A were analysed by fluorescence microscopy after labelling newly incorporated CENP-A molecules. (D) Quantification of TMR-Star fluorescence signal levels associated with the HAC normalized to the average signal at endogenous centromeres. Asterisks indicate a significant difference, with P<0.001 (t-test).
The LSD1K661A catalytically inactive mutant was significantly less efficient at repressing HAC centromere transcription (Figure 6A). During the initial 24 h, transcript levels decreased by only about 30%. Furthermore, 1C7-LSD1K661A cells retained moderate levels (∼30%) of transcriptional activity at the HAC centromere even after 3 days of targeting. These data show that H3K4-demethylating activity facilitates strong transcriptional repression at the HAC centromere. Importantly, repression in 1C7-LSD1K661A cells was paralleled by a gradual loss of centromeric H3K4me2 and H3K36me2 (Figure 3D), consistent with co-transcriptional methylation of H3K4 and H3K36 (Krogan et al, 2003; Ng et al, 2003; Keogh et al, 2005). These data support the hypothesis that transcription through the centromere may contribute to the maintenance of the associated chromatin landscape.
LSD1 activity interferes with incorporation of newly synthesized CENP-A at the HAC kinetochore
The results described above are consistent with LSD1 promoting loss of CENP-A from the HAC kinetochore either by destabilizing the pre-existing structure or by interfering with the incorporation of newly synthesized CENP-A molecules. To distinguish between these hypotheses, we performed SNAP-tag quench-pulse-chase experiments (Jansen et al, 2007) to assess the incorporation of newly synthesized CENP-A at the HAC kinetochore. These experiments were performed by co-transfecting 1C7 cells with a plasmid expressing a CENP-A-SNAP-3xHA fusion plus constructs expressing either tetR-EYFP or tetR-EYFP-LSD1WT. Following quenching of existing SNAP-tagged CENP-A with non-fluorescent BG substrate and subsequent labelling of newly synthesized CENP-A molecules using the fluorescent TMR-Star substrate (Figure 6B; Supplementary data), we analysed incorporation of TMR-Star-labelled SNAP-tagged CENP-A in the subsequent G1 phase by fluorescence microscopy (Figure 6C).
Tethering LSD1 to the HAC centromere resulted in a statistically significant reduction of HAC-associated TMR-Star signal compared with tethering tetR-EYFP alone (Figure 6D). In controls, tethering of the LSD1K661A fusion construct had no statistically significant effect on new CENP-A assembly (Figure 6C and D). Importantly, at this time point 2 days after targeting, HACs tethered with the wild-type LSD1 construct, but not the mutant LSD1K661A, display loss of H3K4me2 (Supplementary Figure S2). These data are therefore consistent with the hypothesis that H3K4me2 demethylation interferes with efficient incorporation of newly synthesized CENP-A into the HAC kinetochore.
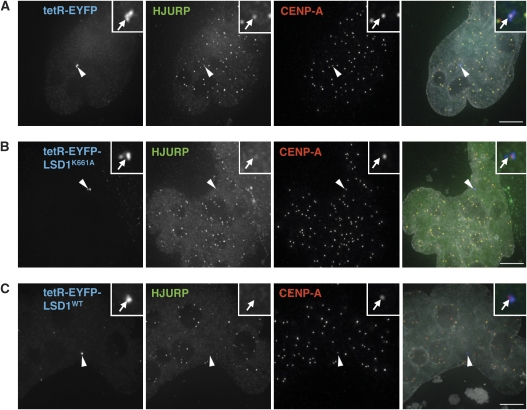
In order to determine the mechanism by which LSD1 interferes with CENP-A incorporation, we examined the recruitment of HJURP, the recently identified histone chaperone that specifically mediates deposition of CENP-A at centromeres (Dunleavy et al, 2009; Foltz et al, 2009). We co-transfected the individual tetR fusion constructs along with a construct expressing HJURP-mRFP and imaged cells in which recruitment of HJURP was evident at endogenous centromeres 2 days after transfection. While HJURP was readily detected at the centromere of HACs targeted by tetR-EYFP, HAC kinetochores targeted with tetR-EYFP-LSD1WT reproducibly displayed reduced or undetectable recruitment of HJURP (Figure 7A and B). In contrast, HJURP generally remained detectable at HAC kinetochores targeted with tetR-EYFP-LSD1K661A (Figure 7C). These findings thus suggest that the chromatin state and/or transcription of centromeres regulates targeting of the CENP-A deposition machinery and therefore, the efficiency of CENP-A incorporation.
Figure 7.
LSD1 perturbs HJURP localization to the HAC centromere. 1C7 cells were co-transfected with plasmids expressing (A) tetR-EYFP, (B) tetR-EYFP-LSD1WT or (C) tetR-EYFP-LSD1K661A plus a C-terminally LAP-tagged (mRFP) HJURP fusion. Cells displaying centromeric HJURP localization (corresponding to G1 phase) were imaged 2 days after transfection. Arrowheads indicate the HAC. Scale bars: 5 μm.
Discussion
Kinetochore assembly and activity are determined by epigenetic factors (Earnshaw and Migeon, 1985; Steiner and Clarke, 1994; Karpen and Allshire, 1997), commonly believed to be combinations of specialized histones and histone modifications. Here, we report the targeted engineering of the chromatin environment within a single active kinetochore. This has enabled us to directly test the idea that histone H3 dimethylated on lysine 4 (H3K4me2) within kinetochore chromatin might be required for kinetochore assembly and activity (Sullivan and Karpen, 2004).
Our analysis of the centromere chromatin of the alphoidtetO HAC in human cells reveals, in addition to the kinetochore histone CENP-A, a pattern of histone modifications diagnostic of that found in the body of many actively transcribed genes (Schneider et al, 2004; Vakoc et al, 2006; Barski et al, 2007; Mikkelsen et al, 2007). Notable features include histone hypoacetylation, mono- and di-methylation of H3K4 and hypermethylation of H3K36.
Genomic chromatin maps, most notably of H3K36 methylation, have recently facilitated the prediction and discovery of numerous long non-coding transcripts (Guttman et al, 2009; Khalil et al, 2009). Thus, our discovery of H3K36me2 and H3K36me3 at the HAC and endogenous centromeres is consistent with our observed transcription of centromeric type I α-satellite repeats. Interestingly, the presence of H3K36 methylation places centromere chromatin in the ‘YELLOW' chromatin class identified in a recent study of Drosophila chromatin (Filion et al, 2010). This class contains genes with a broad expression pattern characteristic of universal cellular functions. Low levels of minor satellite transcripts were previously detected in mouse cells (Kanellopoulou et al, 2005; Martens et al, 2005; Bouzinba-Segard et al, 2006; Efroni et al, 2008) and suggested to be processed by the RNAi machinery (Kanellopoulou et al, 2005). However, evidence for a functional role of these processed transcripts at centromeres is sparse. It will clearly be important, though challenging, in the future to characterize these centromeric transcripts in greater detail.
Our data show that repression of transcription at the HAC centromere following tethering of either wild-type or mutant LSD1 is paralleled by a gradual decrease in the levels of the elongation-associated H3K36me2 mark. Recent studies in yeast highlight an important role for H3K36 methylation in the maintenance of chromatin architecture: co-transcriptional methylation of H3K36 is linked to the recruitment of a HDAC-containing complex that maintains a hypoacetylated state, antagonizes H3K4 trimethylation and suppresses spurious intragenic transcription (Carrozza et al, 2005; Keogh et al, 2005; Li et al, 2007b). A recent report indicates that this process is most important at long and infrequently transcribed genes (Li et al, 2007c), suggesting a particular relevance for centromeres. In the light of the hypoacetylated state and depletion of the H3K4me3 mark from centromeric nucleosomes, local H3K36 methylation may likely be integral to a similar pathway acting to maintain local chromatin architecture. Our data suggest that perhaps in cooperation with interspersed H3K4me2 nucleosomes, methylated H3K36 may form a chromatin environment that directly or indirectly facilitates interaction with the CENP-A deposition machinery. It will be interesting to investigate the consequences of specific removal of H3K36 methylation on centromeric structure in the future.
The landmark discovery of H3K4me2 within the CENP-A chromatin domain (Sullivan and Karpen, 2004) inspired several models that attributed to H3K4me2 a potential key role in maintaining kinetochore function (Allshire and Karpen, 2008). Our present study establishes that centromeric H3K4me2 is indeed required for long-term kinetochore maintenance. We show that loading of de novo synthesized CENP-A molecules is impaired when levels of this mark are strongly reduced. Defective loading of newly synthesized CENP-A is paralleled by (and may result from) a reduced recruitment of the CENP-A-specific histone chaperone HJURP. Perhaps surprisingly, kinetochores depleted of H3K4me2 can still function over several divisions despite the fact that they progressively lose CENP-A. Indeed, we failed to detect defects in HAC segregation in cells where the HAC kinetochore contained only 40–50% of its normal CENP-A complement. Thus, human centromeres apparently contain more CENP-A than absolutely required to assemble a kinetochore. Importantly, as CENP-A levels continue to fall below a threshold value, the kinetochores eventually fail to function and missegregation of the HAC predominates.
Our data further indicate that levels of centromeric transcription rapidly fall in the absence of H3K4me2. This is consistent with a suggested role of H3K4 methylation in transcriptional memory (Muramoto et al, 2010), which in the case of H3K4me2 may in part depend on the direct binding of factors positively mediating transcription elongation (Sims et al, 2005; Kim and Buratowski, 2009). In this respect, it is interesting to note that knockdown of the transcription elongation-associated chromatin remodelling factor CHD1, which can directly bind to H3K4me2 (Sims et al, 2005), was shown to cause a reduction of centromeric CENP-A levels (Okada et al, 2009). Whether the open chromatin conducive to transcription arises from a planar boustrophedon fold as recently proposed for the chicken kinetochore (Ribeiro et al, 2010) rather than a compact solenoidal structure remains for future experiments to determine.
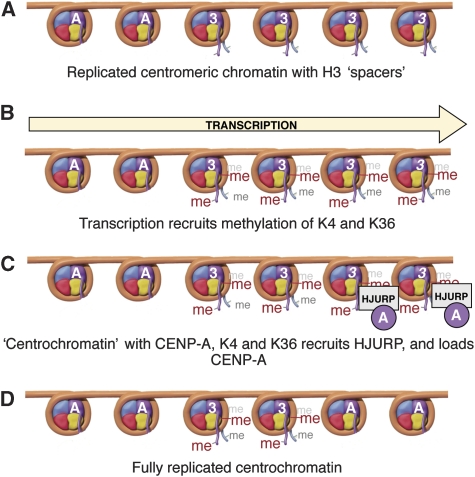
Together, our experiments are consistent with two possible mechanisms for the relationship between H3K4me2 and CENP-A assembly at kinetochores. In the first, components of the RNA polymerase elongation complex could cooperate with HJURP in direct transcription-linked deposition of CENP-A nucleosomes, perhaps in a similar manner discussed for the deposition of other histone variants including H3.3 (Henikoff et al, 2004). Alternatively, RNA polymerase activity may indirectly maintain a chromatin state that promotes the binding of HJURP, and therefore the subsequent deposition of newly synthesized CENP-A (depicted in Figure 8).
Figure 8.
One model explaining the role of transcription and centromere chromatin in kinetochore maintenance. (A) Following DNA replication, histone H3 (italic ‘3') is inserted in place of CENP-A (‘A'). (B) Transcription through the alphoid array results in methylation of H3 on lysines (4 and 36). (C) During mitotic exit, the ‘centrochromatin' environment, with CENP-A, H3K4me2 and H3K36me2 recruits HJURP, which inserts CENP-A in place of some of the H3 molecules, maintaining the identity of the centromeric chromatin in the course of ongoing cell divisions (D). We have shown that removal of H3K4me2 both lowers centromeric transcription and inhibits the targeting of HJURP to centromere chromatin. Nucleosome drawings by Graham T Johnson.
Our findings describing the first directed manipulation of a specific histone modification within a defined centromere strongly support the hypothesis that H3K4me2 is an essential part of the chromatin environment of vertebrate kinetochores required for long-term kinetochore maintenance and function.
Materials and methods
Expression constructs, generation of stable cell lines as well as detailed descriptions of CENP-A-SNAP quench-pulse-chase experiments and ChIP analysis are provided in Supplementary data.
Immunostaining, cytological analysis and fluorescence signal quantification
Preparation and staining of unfixed mitotic chromosomes was essentially performed as described in Keohane et al (1996). In brief, cells were blocked in 100 ng/ml colcemid (KaryoMax, Gibco) for 2 h, and mitotic cells collected by shake-off. Cells were subject to hypotonic treatment, cytospun on glass slides and incubated in KCM buffer (10 mM Tris pH 8.0; 120 mM KCl; 20 mM NaCl; 0.5 mM EDTA; 0.1% Triton X-100) for 10 min prior to labelling with antibodies in KCM buffer, fixation in 4% PFA/KCM and counter staining in DAPI. Cells were subsequently mounted in VectaShield (Vector Labs).
Indirect immunofluorescence staining of cells fixed in 4% PFA/PBS was performed using standard protocols. Antibodies used were mouse anti-CENP-A (AN1), rabbit anti-CENP-A (Valdivia et al, 1998), rabbit anti-CENP-C (R554) and mouse anti-H3K36me2 (2C3). Fluorophore-conjugated secondary antibodies were purchased from Jackson Labs.
Images were acquired on a DeltaVision Core system (Applied Precision) using an inverted Olympus IX-71 stand, with an Olympus UPlanSApo × 100 oil immersion objective (NA 1.4) and a 250W Xenon light source. Camera (Photometrics Cool Snap HQ), shutter and stage where controlled through SoftWorx (Applied Precision). Z-series were collected with a spacing of 0.2 μm, and image stacks were subsequently deconvolved in SoftWorx.
For stable 1C7-LSD1WT cells, a custom-written macro was used for signal quantification in ImagePro software (Media Cybernetics, Inc), details of which are available upon request. In brief, HAC-associated EYFP and Texas Red signals were determined in each HAC-containing section within a circular region of interest nine pixels in diameter. For each section, the average nuclear background for each channel was determined within three regions of interest of the same size and was subtracted from the specific signal. For display purposes only, images are represented as maximum intensity projections.
ChIP experiments
ChIP experiments were performed using a protocol adapted and modified from Kimura et al (2008) described in detail in Supplementary data. ChIP'ed and input DNA was subject to real-time PCR analysis using a SYBR Green Mastermix (Sigma) on a LightCycler480 system (Roche). For each primer pair, a standard curve was prepared from the input material and included on every plate to calculate the percent of precipitated DNA relative to the input material. Oligonucleotide primer pairs were described previously (Cardinale et al, 2009). Histone antibodies used for ChIP were described previously (Kimura et al, 2008). Other antibodies were mouse anti-CENP-A (AN1) and mouse anti-RNA polymerase II [8WG16] (Abcam). Normal mouse IgG was used as control.
Real-time RT–PCR analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. A measure of 2 μg RNA were subsequently used for RT with random hexamer primers, using the Roche Transcriptor High Fidelity cDNA Synthesis Kit. Real-time PCR analysis of cDNA equivalent to ∼70 ng (alphoidtetO, alphoidchr.21) or ∼0.7 ng (Bsr, β-actin) input RNA was then performed using a SYBR Green Mastermix (JumpStart, Sigma) on a LightCycler480 system (Roche). Oligonucleotide primer pairs were described previously (Nakano et al, 2008), except tetO_F/R2 (5′-CCACTCCCTATCAGTGATAGAGAA-3′ and 5′-GTTAAACTCAGTCGTCACCAAGAG-3′) for the alphoidtetO non-coding transcripts, and actin_F/R (5′-GCCGGGACCTGACTGACTAC-3′ and 5′-AGGCTGGAAGAGTGCCTCAG-3′) for β-actin transcripts. For each oligonucleotide primer pair and every plate, a standard curve was created from genomic DNA derived from the corresponding cell line, thereby calculating the transcript copy numbers relative to the genomic locus copy number. Background values (no reverse transcriptase) were subtracted, and all values were normalized to β-actin expression. For time course experiments, the transcript levels were expressed relative to the +Dox values of the alphoidtetO, which was arbitrarily set to 100.
Supplementary Material
Acknowledgments
We thank Graham Johnson for letting us use the nucleosome drawings shown in Figure 8, and Natalay Kouprina, Alison L Pidoux and Irina Stancheva for critical reading of the paper. JHB was funded by a PhD studentship from The Wellcome Trust and NMCM and MGR by studentships from the Fundação para a Ciência e a Tecnologia (FCT). This work was supported by the Fundação Calouste Gulbenkian, FCT grant BIA-PRO/100537/2008, the European Commission FP7 programme and an EMBO installation grant (LETJ); the intramural research program of the NIH, National Cancer Institute, Center for Cancer Research (VL); a grant-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (HM) and the Genome Network project and Grants-in-aid from the MEXT of Japan (HK). Work in the WCE laboratory is funded by The Wellcome Trust, of which he is a Principal Research Fellow.
Author contributions: JHB, LETJ and WCE designed the experiments; JHB, MGR and NMCM performed the experiments; HK, DAK, HM, VL and LETJ contributed new experimental and analytical tools; JHB and LETJ analysed the data; JHB and WCE wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allshire RC, Karpen GH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Fritz B, Hasson D, Abrusan G, Cheung F, Yoda K, Radlwimmer B, Ladurner AG, Warburton PE (2007) Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol 8: R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzinba-Segard H, Guais A, Francastel C (2006) Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA 103: 8709–8714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale S, Bergmann JH, Kelly D, Nakano M, Valdivia MM, Kimura H, Masumoto H, Larionov V, Earnshaw WC (2009) Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell 20: 4194–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Straight AF (2006) Centromere formation: from epigenetics to self-assembly. Trends Cell Biol 16: 70–78 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2008) Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9: 33–46 [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon B (1985) A family of centromere proteins is absent from the latent centromere of a stable isodicentric chromosome. Chromosoma (Berl) 92: 290–296 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H, Stetten G (1989) Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma (Berl) 98: 1–12 [DOI] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, Gingeras TR, Misteli T, Meshorer E (2008) Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2: 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR III, Bassett EA, Wood S, Black BE, Cleveland DW (2009) Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R (2002) A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA 99: 7420–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Furuyama T, Ahmad K (2004) Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet 20: 320–326 [DOI] [PubMed] [Google Scholar]
- Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH (2001) Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem 276: 6817–6824 [DOI] [PubMed] [Google Scholar]
- Ikeno M, Masumoto H, Okazaki T (1994) Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Hum Mol Genet 3: 1245–1257 [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC (1997) The case for epigenetic effects on centromere identity and function. Trends Genet 13: 489–496 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Keohane AM, O'Neill LP, Belyaev ND, Lavender JS, Turner BM (1996) X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol 180: 618–630 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Buratowski S (2009) Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N (2008) The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct 33: 61–73 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R (2005) An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437: 432–435 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007a) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL (2007b) Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL (2007c) Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Deluca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore-microtubule interface. J Cell Sci 117 (Part 23): 5461–5477 [DOI] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T (2005) The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24: 800–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Ikeno M, Nakano M, Okazaki T, Grimes B, Cooke H, Suzuki N (1998) Assay of centromere function using a human artificial chromosome. Chromosoma 107: 406–416 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109: 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AR (1996) The mammalian centromere: its molecular architecture. Mutat Res 372: 153–162 [DOI] [PubMed] [Google Scholar]
- Muramoto T, Muller I, Thomas G, Melvin A, Chubb JR (2010) Methylation of H3K4 is required for inheritance of active transcriptional states. Curr Biol 20: 397–406 [DOI] [PubMed] [Google Scholar]
- Musich PR, Brown FL, Maio JJ (1980) Highly repetitive component alpha and related alphoid DNAs in man and monkeys. Chromosoma 80: 331–348 [DOI] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H (2008) Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 14: 507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Okada M, Okawa K, Isobe T, Fukagawa T (2009) CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell 20: 3986–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC (2010) A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA 107: 10484–10489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffery R, Earle E, Irvine DV, Kalitsis P, Choo KH (1999) Conservation of centromere protein in vertebrates. Chromosome Res 7: 261–265 [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2004) Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 6: 73–77 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D (2005) Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem 280: 41789–41792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos P, Blobel G, Hoelz A (2006) Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol 13: 626–632 [DOI] [PubMed] [Google Scholar]
- Steiner N, Clarke L (1994) A novel epigenetic effect can alter centromere function in fission yeast. Cell 79: 865–874 [DOI] [PubMed] [Google Scholar]
- Sugata N, Li S, Earnshaw WC, Yen TJ, Yoda K, Masumoto H, Munekata E, Warburton PE, Todokoro K (2000) Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere—kinetochore complexes. Hum Mol Genet 9: 2919–2926 [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S (1995) Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet 4: 2189–2197 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA (2005) Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell 19: 381–391 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA (2006) Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 26: 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia MM, Figueroa J, Iglesias C, Ortiz M (1998) A novel centromere monospecific serum to a human autoepitope on the histone H3-like protein CENP-A. FEBS Lett 422: 5–9 [DOI] [PubMed] [Google Scholar]
- Warburton PE, Cooke C, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, Poirier GG, Earnshaw WC (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol 7: 901–904 [DOI] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR (1991) The centromere-kinetochore complex: a repeat subunit model. J Cell Biol 113: 1091–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.