Abstract
Compounds that inhibit signalling upstream of ERK (extracellular-signal-regulated kinase) are promising anticancer therapies, motivating research to define how this pathway promotes cancers. In the present study, we show that human capicúa represses mRNA expression for PEA3 (polyoma enhancer activator 3) Ets transcription factors ETV1, ETV4 and ETV5 (ETV is Ets translocation variant), and this repression is relieved by multisite controls of capicúa by ERK, p90RSK (p90 ribosomal S6 kinase) and 14-3-3 proteins. Specifically, 14-3-3 binds to p90RSK-phosphorylated Ser173 of capicúa thereby modulating DNA binding to its HMG (high-mobility group) box, whereas ERK phosphorylations prevent binding of a C-terminal NLS (nuclear localization sequence) to importin α4 (KPNA3). ETV1, ETV4 and ETV5 mRNA levels in melanoma cells are elevated by siRNA (small interfering RNA) knockdown of capicúa, and decreased by inhibiting ERK and/or expressing a form of capicúa that cannot bind to 14-3-3 proteins. Capicúa knockdown also enhances cell migration. The findings of the present study give further mechanistic insights into why ETV1 is highly expressed in certain cancers, indicate that loss of capicúa can desensitize cells to the effects of ERK pathway inhibitors, and highlight interconnections among growth factor signalling, spinocerebellar ataxias and cancers.
Keywords: cancer, capicúa, Ets translocation variant 1 (ETV1), 14-3-3 protein, spinocerebellar ataxia type 1 (SCA1)
Abbreviations: B2M, β2 microglobuluin; CRE, CIC-responsive element; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagle's medium; DUX4, Double homeobox 4; ECL, enhanced chemiluminescence; EGF, epidermal growth factor; EMSA, electrophoretic mobility-shift assay; ERK, extracellular-signal-regulated kinase; ETV, Ets translocation variant; EWS, Ewing sarcoma protein; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; GIST, gastrointestinal stromal tumour; HA, haemagglutinin; HEK, human embryonic kidney; HMG, high-mobility group; IGF1, insulin-like growth factor 1; KPNA3, importin α4/karyopherin α3; LC, liquid chromatography; MS/MS, tandem MS; NLS, nuclear localization sequence; p90RSK, p90 ribosomal S6 kinase; PEA3, polyoma enhancer activator 3; PDK1, phosphoinositide-dependent kinase 1; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; PKC, protein kinase C; RT, reverse transcription; SCA, spinocerebellar ataxia; siRNA, small interfering RNA
INTRODUCTION
Capicúa is a transcriptional repressor discovered in several developmental contexts in Drosophila [1]. Signalling via specific receptor tyrosine kinase/Ras/Raf/ERK (extracellular-signal-regulated kinase) pathways relieves repression by capicúa leading to the transcription of genes that specify differentiation in wing veins, imaginal eye discs, head and tail [1–6], hence the name capicúa meaning head-and-tail in Catalan. Lack of capicúa enables Drosophila cells to grow without Ras function, but does not compensate for growth defects due to mutations in insulin/PKB (protein kinase B, Akt) signalling [6].
Mammalian capicúa (also known as CIC) is highly expressed during development of the granule layers of the cerebellum [7], and has been linked circumstantially to two disorders of neural crest cell origin, namely SCA1 (spinocerebellar ataxia 1) and Ewing's family tumours [8–10].
SCA1 is a motor disorder caused by a polyglutamine expansion mutation of ataxin-1, with phosphorylation of Ser776 potentiating the disease [11,12]. Phosphorylated Ser776 of ataxin-1 binds to 14-3-3 proteins, which are dimeric proteins that regulate many cellular processes by docking on to specific phosphorylated serine and threonine residues on their targets [13,14]. 14-3-3 binding to phosphorylated Ser776 is proposed to regulate the interactions of ataxin-1 with two protein complexes, one including splicing factors and the other containing capicúa [10,15,16]. The polyglutamine mutation of ataxin-1 that underlies SCA1 may disturb the balance between the two complexes [10,15].
The suggestion of a connection between capicúa and cancer comes from two cases of Ewing-like sarcoma that were found to express transforming CIC–DUX4 (Double homeobox 4) fusion proteins, comprising most of capicúa and part of the double homeodomain protein DUX4 [8]. True Ewing-family tumours more commonly arise from fusions of the EWS (Ewing sarcoma protein) gene with genes encoding Ets transcription factors [17], which raised the question of whether the CIC–DUX4 fusion promotes the expression of Ets transcription factors. Indeed, the HMG (high-mobility group) box of the CIC binds to a DNA sequence within the promoters of genes encoding the PEA3 (polyoma enhancer activator 3) subfamily Ets transcription factors [ETV1, ETV4 and ETV5 (ETV is Ets translocation variant), also known as ER81, PEA3 and ERM respectively], whereas the attached DUX4 portion enhances the transcription of ETV1 and ETV5 [8]. One inference of these findings is that normal capicúa might repress expression of ETV1, ETV4 and/or ETV5.
The three PEA3 Ets transcriptional activators have roles in the development of many tissues and also in cancer progression. For example, ETV1 targets include genes needed to synthesize the neurotransmitter dopamine, as well as genes involved in cell migration and cancer metastases [18]. In addition to EWS, chromosomal rearrangements involving gene fusions and amplifications that lead to overexpression of ETV1, ETV4 and ETV5 have also been identified in breast and prostate cancers, with ETV1 in particular being linked to aggressive prostate tumours and pinpointed as a driver mutation in melanomas [17,19–23]. Enhanced growth factor/ERK signalling has also been linked to ETV1, ETV4 and ETV5 mRNA expression in developing tissues, cultured melanoma cells and gastrointestinal stromal tumours that are positive for the receptor tyrosine kinase KIT, but no underlying mechanisms have been described [24–27]. Phosphorylation of the ETV1 protein by the ERK-activated p90RSK (p90 ribosomal S6 kinase) and/or MSK1 (mitogen- and stress-activated kinase 1) have, however, been found to activate this transcription factor [28].
Overall, these findings raise questions about potential regulatory links between ERK signalling, capicúa and expression of the PEA3 Ets transcription factors in human cells. Specifically, does ERK signalling inhibit human capicúa by analogy with the Drosophila protein? Does capicúa repress ETV1, ETV4 and/or ETV5 expression? If so, the sum of these two negatives would be a positive effect of ERK signalling on transcription of ETV1, ETV4 and/or ETV5. We therefore began the present study by testing the effects of ERK-activating growth factors, ERK pathway inhibitors and capicúa knockdown on the levels of ETV1, ETV4 and ETV5 mRNAs in HEK (human embryonic kidney)-293 and melanoma cells.
MATERIALS AND METHODS
Materials
Synthetic peptides were from Dr Graham Bloomberg (School of Medical Sciences, University of Bristol, Bristol, U.K.). Molecular-mass markers were from Bio-Rad. MG132 was from Calbiochem. The ECL (enhanced chemiluminescence) kit was from GE Healthcare. PD184352, PLX4720 and BI-D1870 were from the DSTT (Division of Signal Tranduction Therapy, University of Dundee, Dundee, Scotland, U.K.). SDS/polyacrylamide precast gels and EGF (epidermal growth factor) were from Invitrogen and PMA was from Sigma.
The pan-14-3-3 antibody (K-19), anti-GFP (green fluorescent protein) (sc-8334) and anti-CIC (goat; sc-67528) antibodies were from Santa Cruz Biotechnology. Antibodies that recognize phosphorylated Thr202/Tyr204 on ERK1/2 (#9101) and anti-ERK1/2 (#9102) were from Cell Signaling Technology. The anti-KPNA3 (importin α4/karyopherin α3) antibody is A301-626A from Bethyl Laboratories, and recognizes an epitope between residues 1 and 50 of human KPNA3 (GenBank® accession number NP_002258.2), which is identical with KPNA4. The anti-CIC (rabbit; ab61860) antibody was from Abcam.
Purified recombinant protein kinases generated in the DSTT (Division of Signal Tranduction Therapy, University of Dundee, Scotland, U.K.) were His–PKCα (protein kinase Cα; residues 1–672 of the human protein) expressed in insect cells (constitutively active); His–PKCζ (protein kinase Cζ; residues 2–592 of the human protein) expressed in insect cells (constitutively active); His–RSK1 (p90RSK 1; residues 1–735 of the rat protein) expressed in insect cells and activated by phospho-ERK2 and PDK1 (phosphoinositide-dependent kinase 1); His–RSK2 (p90RSK 2; residues 2–740 of the rat protein) expressed in insect cells and activated by phospho-ERK2 and PDK1.
Plasmids
Full-length human CIC (Q96RK0) was amplified from IMAGE consortium EST (expressed sequence tag) 100014433 and cloned into a derivative of pcDNA5 FRT/TO (Invitrogen) that contains the coding sequence for GFP upstream of the multiple cloning site, to express N-terminally tagged GFP–CIC. C-terminal truncations of CIC were generated from this plasmid by inserting a stop codon at the appropriate position by mutagenesis. N-terminal truncations were made by re-amplifying the appropriate fragments of CIC and cloning into the same vector. Point mutations of CIC were introduced using KOD Hot Start DNA Polymerase (Novagen). GST-tagged CIC fragments were generated by re-amplifying the appropriate fragment from existing plasmids and cloning into the pGEX6P vector (GE Healthcare).
Full-length human KPNA3 (O00505) was amplified from human brain total RNA (Stratagene) and cloned into a derivative of pcDNA5 FRT/TO (Invitrogen) that contains the coding sequence for the HA (haemagglutinin) tag upstream of the multiple cloning site.
All plasmids were sequence-verified and plasmid transfections of human cells were performed using Lipofectamine™ 2000 (Invitrogen). DNA sequencing was performed by The Sequencing Service, College of Life Sciences, University of Dundee, Dundee, Scotland, U.K. (http://www.dnaseq.co.uk).
Cell stimulations, lysis and isolation of GFP-tagged proteins
Human HEK-293 cells cultured on 10-cm-diameter dishes in DMEM (Dulbecco's modified Eagle's medium) containing 10% (v/v) FBS (fetal bovine serum) were used untransfected, or 24–36 h after transfection with the plasmids indicated. Cells were serum-starved for a further 4–8 h (unstimulated), then stimulated with 100 ng/ml EGF for 15 min and 100 ng/ml PMA for 30 min. Where indicated, cells were incubated with PD184352 (2 μM for 1 h) and BI-D1870 (10 μM for 30 min) prior to stimulation with EGF and PMA. After stimulations, the medium was aspirated and cells lysed in 0.3 ml of ice-cold lysis buffer comprising 25 mM Tris/HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 50 mM NaF, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM benzamidine, 0.2 mM PMSF, 0.1% 2-mercaptoethanol, 1 μM microcystin-LR, 0.27 M sucrose and one mini Complete™ proteinase inhibitor cocktail tablet (Roche) per 10 ml of lysis buffer. Lysates were clarified by centrifugation (15000 g for 2 min), snap-frozen and stored at −80 °C. Protein concentrations were determined with Coomassie Protein Assay Reagent (Thermo Scientific).
To isolate GFP-tagged proteins, 5 μl of GFP–Trap®-agarose (http://www.chromotek.com) was mixed with 1–3 mg of lysate on an orbital shaker at 4 °C for 1 h, and washed by centrifugation (15000 g for 30 s) and resuspension, twice in 1 ml of TBS [Tri-buffered saline; 25 mM Tris/HCl (pH 7.5) and 150 mM NaCl], and twice in 1 ml of TBS containing Tween 20 (0.1% by vol.). Proteins were extracted into 20–40 μl of SDS sample buffer (Invitrogen) containing 1% 2-mercaptoethanol.
Western blots and 14-3-3 overlays
Membranes were incubated in 50 mM Tris/HCl (pH 7.5), 0.15 M NaCl and 0.2% Tween 20 containing 5% (w/v) dried skimmed milk powder (Marvel) and immunoblotted at 4 °C for 16 h [or room temperature (22 °C) for 1 h for anti-GFP and anti-HA antibodies] using the antibodies indicated at 1 μg/ml. Detection was performed using HRP (horseradish peroxidase)-conjugated secondary antibodies (Promega) and ECL® (Amersham Biosciences) for Western blots of endogenous proteins, except phospho- and total ERK1/2 for which secondary antibodies were labelled with IRDye and detected by Li-Cor IR imaging. 14-3-3 Far-Western assays are similar to Western blots, but use DIG (digoxygenin)-labelled 14-3-3 in place of primary antibody to test the ability of phosphorylated proteins on the membranes to bind directly to 14-3-3 [13].
In vitro kinase assay
Phosphorylation reactions were performed at 30 °C in a total volume of 20 μl. The protein kinase reaction buffer contained 50 mM Tris/HCl (pH 8.0), 1 mM EGTA, 10 mM EDTA, 100 mM MgAc and 1 mM ATP with or without 50 m-units/μl of protein kinases (where 1 unit is 1 nmol of phosphate incorporated per min at 30 °C into standard substrate peptides). Reactions were stopped with SDS sample buffer (Invitrogen).
Identification of proteins and phosphorylated residues by mass spectrometric analyses
Protein identification by tryptic mass fingerprinting analysis of selected protein gel bands was performed by LC-MS/MS (liquid chromatography tandem MS) using a linear ion trap-orbitrap hybrid mass spectrometer (LTQ-Orbitrap, Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Thermo) and coupled to a Dionex Ultimate 3000 nano-HPLC system. Peptides were typically injected into a PepMap 100 reverse-phase C18 3 μm column (Dionex) with a flow of 300 nl/min and eluted with a 40 min linear gradient of 95% solvent A (2% acetonitrile and 0.1% formic acid in water) to 50% solvent B (90% acetonitrile and 0.08% formic acid in water). The instrument was operated with the ‘lock mass’ option to improve the mass accuracy of precursor ions and data were acquired in the data-dependent mode, automatically switching between MS and MS/MS acquisition. Full scan spectra (m/z 300–1800) were acquired in the orbitrap with resolution R=60000 at m/z 400 (after accumulation to a target value of 500000). The five most intense ions, above a specified minimum signal threshold of 50000, based upon a low-resolution (R=15000) preview of the survey scan, were fragmented by collision-induced dissociation and recorded in the linear ion trap (target value of 10000).
Phosphopeptides were identified by LC-MS and MS/MS on an Applied Biosystems 4000 QTrap coupled to a Dionex/LC Packings Famos/Switchos/Ultimate HPLC. The mass spectrometer was set to use a precursor ion scan of m/z −79 in negative-ion mode followed by an ion-trap high-resolution scan (enhanced resolution scan) and a high-sensitivity MS/MS scan (enhanced product ion scan) in positive mode. Peptides were typically injected on to a PepMap 100 reverse-phase C18 3 μm column with a flow of 300 nl/min and eluted with a 40 min linear gradient of 95% solvent A (2% acetonitrile and 0.1% formic acid in water) to 50% solvent B (90% acetonitrile and 0.08% formic acid in water). A Harvard syringe pump was used to deliver propan-2-ol at a flow of 100 nl/min with mixing occurring at a T-junction after the LC and prior to the MS [29].
Melanoma cell culture and cell migration
The human melanoma cell lines were kindly provided by Dr Richard Marais (The Institute of Cancer Research, London, U.K.). SKMEL13 melanoma cells were maintained in RPMI containing 10% FBS, and SBCL2 and PWMK cells in DMEM containing 10% FBS in a humidified incubator with 5% CO2 at 37 °C. The ability of cells to migrate was tested in a growth-factor-reduced Matrigel™ invasion chamber (BD Biosciences).
Immunofluorescence and microscopy
Cells grown on coverslips were fixed in −20 °C methanol, permeabilized with 0.2% Triton X-100, rinsed with PBS, stained with DAPI (4′,6-diamidino-2-phenylindole) and mounted. The slides were taken on a Zeiss LSM700 microscope using an alphaPlan-Apochromat ×100 NA (numerical aperture) 1.46 objective and digital images were analysed in Adobe Photoshop.
Quantitative RT (reverse transcription)–PCR analysis
Melanoma cells (250000 per well) were seeded in six-well culture plates and total RNA was isolated using the RNeasy minikit (Qiagen) and treated with RNase-Free DNase (Qiagen). RNA (1 μg) was converted into cDNA using Quanta Biosciences qScript™ cDNA SuperMix (95048–500) using the following primer sequences: CIC (Forward) 5′-GGTACTGGCAAGAAGGTGAAGG-3′ and CIC (Reverse) 5′-ACTCAGGCAACTCAGCAAAGC-3′; ETV1 (Forward) 5′-CCCTCCATCGCAGTCCATACC-3′ and ETV1 (Reverse) 5′-CTTGGCATCGTCGGCAAAGG-3′; ETV4 (Forward) 5′-CAGGCGGAGGTTGAAGAAAGG-3′ and ETV4 (Reverse) 5′-AAGGGCAGAAGAAAGGCAAAGG -3′; ETV5 (Forward) 5′-GGGAAATCTCGATCAGAGGACTG-3′ and ETV5 (Reverse) 5′-GGAGCATGAAGCACCAAGTT-3′; B2M (β2 microglobuluin) (Forward) 5′- TGCCGTGTGAACCATGTG -3′ and B2M (Reverse) 5′- ACCTCCATGATGCTGCTTACA-3′; GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Forward) 5′- CAATGACCCCTTCATTGACC-3′ and GAPDH (Reverse) 5′-GACAAGCTTCCCGTTCTCAG-3′; and β-actin (Forward) 5′-TCTACAATGAGCTGCGTGTG-3′ and β-actin (Reverse) 5′- TAGATGGGCACAGTGTGGGT-3′. PCR cycling steps for the Bio-Rad iCycler iQ® of PCR products were 30 s at 95 °C for initial denaturation, 30 s at 60 °C for annealing, and 30 s at 72 °C for extension. All assays were performed with the comparative threshold cycle (Ct) method, the average of three analyses for each gene was calculated and data were normalized to an internal standard gene of B2M.
siRNAs (small interfering RNAs)
ON-TARGET plus SMARTpool siRNA for human capicúa consisting of four individual siRNAs (GCUUAGUGUAUUCGGACAA, CGGCGCAAGAGACCCGAAA, GAGAAGCCGCAAUGAGCGA and CGAGUGAUGAGGAGCGCAU) was from Dharmacon. siRNA oligonucleotides were transfected into cells at 40–60% confluency using Lipofectamine™ 2000 (Invitrogen).
EMSAs (electrophoretic mobility-shift assays)
Biotin-labelled and unlabelled forms of the following oligonucleotide and complementary sequence were synthesized (Invitrogen): biotin–GTCGCGTTTTTTATGAATGAAAAACGTCCTTACATTCATT [octameric CRE (CIC-responsive element) in the ETV4 and ETV5 promoters [8] is shown in bold type]. (The CRE in the ETV1 promoter has a G in place of the A where underlined [8]). The LightShift® Chemiluminescent EMSA Kit (Pierce) was used for the EMSA assays. The binding reactions (30 min at room temperature in 20 μl) were carried out in the presence of 2.5 ng of poly(dI-dC) in 1× binding buffer using 100 pmol of biotin-labelled duplex probe and, where indicated, a 20-fold molar excess of unlabelled probe. Reactions were stopped by adding 5× loading buffer. DNA–protein complexes were separated on 5% Novex DNA retardation gels (Invitrogen). The complexes were blotted on to a positively charged Biodyne nylon membrane (Thermo Scientific), the membrane reacted with a Spectrolinker XL-1500 UV cross-linker (Spectronics Corporation, Westbury, NY) and probes were visualized according to the LightShift® Chemiluminescent EMSA Kit (Pierce).
RESULTS
ETV1, ETV4 and ETV5 expression is blocked by inhibitors of the ERK cascade and enhanced by EGF stimulation and siRNA knockdown of capicúa in melanoma cells
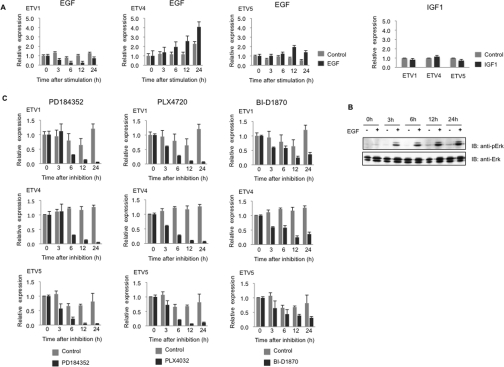
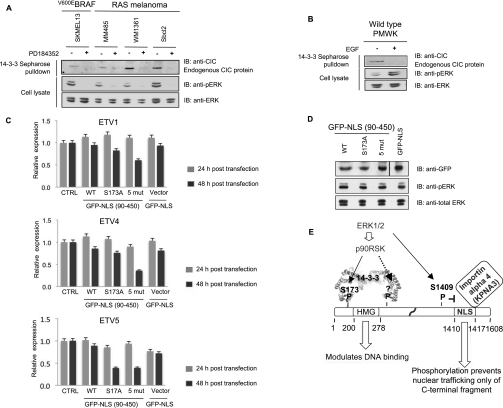
In a wild-type melanoma cell line PMWK, the level of ETV4 and ETV5, but not ETV1, mRNAs was increased in response to EGF stimulation (Figure 1A), which activates prolonged ERK signalling in these cells (Figure 1B), whereas IGF1 (insulin-like growth factor 1) did not affect either ERK phosphorylation or expression of the PEA3 Ets transcription factors (Figure 1A, and results not shown). SKMEL13 melanoma cells carry the BRAF(V600E) mutation rendering the ERK signalling pathway constitutively active. In SKMEL13 cells, the mRNA levels for ETV1, ETV4 and ETV5 were markedly decreased by the ERK pathway inhibitor PD184352 [30] and BRAF(V600E) inhibitor PLX4720 [31], and partially decreased by the p90RSK inhibitor BI-D1870 [32] (Figure 1C). p90RSK is activated downstream of ERK in the melanoma and HEK-293 cells used in the present study (results not shown). Similarly, ETV1, ETV4 and ETV5 expression was inhibited by PD184352 and, to a lesser extent by BI-D1870, in Sbcl2 cells, which are mutant for N-Ras (Supplementary Figure S1 at http://www.BiochemJ.org/bj/433/bj4330515add.htm).
Figure 1. Effect of EGF, PD184352 and capicúa knockdown on expression of ETV1, ETV4 and ETV5 mRNAs in BRAF(V600E) SKMEL13 and/or wild-type PMWK melanoma cells.
(A) qRT-PCR of ETV1, ETV4 and ETV5 mRNA from PMWK cells stimulated with EGF (100 ng/ml) for the number of hours indicated (black bars), IGF1 (50 ng/ml) for 24 h (black bars) and with no stimulus (grey bars). Gene expression is normalized relative to the internal control gene B2M, assigning a value of 1.0 for the zero time point with no stimulus. Values are means±S.D. (four different sets of analyses for each data point). (B) Western blot of whole-cell lysate of PMWK cells stimulated with EGF for the times indicated, and immunoblotted with the antibodies indicated to monitor the phosphorylation status of ERK. (C) The effect of the ERK pathway inhibitor PD184352 (2 μM, [30]), BRAF(V600E) inhibitor PLX4720 (10 μM, [31]) and p90RSK inhibitor BI-D1870 (10 μM, [32]) on expression of ETV1, ETV4 and ETV5 in SKMEL13 cells at the times indicated in hours (black bars). Data for cells with no inhibitor are shown as grey bars. Gene expression is normalized to the internal control gene B2M, assigning a value of 1.0 for the zero time point with no inhibitor. Values are the means±S.D. (four different sets of analyses for each data point).
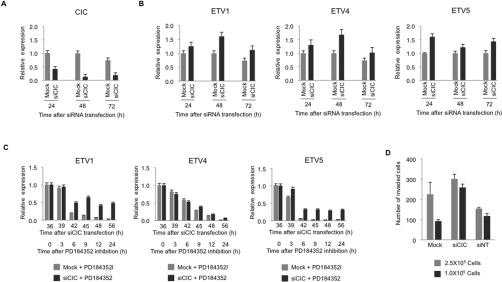
Down-regulating capicúa expression in SKMEL13 melanoma cells (Figure 2A) increased mRNA levels for ETV1, ETV4 and ETV5 (Figure 2B). Furthermore, the knockdown of capicúa lessened the ability of PD184352 to inhibit the expression of ETV1 and ETV5 mRNAs (Figure 2C). Taken together, these data indicate that capicúa suppresses and ERK activity supports the expression of ETV1, ETV4 and ETV5 mRNAs. Furthermore, when Sbcl2 melanoma cells were allowed to migrate in a Matrigel™ invasion chamber after knockdown of capicúa, the knockdown cells displayed a 40–50% increase in migration compared with the control cells (Figure 2D and Supplementary Figure S2 at http://www.BiochemJ.org/bj/433/bj4330515add.htm).
Figure 2. Effect of capicúa knockdown on expression of ETV1, ETV4 and ETV5 mRNAs in BRAF(V600E) SKMEL13 melanoma cells.
(A) The expression of CIC mRNA in SKMEL13 cells treated with CIC siRNAs (siCIC, black bars) compared with mock-transfected controls (grey bars). Values are means±S.D. (four different sets of analyses for each data point). (B) The expression of ETV1, ETV4 and ETV5 mRNA in the same SKMEL13 cells treated with CIC siRNAs that are shown in (A), with data for siRNA-treated cells in black and mock-transfected controls in grey. Values are means±S.D. (four different sets of analysis for each data point). (C) The effect of CIC knockdown followed by exposure to 2 μM PD184352 on ETV1, ETV4 and ETV5 mRNA expression in SKMEL13 cells. Black bars represent data for cells treated with siCIC (siRNA for CIC) and PD184352 for the times indicated, whereas grey bars are for mock-transfected cells treated with PD184352. Values are means±S.D. (four different sets of analyses for each data point). (D) Cells at 30–40% confluency in six-well plates were transfected with siCIC, non-targeting siRNA that does not target any known genes as a control (siNT) and mock transfection. After 24 h, cells were serum-deprived for 2 h, detached with cell dissociation buffer (Gibco), counted, and added in migration buffer (750 μl of DMEM containing 1% BSA) to the upper chambers of a growth factor-reduced Matrigel™ invasion device (BD Biosciences) with chemoattractant (DMEM containing 10% FBS) in the lower wells. The cells that did not migrate were removed from the upper face of the filters using cotton swabs, and cells that had migrated to the lower face of the filters were fixed with Reastain Quick-Diff kit (Reagena). Images (×10) were captured (examples are shown in Supplementary Figure S2 at http://www.BiochemJ.org/bj/433/bj4330515add.htm), migrated cells counted in five random fields, and the histogram shows the number of cells that migrated to the lower face of the filters, presented as means±S.D. for the two experimental conditions (that is experiments starting with 2.5×105 and 1×105 cells respectively).
EGF and phorbol ester inhibit binding of KPNA3 (importin α4) and promote binding of 14-3-3 to capicúa
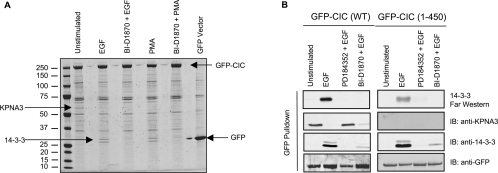
We next explored whether capicúa is a target of regulation by ERK signalling. HEK-293 cells expressing GFP–capicúa were stimulated with EGF and PMA to activate ERK, and the GFP–capicúa protein was immunoprecipitated from lysates. An ~60 kDa protein was co-immunoprecipitated with GFP–capicua from unstimulated cells, but not EGF- and PMA-stimulated cells, and was identified as importin α4/karyopherin α3 (KPNA3) by mass spectrometric analysis of tryptic peptides and Western blotting (Figure 3A). Although inhibiting p90RSK with BI-D1870 had no effect on the capicúa–KPNA3 interaction, inhibiting upstream ERK signalling with PD184352 (Figures 3A and 3B) and UO126 (results not shown) restored the binding of capicúa to KPNA3 in EGF-stimulated cells (Figures 3A and 3B) and phorbol ester-stimulated cells (results not shown).
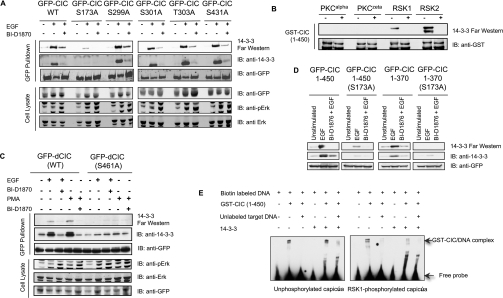
Figure 3. Identification of regulated phosphorylated sites on GFP–capicúa and its regulated interactions with KPNA3 and 14-3-3.
(A) GFP–capicúa (CIC) and GFP (control) proteins were isolated from lysates of transfected cells using GFP–Trap® (an immobilized single-chain antibody) coupled to agarose. Cells were stimulated with EGF (100 ng/ml for 30 min) and PMA (100 ng/ml for 30 min) with or without pre-incubation with the p90RSK inhibitor BI-D1870 (10 μM for 30 min). Captured proteins were subjected to SDS/PAGE, gels were stained with colloidal Coomassie Blue, and proteins and phosphorylated sites were identified by MS as described in the Materials and methods section and Supplementary Figure S3 (at http://www.BiochemJ.org/bj/433/bj4330515add.htm). Arrows point to the proteins identified as GFP–capicúa (GFP–CIC), 14-3-3s and KPNA3. The molecular mass in kDa is indicated on the left-hand side. (B) Effects of inhibitors of the ERK pathway and p90RSK on the EGF-regulated binding of KPNA3 and 14-3-3 to GFP–capicúa. HEK-293 cells were transfected to express GFP–CIC (full-length, WT) and GFP–CIC-(1–450), and stimulated or not with EGF±PD184352 (2 μM inhibitor for 60 min) and±BI-D1870 (10 μM inhibitor for 30 min). EGF was used at 100 ng/ml for 30 min. Lysates (2.5 mg) were subject to GFP–Trap® immunoprecipitation, followed by immunoblotting with the antibodies indicated. In addition, the ability of the GFP–CIC-(1–450) to bind directly to 14-3-3 was tested with a 14-3-3 Far-Western assay. IB, immunoblot.
GFP–capicúa also interacted with 14-3-3 proteins, visualized as two Coomassie-Blue-stained bands of which the upper contained 14-3-3ϵ and the lower comprised 14-3-3θ, γ, α/β, η and δ/ζ (Figure 3A). 14-3-3σ is absent from these cells and was not detected. In contrast with KPNA3, the capicúa–14-3-3 interaction was promoted by EGF and PMA, and was prevented when p90RSK was inhibited directly with BI-D1870 and when upstream ERK signalling was blocked with PD184352 (Figures 3A and 3B) and UO126 (results not shown). The binding of capicúa to 14-3-3 was also promoted by IGF1, although to a lesser extent than with EGF, and the response to IGF1 was blocked by PI3K (phosphoinositide 3-kinase) inhibitors (results not shown).
Tryptic digests of the extracted GFP–capicúa from EGF-stimulated cells were analysed by precursor ion scanning and MS/MS, revealing the presence of several phosphopeptides (Supplementary Figure S3 at http://www.BiochemJ.org/bj/433/bj4330515add.htm). Taken together with previous phosphorylation site data [13] (http://www.phosphosite.org), several phosphorylated residues within RXX(pS) motifs that are potential p90RSK and 14-3-3-binding sites were identified near the HMG box, such as phosphorylated Ser173 (Supplementary Figure S3). In contrast, the C-terminal region of capicúa is phosphorylated mainly on (pS/T)P motifs, which are potential ERK sites, but not the type of motif generally associated with binding to 14-3-3 proteins (Supplementary Figure S3).
ERK phosphorylation of capicúa blocks binding of a C-terminal NLS (nuclear localization sequences) to KPNA3
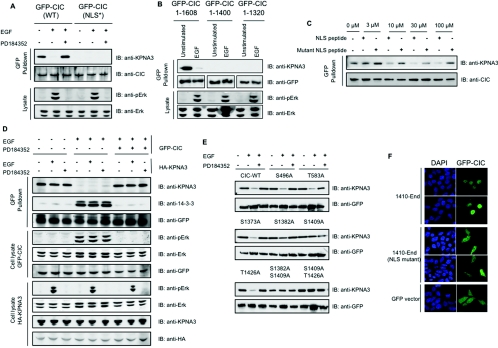
The importin-α proteins bind to NLS in cargo proteins that are then carried into the nucleus. A cluster of basic residues (K1411RKMRR1416) in the C-terminal region of capicúa conformed to a potential monopartite NLS (http://cubic.bioc.columbia.edu/db/NLSdb/). Consistent with KRKMRR binding to KPNA3, GFP–capicúa-(1–450) did not bind to KPNA3 (Figure 3B), but GFP–capicúa-(451-end) did bind to KPNA3 in lysates of unstimulated cells (results not shown). Also, KPNA3 binding was lost when KRKMRR was changed to KAAMRR in full-length GFP–capicúa (Figure 4A), and when the C-terminal region containing this sequence was deleted in GFP–capicúa-(1–1320) and GFP–capicúa-(1–1400) (Figure 4B). The interaction between capicúa and KPNA3 in cell lysates was also blocked by competition with a synthetic peptide containing the canonical NLS sequence from Simian virus 40 large T-antigen, but not a mutant version [33] (Figure 4C).
Figure 4. Mechanism of regulated binding of an NLS in GFP–capicúa to KPNA3.
(A) HEK-293 cells were transfected to express GFP–capicúa (GFP–CIC) and GFP–capicúa with a double R1412A and K1413A substitution in the putative NLS [GFP–CIC (NLS*)], and stimulated or not with EGF±PD184352. Lysates were subjected to GFP–Trap® immunoprecipitation, and immunoprecipitates and whole-cell lysates were immunoblotted with the antibodies indicated. (B) HEK-293 cells were transfected to express the indicated GFP–capicúa proteins and stimulated or not with EGF. Cell lysates and GFP–Trap® immunoprecipitates were analysed with the antibodies indicated. (C) HEK-293 cells were transfected to express GFP–capicúa and cell lysates were incubated with the indicated concentrations of synthetic wild-type (CASPKKKRKV-OH) or mutant (CASPKTKRKV-OH) simian-virus-40 large T-antigen NLS peptides for 30 min at 4 °C followed by GFP–Trap® immunoprecipitation and analysis by immunoblotting. (D) GFP–capicúa and HA–KPNA3 were expressed separately in HEK-293 cells that were stimulated or not with EGF±PD184352. Different combinations of the lysates were mixed as indicated, and GFP–capicúa was immunoprecipitated from the mixtures and immunoblotted with the antibodies indicated. (E) HEK-293 cells were transfected to express wild-type and mutated forms of GFP–capicúa as indicated and stimulated or not with EGF±PD184352 followed by GFP–Trap® immunoprecipitation. Immunoprecitated proteins were analysed by Western blotting using anti-GFP and anti-KPNA3 antibodies. (F) DAPI staining and GFP immunofluorescence of HEK-293 cells transfected with the indicated GFP–capicúa constructs. Scale bar=10 μm. IB, immunoblot.
The results in Figures 3(A) and 3(B) indicated that phosphorylation of capicúa and/or KPNA3 by ERK prevents the interaction between these two proteins. To determine which is the relevant ERK target, GFP–capicúa and HA–KPNA3 were expressed separately in HEK-293 cells that were stimulated or not with EGF±PD184352. When the various cell lysates were mixed and GFP–capicúa extracted, only GFP–capicúa from unstimulated and PD184352-treated cells was bound to HA–KPNA3 (Figure 4D). In contrast, HA–KPNA3 could bind to GFP–capicúa irrespective of whether the HA–KPNA3-expressing cells had been stimulated with EGF (Figure 4D). This means that ERK phosphorylation of capicúa blocks its interaction with KPNA3.
When the 11 (pS/T)P sites identified on capicúa were mutated, only alanine mutations of Ser1382 and Ser1409, which are closest to the NLS, prevented the EGF-induced loss of binding of capicúa to KPNA3 (Figure 4E and results not shown). These results suggest that ERK phosphorylation of Ser1382 and Ser1409 masks the NLS and prevents its binding to KPNA3.
We hypothesized that disruption of the NLS, and EGF-stimulated dissociation of capicúa from KPNA3, would prevent capicúa from entering the nucleus. In contrast, however, full-length GFP–capicúa was nuclear under all conditions tested, even when the NLS was mutated (results not shown). However, the C-terminal part of GFP–capicúa (residue 451 to the end) required the intact NLS for its nuclear localization (Figure 4F and Supplementary Figure S4 at http://www.BiochemJ.org/bj/433/bj4330515add.htm). These data indicate that, although the NLS–importin interaction has the potential to direct localization, capicúa also has other mechanisms for getting into the nucleus. Indeed, the N-terminal half of the protein was also nuclear, even though it does not bind to KPNA3, indicating that this part of the protein also specifies nuclear targeting (Supplementary Figure S4).
14-3-3 binds to the evolutionarily conserved p90RSKphosphorylated Ser173, adjacent to the HMG box of capicúa
Both full-length protein and truncated GFP–capicúa (residues 1–450) bound to 14-3-3 proteins in response to EGF and PMA (Figures 3A, 3B and 5A, and results not shown). However, GFP–capicúa(S173A) showed only a trace of EGF- and PMA-stimulated binding to 14-3-3 proteins (Figure 5A), implicating phospho-Ser173 [KRRTQ(pS173)LS] in 14-3-3 binding. Consistent with the cellular analyses, GST–capicúa-(1–450) that was phosphorylated in vitro on Ser173 and other residues by p90RSK could bind to 14-3-3 proteins (Figure 5B), whereas S173A-substituted protein could not (results not shown).
Figure 5. Identification of the p90RSK-phosphorylated and 14-3-3-binding residues on human and Drosophila capicúa proteins.
(A) HEK-293 cells were transfected with the indicated GFP–capicúa constructs and stimulated or not with EGF±BI-D1870. Lysates were subject to GFP–Trap® immunoprecipitation, and immunoprecipitates and lysates were immunoblotted with the indicated antibodies. The ability of GFP–capicúa proteins to bind to 14-3-3 in vitro was assessed by a 14-3-3 Far Western assay. (B) Purified recombinant constitutively active forms of PKCα, PKCζ, p90RSK1 and p90RSK2 were used for in vitro phosphorylation of GST–capicúa (CIC)-(1–450). The reactions were performed with or without kinase in the presence of Mg-ATP. A 14-3-3 Far Western assay was used to assess the ability of phosphorylated protein to bind to 14-3-3. (C) Lysates from HEK-293 cells transfected to express the indicated Drosophila capicúa proteins, and stimulated or not with EGF±BI-D1870 and PMA±BI-D1870 were subjected to GFP pulldown assay followed by immunoblotting with the antibodies indicated. Direct binding to 14-3-3 was detected in a 14-3-3 Far Western assay. (D) HEK-293 cells were transfected to express the indicated GFP–capicúa proteins and stimulated or not with EGF±BI-D1870 and subjected to GFP–Trap® immunoprecipitation. Immunoprecipitates were immunoblotted with the indicated antibodies. 14-3-3 interaction was detected in 14-3-3 Far Western assay. (E) EMSAs were performed using biotin-labelled DNA probe containing CRE. Where indicated, unlabelled DNA was also included. DNA was incubated with GST–capicúa-(1–450) (1 μM) that was phosphorylated in vitro with p90RSK (right-hand panel) or not (left-hand panel), and used in the presence and absence of 5 μM 14-3-3 dimer. The upper arrow shows the GST–capicúa–DNA complex. Lower bands indicate that the added 14-3-3 has some sequence-non-specific affinity for DNA. IB, immunoblot; WT, wild-type.
Ser173 is conserved in metazoan forms of capicúa (Supplementary Figure S5 at http://www.BiochemJ.org/bj/433/bj4330515add.htm). When expressed in human cells, Drosophila capicúa interacted with 14-3-3 in response to EGF and PMA, and this interaction required Ser461, corresponding to Ser173 in the human protein (Figure 5C). Thus 14-3-3 binding to p90RSK-phosphorylated Drosophila capicúa may help explain how its repressor activity is relieved by ERK signalling in flies [1].
14-3-3 proteins are dimeric and often bind to two phosphorylated sites on their targets [14], and we therefore searched for a second 14-3-3-binding site on human capicúa. Single serine-to-alanine substitution of other phosphorylated sites identified between residues 1 and 450 (Supplementary Figure S3) had no obvious effect on 14-3-3 binding. A truncated GFP–capicúa-(1–370) with S173A did not show even trace binding to 14-3-3 proteins, however (Figure 5D), and we tentatively conclude that phospho-Ser431 [SQRAA(pS431)ED] or a nearby residue provides a second low-affinity binding site for a 14-3-3 dimer.
This proposed configuration of a 14-3-3 dimer straddling phosphorylated Ser173 and possibly phospho-Ser431 on either side of the DNA-binding HMG box (residues 200–268) suggests that the 14-3-3 protein might directly affect the binding of capicúa to DNA. Consistent with this hypothesis, we found that capicúa bound to double-stranded DNA containing the minimal CRE (5′-TGAATGAA-3′), which was previously identified to bind to capicúa [8], and addition of exogenous 14-3-3 to p90RSK-phosphorylated capicúa decreased the capicúa interaction with DNA (Figure 5E). 14-3-3 had no effect on DNA binding of unphosphorylated capicúa (Figure 5E).
Expression of ETV1, ETV4 and ETV5 in melanoma cells is inhibited by a form of capicúa that cannot bind to 14-3-3
Endogenous capicúa also binds to 14-3-3 in lysates of melanoma cells containing constitutively active ERK, and this interaction is prevented by PD184352 (Figure 6A), whereas in wild-type melanoma cells the capicúa–14-3-3 interaction depends on EGF (Figure 6B). This finding suggested that the p90RSK- and 14-3-3-mediated regulation of capicúa might contribute to the ERK-dependent expression of ETV1, ETV4 and ETV5 (Figure 1A), and we tested this hypothesis. Overexpressing capicúa-(90–450) had no obvious effect on mRNA levels for these Ets transcription factors. However, their expression, particularly for ETV5, was decreased by expressing capicúa-(90–450) with Ser173 replaced by alanine, and more-so by a form of capicúa lacking all five of the phosphorylated sites identified in the N-terminal half of the protein (Figures 6C and 6D). These data are consistent with the hypotheses that capicúa contributes to the repression of transcription of ETV1, ETV4 and ETV5, and p90RSK-mediated binding of 14-3-3 to capicúa inhibits its repression of these Ets transcription factors.
Figure 6. Effect of overexpressing capicúa that cannot bind to 14-3-3 on regulation of ETV1, ETV4 and ETV5 mRNA expression in BRAF(V600E) SKMEL13 melanoma cells.
(A) Western blot of endogenous capicúa isolated by its binding to 14-3-3–Sepharose in lysates of BRAF(V600E) SKMEL13 cells, and also the indicated cells with constitutively active RAS proteins. Cells were treated with 2 μM PD184352 for 60 min where indicated, and the phosphorylation status of ERK was monitored in the lysates. (B) Effect of EGF (100 ng/ml for 60 min) on binding of endogenous capicúa to 14-3-3–Sepharose in lysates of PMWK melanoma cells. (C) Effect of expressing the GFP–capicúa constructs indicated in SKMEL13 melanoma cells. ETV1, ETV4 and ETV5 mRNA levels were quantified by qRT-PCR 24 and 48 h post-transfection. All constructs encode added NLS to ensure nuclear targeting of all expressed proteins. Data were normalized to the internal control gene B2M, and results from transfected cells are shown by black bars and untransfected cells by grey bars. (D) For the experiment shown in (C), Western blots of parallel sets of cell lysates were performed to assess the expression levels of the proteins expressed from the constructs 48 h post-transfection. (E) Schematic diagram depicting the regulation of human capicúa. Activated p90RSK phosphorylates capicúa residues either side of the HMG box creating docking sites for a 14-3-3 dimer, and the binding of 14-3-3 affects the binding of the HMG box to the promoters of PEA3 family transcription factors. In addition, ERK phosphorylation of Ser1409 and other sites prevents binding of importin α4/KPNA3 to the NLS of ERK1/2 capicúa. However, experimental disruption of the capicúa–KPNA3 interaction only prevents nuclear import of the C-terminal part of capicúa, and the N-terminal part of capicúa can also enter the nucleus by another mechanism.
DISCUSSION
The ERK cascade regulates cellular proliferation, differentiation and survival, and is commonly activated in human cancers where it is associated with tumour aggression and metastases. In the present paper, we provide mechanistic and functional evidence of a regulatory pathway linking ERK signalling via inhibition of the transcriptional repressor capicúa to expression of the PEA3 Ets transcription factors that promote the transcription of pro-metastatic genes. Our findings could therefore explain why mRNA levels of ETV1 in particular are often elevated in breast, prostate, melanoma, gastrointestinal and other tumours, even when ETV1 overexpression is not driven by gene amplification or fusion to another gene [17,19,20,22–24,27]. In particular, our data are consistent with the identification of ETV1 and ETV5 in screens for mRNAs whose expression was decreased by BRAF(V600E)/MEK [MAPK (mitogen-activated protein kinase)/ERK kinase] inhibitors in melanoma cells [25,26]. It is also tempting to speculate that the mechanism that we describe in the present paper could conceivably underpin the KIT receptor tyrosine kinase/ERK-driven expression of ETV1 in GISTs (gastrointestinal stromal tumours) [27]. High ETV1 expression was found to support growth of GIST cell lines, whether they contained KIT that is sensitive or resistant to imatinib (Gleevec) [27]. Placing capicúa and ETV1/ETV4/ETV5 in a transcriptional axis downstream of ERK therefore raises the pressing question of whether loss or inactivation of capicúa, and hence expression of ETV1, could desensitize tumours to the therapeutic effects of ERK pathway inhibitors (such as Gleevec) as anticancer drugs. Indeed, in the present study we found that capicúa-knockdown caused significant desensitization of melanoma cells to the inhibitory effect of PD184352 on ETV1 and ETV5 mRNA expression. Capicúa-knockdown also enhanced cell invasion in a two-dimensional assay (Figure 2D and Supplementary Figure S2), and how ETV1 and/or other downstream targets mediate the effects of capicúa on proliferation and migration is a critical question for future research.
With these data inferring that deregulated capicúa could conceivably contribute to tumour progression, we focused on the underlying mechanistic details, and our findings are summarized in Figure 6(E). Capicúa was discovered to be a target of concerted controls by ERK itself, and via ERK-activated p90RSK working together with 14-3-3 proteins. The effect of ERK-mediated phosphorylation releasing the importin-α protein KPNA3 from the NLS in the C-terminal region of capicúa is clear, although how this regulation has an impact on subcellular localization of capicúa is not. Our results indicate that the protein has at least one further nuclear-localization mechanism mediated by the N-terminal part of the protein, within or close to the DNA-binding HMG box. We also found that 14-3-3 binds to p90RSK-phosphorylated Ser173 and a second phosphorylated residue that is possibly on the C-terminal side of the DNA-binding HMG box, and initial results indicate that p90RSK phosphorylation and 14-3-3 binding inhibits the binding of capicúa to the minimal CRE (5′-TGAATGAA-3′) [8]. However, it is possible that the effects of 14-3-3 on capicúa may be more complicated than these initial data suggest. In future, a more thorough study of the interplay between capicúa, 14-3-3 and the promoter regions of each of the PEA3 transcription factors will be required. The evolutionary conservation of p90RSK-mediated binding of 14-3-3 proteins to phosphorylated Ser173 in human and Ser461 in Drosophila capicúas respectively, suggests that this mechanism may contribute to the relief of capicúa action triggered by receptor tyrosine kinase signalling during Drosophila development. However, no obvious matches for the NLS or other phosphorylations in the human protein could be identified in the Drosophila protein.
Our findings suggest that 14-3-3s should at least be considered as potential drug targets for tumours that depend on ERK signalling, if the drugs could be sufficiently well targeted to the cancers. As well as capicúa, further proteins bind to 14-3-3 via p90RSK-phosphorylated sites (S. Synowsky, O. Olsson and C. MacKintosh, unpublished work). We also note that 14-3-3η has been reported to contribute to prostate cancer, based on a study in which 14-3-3η enhanced androgen- and EGF-induced androgen receptor transcriptional activity [34]. Prototypic antagonists that block binding of phosphorylated proteins to 14-3-3 proteins promote apoptosis in cell-based experiments [35], which is consistent with indications that 14-3-3 proteins may mediate many downstream effects of PI3K/PKB and ERK/p90RSK signalling.
Our findings also highlight interconnections between SCA and growth-factor signalling networks. Polyglutamine mutations in ataxin-1 cause SCA1, and ataxin-1 protein interacts with capicúa [10,15]. In this connection, it was interesting to note that Drosophila capicúa contains polyglutamine stretches that are absent in the human protein, although whether this is coincidental or has any connection with polyglutamine disease is obscure. It is also intriguing that capicúa represses the ETV1 specifier of dopaminergic neurons [18], given that defects in these neurons underlie other movement disorders, namely Parkinson's disease and dopamine-responsive dystonia. Interestingly, SCA types 14 and 27 were discovered to arise from defects in PKCγ and the FGF14 (fibroblast growth factor 14) growth factor respectively, which have an impact on ERK signalling [36–38]. Thus there are signs of intertwining of the molecular networks underpinning SCAs and certain tumours. Indeed, while the present paper was under review, ataxin-1 was reported to interact with capicúa (CIC) at the promoters of genes, including ETV5 [39]. It would be interesting to dissect further how the gene networks effected by mutations in ataxin-1 overlap with the gene targets of capicúa and ETV1, ETV4 and ETV5. Perhaps faulty growth-factor regulation of the ataxin-1/capicúa/ETV1 axis is a common denominator that interconnects SCAs, dopaminergic disorders and cancers derived from neural crest progenitor cells?
AUTHOR CONTRIBUTION
Carol MacKintosh, Kumara Dissanayake and Olof Olsson conceived the study. Carol MacKintosh and Kumara Dissanayake analysed data and wrote the paper. Kumara Dissanayake, Rachel Toth and Jamie Blakey performed cloning, biochemistry and cell-based experiments. David Campbell generated and interpreted MS data. Alan Prescott contributed to experiments involving microscopy.
ACKNOWLEDGEMENTS
We thank Professor Richard Marais (The Institute of Cancer Research, London) for melanoma cells; Dr Thomas Macartney [DSTT (Division for Signal Transduction Therapy), College of Life Sciences, University of Dundee, Dundee, Scotland, U.K.] for generating a construct to express capicúa; the DSTT team co-ordinated by Dr James Hastie and Dr Hilary McLauchlan for protein production; Kirsten Mcleod and Janis Stark for tissue culture support; and Paul Appleton for bright-field images.
FUNDING
We thank the U.K. Medical Research Council and pharmaceutical companies that support the Division of Signal Transduction Therapy (DSTT) at Dundee (AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck-Serono and Pfizer) for financial support.
References
- 1.Jimenez G., Guichet A., Ephrussi A., Casanova J. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14:224–231. [PMC free article] [PubMed] [Google Scholar]
- 2.Atkey M. R., Lachance J. F., Walczak M., Rebello T., Nilson L. A. Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror. Development. 2006;133:2115–2123. doi: 10.1242/dev.02369. [DOI] [PubMed] [Google Scholar]
- 3.Goff D. J., Nilson L. A., Morisato D. Establishment of dorsal-ventral polarity of the Drosophila egg requires capicua action in ovarian follicle cells. Development. 2001;128:4553–4562. doi: 10.1242/dev.128.22.4553. [DOI] [PubMed] [Google Scholar]
- 4.Lohr U., Chung H. R., Beller M., Jackle H. Antagonistic action of Bicoid and the repressor Capicua determines the spatial limits of Drosophila head gene expression domains. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21695–21700. doi: 10.1073/pnas.0910225106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roch F., Jimenez G., Casanova J. EGFR signalling inhibits Capicuadependent repression during specification of Drosophila wing veins. Development. 2002;129:993–1002. doi: 10.1242/dev.129.4.993. [DOI] [PubMed] [Google Scholar]
- 6.Tseng A. S., Tapon N., Kanda H., Cigizoglu S., Edelmann L., Pellock B., White K., Hariharan I. K. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr. Biol. 2007;17:728–733. doi: 10.1016/j.cub.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee C. J., Chan W. I., Cheung M., Cheng Y. C., Appleby V. J., Orme A. T., Scotting P. J. CIC, a member of a novel subfamily of the HMG-box superfamily, is transiently expressed in developing granule neurons. Brain Res. Mol. Brain Res. 2002;106:151–156. doi: 10.1016/s0169-328x(02)00439-4. [DOI] [PubMed] [Google Scholar]
- 8.Kawamura-Saito M., Yamazaki Y., Kaneko K., Kawaguchi N., Kanda H., Mukai H., Gotoh T., Motoi T., Fukayama M., Aburatani H., et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 9.Lee C. J., Chan W. I., Scotting P. J. CIC, a gene involved in cerebellar development and ErbB signaling, is significantly expressed in medulloblastomas. J. Neurooncol. 2005;73:101–108. doi: 10.1007/s11060-004-4598-2. [DOI] [PubMed] [Google Scholar]
- 10.Lim J., Crespo-Barreto J., Jafar-Nejad P., Bowman A. B., Richman R., Hill D. E., Orr H. T., Zoghbi H. Y. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H. K., Fernandez-Funez P., Acevedo S. F., Lam Y. C., Kaytor M. D., Fernandez M. H., Aitken A., Skoulakis E. M., Orr H. T., Botas J., Zoghbi H. Y. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- 12.Emamian E. S., Kaytor M. D., Duvick L. A., Zu T., Tousey S. K., Zoghbi H. Y., Clark H. B., Orr H. T. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–387. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- 13.Dubois F., Vandermoere F., Gernez A., Murphy J., Toth R., Chen S., Geraghty K. M., Morrice N. A., MacKintosh C. Differential 14-3-3 affinity capture reveals new downstream targets of phosphatidylinositol 3-kinase signaling. Mol. Cell. Proteomics. 2009;8:2487–2499. doi: 10.1074/mcp.M800544-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson C., Crowther S., Stafford M. J., Campbell D. G., Toth R., MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Chiara C., Menon R. P., Strom M., Gibson T. J., Pastore A. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS One. 2009;4:e8372. doi: 10.1371/journal.pone.0008372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam Y. C., Bowman A. B., Jafar-Nejad P., Lim J., Richman R., Fryer J. D., Hyun E. D., Duvick L. A., Orr H. T., Botas J., Zoghbi H. Y. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 17.Jeon I. S., Davis J. N., Braun B. S., Sublett J. E., Roussel M. F., Denny C. T., Shapiro D. N. A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–1234. [PubMed] [Google Scholar]
- 18.Flames N., Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver B. S., Tran J., Chen Z., Carracedo-Perez A., Alimonti A., Nardella C., Gopalan A., Scardino P. T., Cordon-Cardo C., Gerald W., Pandolfi P. P. ETS rearrangements and prostate cancer initiation. Nature. 2009;457:E1. doi: 10.1038/nature07738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermans K. G., Van Der Korput H. A., van Marion R., van de Wijngaart D. J., Ziel-van der Made A., Dits N. F., Boormans J. L., Van Der Kwast T. H., van Dekken H., Bangma C. H., et al. Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res. 2008;68:7541–7549. doi: 10.1158/0008-5472.CAN-07-5930. [DOI] [PubMed] [Google Scholar]
- 21.Jane-Valbuena J., Widlund H. R., Perner S., Johnson L. A., Dibner A. C., Lin W. M., Baker A. C., Nazarian R. M., Vijayendran K. G., Sellers W. R., et al. An oncogenic role for ETV1 in melanoma. Cancer Res. 2010;70:2075–2084. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlins S. A., Laxman B., Dhanasekaran S. M., Helgeson B. E., Cao X., Morris D. S., Menon A., Jing X., Cao Q., Han B., et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 23.Tomlins S. A., Rhodes D. R., Perner S., Dhanasekaran S. M., Mehra R., Sun X. W., Varambally S., Cao X., Tchinda J., Kuefer R., et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 24.Lunn J. S., Fishwick K. J., Halley P. A., Storey K. G. A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev. Biol. 2007;302:536–552. doi: 10.1016/j.ydbio.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Packer L. M., East P., Reis-Filho J. S., Marais R. Identification of direct transcriptional targets of (V600E)BRAF/MEK signalling in melanoma. Pigment Cell Melanoma Res. 2009;22:785–798. doi: 10.1111/j.1755-148X.2009.00618.x. [DOI] [PubMed] [Google Scholar]
- 26.Pratilas C. A., Taylor B. S., Ye Q., Viale A., Sander C., Solit D. B., Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi P., Chen Y., Zhang L., Guo X., Wongvipat J., Shamu T., Fletcher J. A., Dewell S., Maki R. G., Zheng D., et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467:849–853. doi: 10.1038/nature09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janknecht R. Regulation of the ER81 transcription factor and its coactivators by mitogen- and stress-activated protein kinase 1 (MSK1) Oncogene. 2003;22:746–755. doi: 10.1038/sj.onc.1206185. [DOI] [PubMed] [Google Scholar]
- 29.Williamson B. L., Marchese J., Morrice N. A. Automated identification and quantification of protein phosphorylation sites by LC/MS on a hybrid triple quadrupole linear ion trap mass spectrometer. Mol. Cell. Proteomics. 2006;5:337–346. doi: 10.1074/mcp.M500210-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai J., Lee J. T., Wang W., Zhang J., Cho H., Mamo S., Bremer R., Gillette S., Kong J., Haass N. K., et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rexach M., Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 34.Titus M. A., Tan J. A., Gregory C. W., Ford O. H., Subramanian R. R., Fu H., Wilson E. M., Mohler J. L., French F. S. 14-3-3η amplifies androgen receptor actions in prostate cancer. Clin. Cancer Res. 2009;15:7571–7581. doi: 10.1158/1078-0432.CCR-08-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masters S. C., Fu H. 14-3-3 proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 2001;276:45193–45200. doi: 10.1074/jbc.M105971200. [DOI] [PubMed] [Google Scholar]
- 36.Shakkottai V. G., Xiao M., Xu L., Wong M., Nerbonne J. M., Ornitz D. M., Yamada K. A. FGF14 regulates the intrinsic excitability of cerebellar Purkinje neurons. Neurobiol. Dis. 2009;33:81–88. doi: 10.1016/j.nbd.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verbeek D. S., Goedhart J., Bruinsma L., Sinke R. J., Reits E. A. PKCγ mutations in spinocerebellar ataxia type 14 affect C1 domain accessibility and kinase activity leading to aberrant MAPK signaling. J. Cell Sci. 2008;121:2339–2349. doi: 10.1242/jcs.027698. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Snider A., Willard L., Takemoto D. J., Lin D. Loss of Purkinje cells in the PKCγ H101Y transgenic mouse. Biochem. Biophys. Res. Commun. 2009;378:524–528. doi: 10.1016/j.bbrc.2008.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crespo-Barreto J., Fryer J. D., Shaw C. A., Orr H. T., Zoghbi H. Y. Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis. PLoS Genet. 2010;6:e1001021. doi: 10.1371/journal.pgen.1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]