Abstract
Background information. CDC25 (cell division cycle 25) phosphatases function as activators of CDK (cyclin-dependent kinase)–cyclin complexes to regulate progression through the CDC. We have recently identified a pool of CDC25B at the centrosome of interphase cells that plays a role in regulating centrosome numbers.
Results. In the present study, we demonstrate that CDC25B forms a close association with Ctn (centrin) proteins at the centrosome. This interaction involves both N- and C-terminal regions of CDC25B and requires CDC25B binding to its CDK–cyclin substrates. However, the interaction is not dependent on the enzyme activity of CDC25B. Although CDC25B appears to bind indirectly to Ctn2, this association is pertinent to CDC25B localization at the centrosome. We further demonstrate that CDC25B plays a role in maintaining the overall integrity of the centrosome, by regulating the centrosome levels of multiple centrosome proteins, including that of Ctn2.
Conclusions. Our results therefore suggest that CDC25B associates with a Ctn2-containing multiprotein complex in the cytoplasm, which targets it to the centrosome, where it plays a role in maintaining the centrosome levels of Ctn2 and a number of other centrosome components.
Keywords: CDC25B, centrin (Ctn), centrosome, cyclin-dependent kinase (CDK), fluorescence lifetime imaging microscopy (FLIM)
Abbreviations: CaM, calmodulin; CCD, charge-coupled device; CDC25, cell division cycle 25; CDK, cyclin-dependent kinase; Ctn1, centrin 1; DMEM, Dulbecco's modified Eagle's medium; FLIM, fluorescence lifetime imaging microscopy; FRET, fluorescence resonance energy transfer; GFP, green fluorescent protein; GST, glutathione transferase; MCP-PMT, microchannel plate-photomultiplier tube; NER, nucleotide excision repair; PCM, pericentriolar material; siRNA, small interfering RNA; WT, wild-type; XPC, xeroderma pigmentosa group C
Introduction
CDC25 (cell division cycle 25) phosphatases play key roles in regulating the CDC through the localized and timely activation of their CDK (cyclin-dependent kinase)–cyclin substrates. Although we have some knowledge of the contributions of nuclear and cytoplasmic pools of CDC25 proteins, in terms of their upstream regulators and downstream targets, much less is known about their collaborators at the centrosome.
The primary role of the centrosome in non-dividing mammalian cells is to assemble and organize microtubules in order to maintain the shape and polarity of the cell. In the dividing cell, the centrosome takes on a more dynamic role as it transforms into the bipolar mitotic spindle required for accurate segregation of duplicated chromosomes between the two daughter cells (Doxsey, 2001). For this, the G1 phase centrosome, consisting of a mother and daughter centriole pair surrounded by a dense network of proteins, known as the PCM (pericentriolar material) must duplicate itself once before mitotic entry. This process begins in the early S phase and usually occurs in a semi-conservative manner, in that a new centriole (or procentriole) forms perpendicular to each existing mother or daughter centriole. It then elongates by recruiting essential components from the surrounding PCM (Doxsey, 2001; Azimzadeh and Bornens, 2007).
We have recently demonstrated that a centrosomal pool of CDC25B participates in the regulation of two important processes during interphase: (i) nucleation of microtubules and (ii) assembly of centrosomes (Boutros et al., 2007b). Further, cells depleted of CDC25B and consequently arrested in the G2 phase of the cell cycle have a reduced number of Ctn (centrin)-positive centrioles within each centrosome (Boutros and Ducommun, 2008). This loss of centrioles and/or Ctn protein following CDC25B depletion is not the result of premature centrosome separation (Boutros and Ducommun, 2008). Thus, it is most likely caused either by a defect in centriole duplication or a defect in the incorporation of Ctn itself into replicating centrioles.
Ctns are small calcium-binding proteins, and although more than 90% of the protein is partitioned between the cytoplasm and nucleus in human cells, the remaining pool is concentrated at the centrosome (Baron et al., 1994; Paoletti et al., 1996). Ctns are localized within multiple regions of the centrosome, including the distal lumen of the centriole core structure, the interconnecting bridge between the mother and daughter centriole pair, and in the PCM (Paoletti et al., 1996; Salisbury, 2007). In addition to making up an important structural component of the centrosome, Ctns have been reported to be involved in either centriole assembly or centriole segregation, depending on the organism in question. In Saccharomyces cerevisiae, the Ctn homologue CDC31p has been found to be essential for initiating spindle pole body duplication (Baum et al., 1986; Paoletti et al., 2003), whereas two Ctn orthologues found in Paramecium were reported to be involved in centriole positioning (Ruiz et al., 2005). In mammalian cells, determining the role(s) of Ctns in centriole biogenesis has proven to be more complicated. Four Ctn isoforms are known to exist, which are differentially expressed. Ctn1 and Ctn4 appear to be restricted to terminally differentiate ciliated cells, whereas Ctn2 and Ctn3 are expressed in all somatic cells (Salisbury, 2007). The role of Ctn2 in centriole duplication in human cells remains unclear. Although siRNA (small interfering RNA) depletion of Ctn2 was first reported to cause a failure in centriole duplication in HeLa cells (Salisbury et al., 2002), these results were challenged by the finding that centrioles can continue to duplicate normally in the absence of both Ctn2 and Ctn3 in PLK4-overexpressing U2OS cells (Kleylein-Sohn et al., 2007). However, other evidence exists that suggests Ctn2 can contribute to centrosome duplication, through interactions with CaM (calmodulin) and CP110 (Tsang et al., 2006), hSfi1 (Kilmartin, 2003; Martinez-Sanz et al., 2006) and hPOC5 (Azimzadeh et al., 2009). Thus, it appears that the involvement of Ctns in the centrosome assembly pathway in mammalian cells is most likely exerted through their association(s) with other proteins and protein complexes.
In the present study, we have investigated the relationship between CDC25B and Ctn at the centrosome. We found that CDC25B can associate with multiple Ctn isoforms, expressed either endogenously or exogenously in HeLa and U2OS cells, although this association appears to occur indirectly. Nonetheless, we found that Ctn2 depletion results in a loss of CDC25B from the centrosome. Similarly, CDC25B depletion causes a loss of centrosomal Ctn2, in addition to γ-tubulin, Nedd1, and the pericentriolar matrix protein PCM1. Together, our results suggest that CDC25B forms a complex with Ctn2, which is necessary for its localization to the centrosome, where it is involved in regulating the local levels of multiple centrosome proteins.
Results
CDC25B localization to the centrosome is dependent on both N- and C-terminal domains
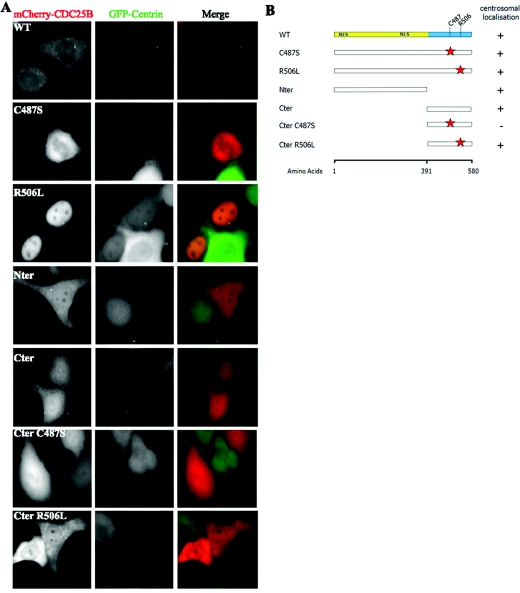
We have demonstrated using fixed human cell lines that CDC25B localizes to the centrosome throughout the cell cycle (Schmitt et al., 2006; Boutros et al., 2007b; Boutros and Ducommun, 2008). In live cells, however, in which cytoplasmic CDC25B protein cannot be pre-extracted, the detection of the centrosomal pool of CDC25B against a strong cytoplasmic background has proven technically challenging, and CDC25B was almost never discernible in cells transiently expressing fluorescently tagged CDC25B. To overcome this, we attempted to transiently express low levels of an mCherry-tagged CDC25B in a HeLa cell line that stably expresses human GFP (green fluorescent protein)-Ctn1 (HeLa-Ctn1, generously given by Dr Michel Bornens, Curie Institute, Paris, France), as a centrosome marker. Surprisingly, we observed that the visibility of CDC25B at the centrosome was enhanced in cells overexpressing Ctn (Figure 1A). In fact, when transfected into HeLa-Ctn1 cells, mCherry-CDC25B was readily detectable at the centrosome(s) in approx. 25% of interphase cells, in comparison with less than 1% when expressed in parental HeLa cells [Figure 1A; WT (wild-type) and results not shown].
Figure 1. Both N- and C-terminal domains of CDC25B are involved in its centrosome targeting.
Various mutant forms of mCherry-CDC25B were derived, transfected into HeLa-Ctn1 cells and analysed for their ability to localize to the centrosome. (A) Examples of HeLa-Ctn1 cell transfected with mCherry-WT, -R506L, -Nter or -Cter CDC25B constructs. (B) Schematic diagram representing various N- (regulatory, yellow) and C- (catalytic, blue) domain mutant forms of CDC25B, in addition to mutations of specific amino acids within the catalytic domain (red stars), representing the mutation of Cys487 [critical for CDC25B catalytic activity (Gabrielli et al., 1996)] to serine (C487S), or Arg506 [critical for substrate binding (Sohn et al., 2004)] to leucine (R506L). The right-hand column indicates the presence (+) or absence (−) of each mutant at the centrosome 24 h post-transfection in HeLa-Ctn1 cells.
This permitted us to use live cell imaging of various mutant forms of CDC25B to try to gain some insight into which region(s) of the protein may be important for its centrosomal localization. As illustrated in Figure 1(B), a series of CDC25B mutants were generated that lack a specific region of the gene and/or have mutations in specific residues required for the catalytic activity of CDC25B (C487S and Cter C487S) (Gabrielli et al., 1996) or CDK–cyclin substrate-binding capacity (R506L and Cter R506L) (Sohn et al., 2004). These CDC25B mutants were transiently expressed as mCherry fusion proteins in HeLa-Ctn1 cells and assessed for their abilities to localize to centrosomes in live cells. As summarized in Figure 1, full-length CDC25B forms localized to the centrosome, even when either the critical catalytic residue Cys487 or substrate-binding residue Arg506 were mutated. Furthermore, both Nter (regulatory region only) and Cter (catalytic domain only) mutants were capable of localizing to the centrosome (Figure 1). However, when the catalytic site Cys487 residue was mutated (Cter C487S), the Cter domain alone was no longer sufficient for CDC25B localization to the centrosome. As the levels of CDC25B expressed showed significant cell-to-cell variations for all mutants examined, we were unable to determine with any certainty whether any of the various CDC25B mutants examined preferentially localize to the centrosome in comparison with the WT protein.
Together, these results indicate that CDC25B localization to the centrosome may be mediated by either N- or C-terminal residues and involves the catalytic site, but not the CDK–cyclin-binding domain of CDC25B.
Centrosomal CDC25B protein levels are regulated by Ctn
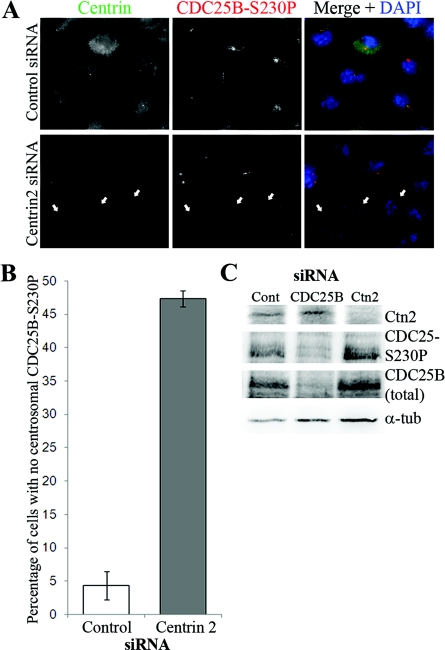
Since Ctn1 overexpression enhances the detection of CDC25B at the centrosome in live cells (Figure 1), we next investigated the effect of its depletion on centrosomal CDC25B levels. As Ctn1 (which shares 85% sequence homology with Ctn2) is restricted to differentiated cells, we targeted Ctn2, which is ubiquitously expressed at the centrioles of all somatic cells (Salisbury et al., 2002; Salisbury, 2007). To detect the centrosomal pool of CDC25B, immunofluorescence analyses were performed using an antibody that specifically detects CDC25B phosphorylated on Ser230 (CDC25B-S230P), which is mediated by Chk1 throughout the cell cycle (Schmitt et al., 2006; Boutros et al., 2007b), as this is currently the only reliable antibody available for the reproducible detection of endogenous CDC25B at the centrosome in all cells. We observed a significant loss in the centrosomal pool of CDC25B in approx. 50% of HeLa (Figure 2) and 40% of U2OS (Supplementary Figure S1 at http://www.biolcell.org/boc/103/boc1030055add.htm) cells, in which Ctn2 was efficiently depleted by 48 h siRNA treatment (Figure 2C). However, Western-blot analyses showed no effect of Ctn2 siRNA depletion on either total or S230-phosphorylated pools of CDC25B protein (Figure 2C).
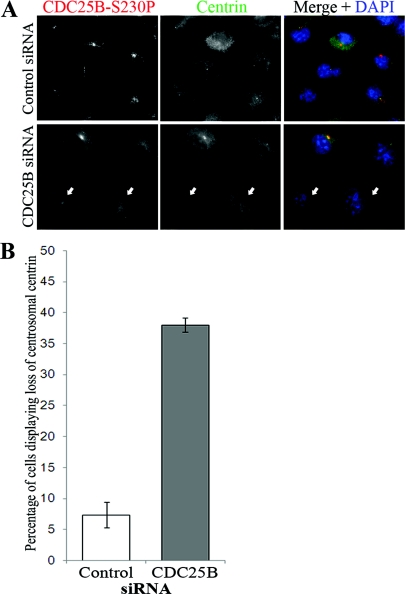
Figure 2. Ctn2 depletion causes loss of CDC25B from the centrosome.
HeLa cells were treated with scrambled control or Ctn2 siRNA duplexes for 48 h before immunofluorescence detection of Ctn (green) and CDC25B (CDC25B-S230P, red). (A) Examples of Ctn2-depleted cells also demonstrating a detectable loss of centrosomal CDC25B (arrows) in comparison with cells treated with control siRNA duplexes. (B) Percentage of HeLa cells in which CDC25B-S230P and Ctn2 (Ctn2 siRNA only) were not detectable or substantially diminished from the centrosomes. In Ctn2 siRNA-treated cells, only the cells displaying a loss of centrosomal Ctn2 were scored for the presence or significant loss of centrosomal CDC25B-S230P signal. Bars represent mean results for at least 200 cells counted from three independent experiments ±S.D. (C) Western blots of lysates from control (Cont), Ctn2 and CDC25B siRNA-depleted cells immunoblotted for Ctn2, total CDC25B (using the C20 antibody from Santa Cruz), CDC25B-S230P and α-tubulin (α-tub) as loading control.
To ensure that observed effects of Ctn2 depletion on centrosomal CDC25B were not an indirect effect on centrosomal Chk1 levels (as a loss of Chk1 would also cause a loss of CDC25B-S230P signal; Schmitt et al., 2006), co-immunofluorescence analyses of Chk1 and Ctn were performed. We observed no significant effect on centrosomal Chk1 levels in Ctn2-depleted cells (Supplementary Figure S2 at http://www.biolcell.org/boc/103/boc1030055add.htm). These results therefore suggest that Ctn2 controls either the localization or the levels of CDC25B at the centrosome.
CDC25B can associate with Ctns at the centrosome and in the cytoplasm
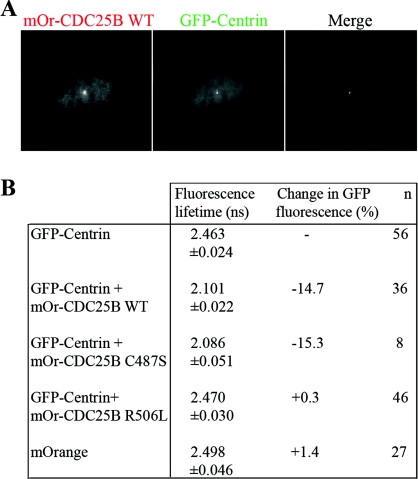
The observation that CDC25B levels at the centrosome can be regulated by Ctn led us to question whether CDC25B and Ctn proteins physically associate. We first addressed this question using a FRET (fluorescence resonance energy transfer)-based approach in HeLa-Ctn1 cells, in which GFP-Ctn1 acted as the donor and mOrange-CDC25B as the acceptor fluorophore. In the case of an interaction between the two proteins, the fluorescence t1/2 of GFP is shortened in proportion with the intensity of energy transfer to the mOrange fluorophore, directly reflecting the strength of the interaction between GFP-Ctn1 and mOrange-CDC25B. We used multiphoton FLIM (fluorescence lifetime imaging microscopy) analyses to characterize the spatial organization and the kinetics of the interaction. Co-localization of Ctn1 and CDC25B proteins were detected at the centrosomes of live HeLa-Ctn1 cells transiently transfected with mOrange-CDC25B WT, C487S and R506L (Figure 3A and results not shown). The lifetime of GFP fluorescence was found to be 2.463 ns for untransfected HeLa-Ctn1 cells (Figure 3B). However, when co-transfected with mOrange-CDC25B-WT or mOrange-CDC25B C487S mutant, this value was reduced to 2.101 and 2.086 ns respectively (representing 14.7 and 15.3% variations in fluorescence lifetime). However, no variation in GFP fluorescence lifetime was observed in cells co-transfected with the mOrange-CDC25B R506L mutant, or mOrange vector alone (Figure 3B and Supplementary Figure S3 at http://www.biolcell.org/boc/103/boc1030055add.htm). These results indicate that CDC25B interacts with Ctn1 at the centrosome, and that this interaction is dependent on CDC25B substrate-binding capacity, but not its catalytic activity. The interaction between Ctn1 and CDC25B was confirmed using co-immunoprecipitation analyses of endogenous CDC25B and GFP-Ctn1 from soluble/cytoplasmic extracts of HeLa-Ctn1 cells and was found to be independent of cell-cycle phase (Supplementary Figure S4 at http://www.biolcell.org/boc/103/boc1030055add.htm).
Figure 3. WT CDC25B interacts with Ctn1 at the centrosome in live cells.
FRET–FLIM analyses of HeLa-Ctn1 cells co-expressing mOrange-CDC25B. (A) Example of a HeLa-Ctn1 cell co-transfected with mOrange (mOr)-CDC25B WT, demonstrating co-localization with GFP-Ctn1. (B) GFP fluorescent lifetime values in HeLa-Ctn1 cells that were either mock-transfected (GFP-Ctn) or co-transfected with mOrange vector control, WT CDC25B (mOr-CDC25B WT), the catalytically inactive mutant mOr-CDC25B C487S, or the substrate-binding mutant mOr-CDC25B R506L. Fluorescence lifetime measurements are in nanoseconds (ns) and n represents the total number of cells analysed from three to five independent experiments.
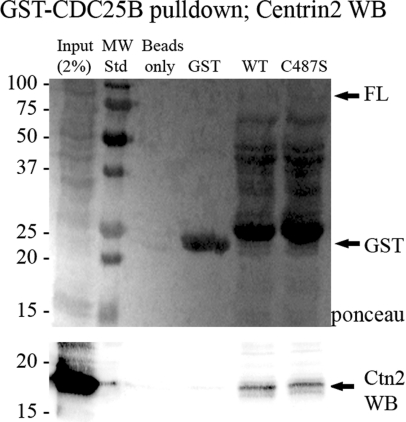
We next addressed whether endogenous Ctn2 could associate with CDC25B using GST (glutathione transferase)-tagged recombinant CDC25B incubated with HeLa or U2OS cell protein extracts. As shown in Figure 4, both WT and the catalytically inactive (C487S) full-length GST-tagged CDC25B pulled down endogenous Ctn2, whereas GST alone did not.
Figure 4. Endogenous Ctn2 can be pulled down with GST–CDC25B.
GST–CDC25B fusion proteins produced in bacteria were bound to GST beads and incubated with protein extracts from HeLa cells. Proteins attached to the GST beads were denatured and separated by SDS/PAGE. (Upper panel) Ponceau stain of nitrocellulose showing levels of GST or the various GST–CDC25B fusion proteins in each reaction. (Lower panel) Western-blot analysis of the same nitrocellulose membrane, immunostained for Ctn2.
Finally, to determine whether CDC25B could bind Ctn1, Ctn2 or Ctn3 directly, we incubated the full-length GST–CDC25B WT or C487S fusion proteins with in vitro translated 35S-labelled Ctn1, Ctn2 or Ctn3. However, we failed to pull down any Ctn isoform with CDC25B (Supplementary Figure S5 at http://www.biolcell.org/boc/103/boc1030055add.htm; results not shown). In agreement with this result, small peptides encoding putative Ctn-binding domains (Charbonnier et al., 2006; Yang et al., 2006; Azimzadeh et al., 2009) from within CDC25B failed to bind Ctn2 (S. Miron, personal communication).
Together, these results indicate that CDC25B and Ctn proteins are within very close proximity at the centrosome (Figure 3 and Supplementary Figure S3) and can be found in the same protein complex (Figure 4 and Supplementary Figures S4 and S5), they are likely to associate via an indirect mechanism.
CDC25B is required for centrosome integrity
Since Ctns and CDC25B proteins co-localized and associated at the centrosome (Figures 1, 3 and 4), and the centrosomal localization of CDC25B appeared to be dependent on Ctn protein levels (Figures 1 and 2), we questioned whether the localization of Ctn2 to the centrosome might be similarly dependent on CDC25B. We therefore depleted CDC25B from HeLa and U2OS cells by siRNA, and examined the centrosomal levels of Ctn2. We observed that CDC25B-depleted cells displayed a significant loss of Ctn2 from the centrosome in approx. 30–40% of both HeLa (Figure 5) and U2OS (Supplementary Figure S6 at http://www.biolcell.org/boc/103/boc1030055add.htm) cells, although no effect on total Ctn2 protein levels were detected by Western-blot analyses (Figure 2C). These results are consistent with our previous observation that CDC25B depletion in HeLa cells results in a significant proportion of cells in the G2 phase displaying only one detectable (or barely detectable) Ctn-positive centriole, rather than two (Boutros and Ducommun, 2008).
Figure 5. CDC25B and Ctn2 centrosomal localizations are interdependent.
HeLa cells were treated with scrambled control or CDC25B siRNA duplexes for 48 h before immunofluorescence detection of CDC25B (CDC25B-S230P) and Ctn. (A) Examples of siRNA-treated cells from which CDC25B was successfully depleted which also show a loss of centrosomal Ctn2 (arrows). (B) Percentage of HeLa cells displaying loss of both CDC25B-S230P (CDC25B siRNA only) and Ctn2 from the centrosomes. Bars represent means for at least 200 cells counted from three independent experiments ±S.D.
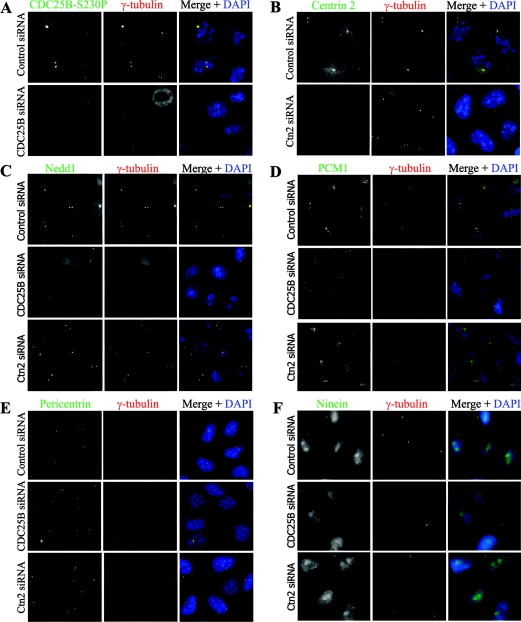
Since we had previously observed a similar effect of CDC25B depletion on centrosomal γ-tubulin levels (Boutros et al., 2007b), we questioned whether CDC25B may have a role in maintaining the overall integrity of the centrosome, or the PCM cloud, perhaps by controlling the centrosome levels of multiple centrosome proteins. CDC25B siRNA and subsequent immunofluorescence analyses revealed that depletion of CDC25B caused a visible reduction in, or a dispersal of, centrosomally localized γ-tubulin [in agreement with our previous finding (Boutros et al., 2007b)], in addition to Nedd1, ninein and PCM1 proteins, but had no effect on pericentrin (Figure 6). In contrast, Ctn2 depletion had no significant effect on any of the centrosome proteins examined (Figure 6).
Figure 6. CDC25B depletion causes a loss of centrosome integrity in HeLa cells.
HeLa cells were treated with scrambled control, CDC25B or Ctn2 siRNA duplexes, and co-stained for γ-tubulin (red) and 4′, 6-diamino-2-phenylindole (DAPI, blue), along with (A) CDC25B (CDC25B-S230P), (B) Ctn2, (C) Nedd1, (D) PCM1, (E) pericentrin or (F) ninein (green).
Together, these results demonstrate that CDC25B forms a tight association with a Ctn2-containing complex that is required for CDC25B localization to the centrosome. Here, CDC25B plays a role in maintaining overall centrosome integrity, probably by controlling the local levels of key centrosome components.
Discussion
The centrosome duplication cycle must be tightly regulated and synchronized with the CDC to ensure the accurate segregation of chromosomes among dividing cells. In recent years, a small subset of centrosome proteins have been identified as being essential for centriole duplication, including PLK4 (Bettencourt-Dias et al., 2005; Kleylein-Sohn et al., 2007), hSAS-6 (Rodrigues-Martins et al., 2007a, 2007b), CPAP (Rodrigues-Martins et al., 2007b), CP110 (Chen et al., 2002; Kleylein-Sohn et al., 2007) and Cep135 (Kleylein-Sohn et al., 2007), in addition to CDK2 (Hinchcliffe et al., 1999; Meraldi et al., 1999) and γ-tubulin (Ruiz et al., 1999; Kleylein-Sohn et al., 2007). However, many other proteins that are present at the centrosome also contribute to centrosome biogenesis. One such protein is Ctn, a member of the EF-hand superfamily of calcium-binding proteins that is recruited to the site of centriole assembly from the very early S phase (Klink and Wolniak, 2001; Koblenz et al., 2003; Stemm-Wolf et al., 2005; Kleylein-Sohn et al., 2007), but whose exact role in centriole assembly in mammalian cells remains unclear. A number of Ctn2-binding partners have been identified to date which together suggest that it functions primarily through modifying the activities of its binding partners.
At the centrosome, Ctn2 has been shown to associate with its highly related EF-hand family member CaM, which simultaneously associates with CP110, in a high-molecular-mass multiprotein complex, involved in centriole duplication and cytokinesis (Chen et al., 2002; Tsang et al., 2006). Two other known direct-binding partners for human Ctn2 are involved in centrosome duplication. The human homologue of the Sfi1 protein, identified in the yeast S. cerevisiae, is an essential component of centrosome duplication and the G2–M transition (Kilmartin, 2003; Martinez-Sanz et al., 2006). The recently identified centriole protein hPOC5, which contains Ctn-binding repeats similar to those found in Sfi1p and is recruited to the distal ends of centrioles in the G2 phase of the cell cycle, to regulate centriole elongation (Azimzadeh et al., 2009). In the nucleus, Ctn2 interacts with the XPC (xeroderma pigmentosa group C gene product), which is involved in the early steps of NER (nucleotide excision repair) of DNA lesions (Araki et al., 2001; Nishi et al., 2005), and enhances the XPC-stabilizing activity of a second NER complex protein HR23B via an indirect association (Araki et al., 2001). The nuclear pool of Ctn2 is modified by the SUMO (small ubiquitin-related modifier) machinery to regulate its nuclear localization and its interaction with XPC (Klein and Nigg, 2009). Nuclear Ctn2 has also been shown to play a role in mRNA export, through its interaction with the nucleoporin Nup107–160 complex (Resendes et al., 2008). Thus, in a fashion similar to CaM, with which it shares at least 50% amino acid sequence homology (Friedberg, 2006), Ctn2 appears to exert its effect on centrosome duplication and other cellular processes through interactions with other proteins.
Here, we have investigated the relationship between Ctn and the CDC25B phosphatase at the centrosome, and found that the depletion of one results in the loss of the other protein from the centrosome. This appears to be due to a common association with an as yet unidentified protein or protein complex. The formation of this CDC25B/Ctn-containing complex controls centrosomal concentrations of both CDC25B and Ctn2, albeit through different mechanisms. CDC25B appears to associate with a Ctn-containing complex for targeting to the centrosome, where it is involved in regulating centrosome numbers and microtubule nucleation (Boutros et al., 2007b). Notably, we had observed that CDC25B depletion disrupts the microtubule network by reducing the level of γ-tubulin at the centrosome (Boutros et al., 2007b). We now observe a loss of other centrosomal components following CDC25B depletion, including Ctn2 itself, and a more dispersed organization of PCM1, which is required for microtubule anchoring (Dammermann and Merdes, 2002). Together, these results suggest that the overall integrity of the centrosome is compromised in the absence of CDC25B and that the associated abnormalities in the microtubule network are probably caused by the combined loss or dispersal of multiple centrosome proteins required for anchoring and assembling tubulin polymers.
We have previously observed that CDC25B, along with a number of other cell cycle regulators that localize to the centrosome, are found predominantly around the mother centriole during S and early G2 phases of the cell cycle (Boutros and Ducommun, 2008). Moreover, it has been suggested that the PCM promotes the formation of daughter centrioles by concentrating γ-tubulin around the mother centriole (Dammermann et al., 2004). It is therefore tempting to postulate that overexpression of CDC25B, which is common in many human cancers (Boutros et al., 2007a), contributes to a local increase in centrosomal levels of Ctn, γ-tubulin, and perhaps other PCM components, to promote the assembly of excess centrioles. Indeed, CDC25B overexpression alone is sufficient to cause centrosome overduplication during the S phase (Boutros et al., 2007b).
Our combined results from live cell imagining and FRET analyses suggest that it may be a CDK–cyclin substrate that mediates an indirect association between CDC25B and Ctn. The R506L substrate-binding mutant of CDC25B failed to interact with Ctn at the centrosome in FRET analyses, whereas the catalytically inactive C487S form of CDC25B could interact with Ctn1. This interaction was also true for GST–CDC25B C487S and endogenous Ctn2 protein from both HeLa and U2OS cells, in pull-down analyses. Our results suggest that once at the centrosome, CDC25B must also be capable of binding to its CDK–cyclin substrates to remain in close proximity to Ctn. One possibility is that CDC25B controls the centrosomal levels of Ctn through a CDK–cyclin-mediated phosphorylation of Ctn, which protects the protein from recognition by the proteasome machinery. Indeed, Ctn2 has been shown in vitro to be a CDK substrate (Lutz et al., 2001). This model would be analogous to CDK2-cyclin E phosphorylation of centrosomal Mps1 (Kasbek et al., 2007).
Finally, it is possible that the association between CDC25B and Ctn requires prior phosphorylation of Ctn, since endogenous Ctn was found within the same protein complex as CDC25B in both FRET and pull-down analyses, whereas in vitro translated Ctn could not associate with recombinant CDC25B. This is a possibility that will require further exploration.
In the present study, we have identified an association between Ctn and a CDC25B-containing complex that is involved in maintaining centrosome integrity. We propose that CDC25B and Ctn are targeted to the centrosome together in a multiprotein complex, where the levels of Ctn and other PCM components are regulated at least in part, by CDC25B. Excess quantities of these PCM components, including Ctn and γ-tubulin, around the mother centriole appear to be sufficient for the formation of excess daughter centrioles (Dammermann et al., 2004; Prosser et al., 2009). Thus, this may be a common mechanism by which CDC25B, through its CDK-cyclin substrates, controls the local concentrations of several centrosome proteins to control centrosome numbers.
Materials and methods
Cell culture
U2OS and HeLa cells obtained from the American Type Culture Collection were maintained at 37°C/5% CO2 in DMEM (Dulbecco's modified Eagle's medium) containing 10% fetal calf serum or serum supreme respectively. HeLa cells stably expressing GFP-Ctn1 (HeLa-Ctn1) were kindly given by Dr Michel Bornens (Curie Institute) and were maintained in DMEM media with 10% fetal calf serum.
Plasmids
For the derivation of mCherry- and mOrange-tagged CDC25B variants, single-base mutations C487S and R506L were performed using site-directed mutagenesis (Stratagene). N- and C-terminal deletions were created by PCR amplification of the region of interest, flanked by HindIII (N-terminal) and BamHI (C-terminal) restriction sites. mCherry- and mOrange-tagged CDC25B WT, C487S and R506L variants were all cloned as HindIII–BamHI inserts into pmCherry-C1 and pmOrange-C1 vectors respectively. GST and GST-tagged CDC25B variants were expressed from pGEX-2T vectors as described previously (Gabrielli et al., 1996; Forrest and Gabrielli, 2001).
DNA transfection
For transient transfections of HeLa-Ctn1 cells with mCherry or mOrange plasmids for live cell imaging, cells were seeded in two-well chamber slides 24 h before transfection with 1 μg of DNA using JetPEI™ reagent (Polyplus-Transfection Inc.), according to the manufacturer's instructions, and cells were imaged 20 h later.
RNA silencing
siRNA duplexes were transfected into HeLa or U2OS cells using Lipofectamine™ 2000 (Invitrogen), according to the manufacturer's instructions. Briefly, cells were seeded into 60-mmdiameter culture dishes at a density of 300000 cells per plate, and grown for 16 h before transfection with 60 pmol of siRNA/0.5 ml for 48 h. Control T12N scramble (Invitrogen), CDC25B (Kramer et al., 2004) and Ctn2 (Salisbury et al., 2002) siRNA duplexes were purchased from Invitrogen, and Ctn2 Smart-pool siRNA duplexes were purchased from Dharmacon.
Immunofluorescence microscopy
For immunofluorescence studies, HeLa or U2OS cells seeded on to glass coverslips and treated as indicated in the text were fixed either in ice-cold methanol for 5 min, or in 4% (w/v) paraformaldehyde at room temperature (21°C) for 20 min and then permeabilized for 5 min in 0.5% Triton X-100. Cells were blocked in 5% serum for 1 h at room temperature before antibody incubations. Centrosomal CDC25B was detected using the antibody against CDC25B-S230P that has already been described (Schmitt et al., 2006). Anti-γ-tubulin (GTU-88) and -Chk1 (DCS-310) monoclonal antibodies were purchased from Sigma–Aldrich, anti-Ctn2 (N-17) from Santa Cruz Biotechnology and anti-pericentrin from Abcam. The monoclonal antibody against Ctn2 and Ctn3 (20H5) was a gift from Dr Jeffrey Salisbury (Mayo Clinic, Rochester, NY, U.S.A.), rabbit polyclonal antibodies against Nedd1 and PCM1 were gifts from Dr Andreas Merdes (CNRS, Toulouse, France) and anti-ninein was a gift from Dr Michel Bornens (Curie Institute, Paris, France). Secondary antibodies labelled with Alexa Fluor® 488, 555 or 647 were purchased from Invitrogen. DNA was visualized using 4′,6-diamino-2-phenylindole staining. Images were acquired using an Apotome microscope (Carl Zeiss), and subsequently processed using AxioVision or Photoshop software packages.
FRET–FLIM
The light source used in the FRET experiments was a mode-locked Ti:sapphire laser (Tsunami, model 3941, Spectra-Physics) pumped by a 10 W diode laser (Millennia Pro, Spectra-Physics) and delivering ultrafast femtosecond pulses of light with a fundamental frequency of 80 MHz. A pulsepicker (model 3980, Spectra-Physics) was used to reduce the repetition rate to 2 MHz to satisfy the requirements of the triggering unit working at 2 MHz. The experiments were performed at λ = 880 nm, the optimal wavelength to excite GFP in multiphoton mode (Chen and Periasamy, 2004). The power delivered at the entrance of the FLIM optics was 14 mW.
All images were acquired with a ×60 oil-immersion lens (Plan Apo 1.4 NA, IR) mounted on an inverted microscope (Eclipse TE2000E; Nikon) coupled to the FLIM system. In this set-up the femtosecond-pulsed laser light is scanned into the left camera port of the microscope through the FLIM optics. The fluorescence emission is directed back into the detection unit through the same camera port and a short-pass filter (λ < 750 nm). The FLIM unit was composed of two galvano mirrors, scanning along the x and y axes, a relay lens to focus the laser beam on to the dichroic (with a cut-off at 750 nm), and the streak camera (Streakscope C4334, Hamamatsu Photonics) coupled to a fast and high-sensitivity CCD (charge-coupled device) camera (model C8800-53C, Hamamatsu Photonics). The streak camera consisted of a photocathode surface, a sweep electrode to deflect the photoelectrons according to their arrival time, an MCP-PMT (microchannel plate-photomultiplier tube) to amplify photoelectrons and a phosphor screen to detect the amplified photoelectrons. The images on the phosphor screen were read-out by the CCD camera (for a comprehensive description of the streak-FLIM system, see Krishnan et al., 2003). The images were acquired with a lateral resolution of 0.246 nm. For each y position in the object, an (x,t) image was produced by recording the fluorescence decay during a given time interval (full timescale of 10 ns with a temporal resolution of 60 ps). Scanning the entire optical image in the y-direction results in a stack of (x,t) images from which fluorescence decays at each (x,y) position can be extracted. The intensity image of the object plane was obtained by integrating fluorescence intensities over time. The CCD camera gain was set to 0 to minimize electronic noise. The MCP-PMT gain (6–8) and the exposure time of the CCD (100 ms) were adjusted to avoid overexposure of the CCD (max value of 4096 grey levels). Frame accumulation (up to 20 frames) was used to increase the signal/noise ratio. The total acquisition time per image ranged from 60 to 80 s (imaging field of 15 μm).
For each image, average fluorescence decay profiles were calculated in the region of interest corresponding to the centrosome (4 or 6 pixels). Then, lifetimes were estimated by fitting these results with a mono-exponential function using a non-linear least-squares estimation procedure with Origin 7.5 software (OriginLab, Northampton, MA, U.S.A.) or the profile fitting function of the Hamamatsu FLIM analysis software.
Statistical and image analyses
All statistical analyses were performed using paired Student's t tests and Microsoft Excel or Prism software packages. Image analyses were performed using the Image J imaging software package, as outlined in the Figure legends.
Recombinant protein expression
PGEX-2T and PGEX-2T-CDC25B constructs were transformed into Escherichia coli BL21 cells, and protein expression induced by 0.5 mM IPTG (isopropyl β-D-thiogalactoside) for 4 h at 37°C. Fusion proteins were extracted by Sarkosyl lysis and affinity-purified on glutathione-agarose, as described by Gabrielli et al. (1996).
In vitro transcription/translation
Ctn1, Ctn2 and Ctn3 ORFs (open reading frames; clones 11074, 11076 and 11043 respectively) were obtained from the Orfeome1.1 human DNA library (Open Biosystems). Coupled in vitro transcription/translation was performed using the TNT T7 rabbit reticulocyte lysate system (Promega), following the manufacturer's instructions, and proteins were radiolabelled with [35S]methionine (PerkinElmer).
Pull-down assays
For pull-down assays performed on endogenous Ctn2 protein from HeLa or U2OS cell extracts, cells were lysed into NETN buffer containing 0.1% SDS, 30 mM NaCl, phosphatase inhibitors and protease inhibitors and 5 mg of soluble protein extracts incubated with approx. 50 μg of purified GST or GST–CDC25B fusion proteins bound to glutathione beads for 2 h at 4°C. Beads were washed five times in lysis buffer and bound protein complexes separated on SDS/15% PAGE gels and transferred to nitrocellulose membranes. For comparison of GST and GST–CDC25B fusion protein quantities used in each reaction, membranes were stained with Ponceau S before Western-blot analyses.
For pull-down assays performed using in vitro translated Ctn proteins, 40 μl (80% of the reaction) in vitro translated protein was incubated with approx. 50 μg purified GST or GST–CDC25B recombinant proteins bound to glutathione beads, in NETN lysis buffer on a rotor at 4°C for 6 h. Beads were washed five times in NETN buffer and once in PBS. Bound protein complexes were separated on SDS/12.5% PAGE gels. The gels were Coomassie Brilliant Blue-stained, dried and exposed to a phosphorimager.
Co-immunoprecipitation assays
Soluble protein extracts were prepared by lysing cell pellets in 1 mM Tris (pH 6.8) containing 0.1% saponin, in the presence of phosphatase and protease inhibitors. A 5 mg portion of protein extract was pre-cleared by incubation with Protein G or A Sepharose beads for 2 h on a rotary mixer. A 5 mg portion of GFP or CDC25B antibodies were incubated on ice for 30 min with Protein G or A Sepharose respectively before the addition of pre-cleared protein extracts. Immunoprecipitation reactions were performed overnight at 4°C. Beads were washed four times with lysis buffer and immunoprecipitated proteins eluted, separated on SDS/12.5% PAGE gels and transferred to nitrocellulose membranes for Western-blot analyses.
Western-blot analyses
Cell pellets were resuspended into lysis buffer containing 50 mM Tris (pH 7.5), 150 mM sodium chloride, 1% deoxycholic acid, 1% SDS, 1% NP40 (Nonidet P40), 1 mM dithiothreitol and phosphatase and protease inhibitors, and sonicated. Proteins were separated on SDS/12.5–15% PAGE gels and transferred on to nitrocellulose membranes. Western-blot analyses were performed with antibodies against phosphorylated CDC25B (CDC25B-S230P; Schmitt et al., 2006), total CDC25B (C-20) from Santa Cruz Biotechnology, Ctn2 (N-17) from Santa Cruz Biotechnology, α-tubulin (B-5-1-2) and γ-tubulin (GTU-88) from Sigma–Aldrich, and GFP from Roche. Secondary antibodies conjugated with horseradish peroxidase were purchased from Zymed.
Online data
Author contribution
Rose Boutros, Bernard Ducommun, Corinne Lorenzo and Brian Gabrielli designed and conceived the experiments. Odile Mondesert and Christine Dozier cloned the CDC25B mutants. Odile Mondesert performed live-cell imaging on these, and, together with Corinne Lorenzo and Alain Jauneau, performed the FRET analyses. Vanessa Oakes assisted Rose Boutros with in vitro translation of Ctn proteins. Rose Boutros performed all other experiments and wrote the first draft of the manuscript. All other authors contributed to the final manuscript.
Acknowledgements
We are sincerely grateful to Dr Megan Chircop for critical reading of this manuscript prior to submission. We thank Dr Michel Bornens (Curie Institute) for the HeLa-Ctn1 cell line and the ninein polyclonal antibody, Dr Andreas Merdes (CNRS, Toulouse, France) for the Nedd1 and PCM1 polyclonal antibodies and Dr Jeffrey Salisbury (Mayo Clinic, Rochester NY, U.S.A.) for the Ctn 20H5 monoclonal antibody.
Funding
Work in the Ducommun laboratory was supported by Centre National de la Recherche Scientifique, l'Université Paul Sabatier, la région Midi-Pyrénées, le Cancéropôle Grand-Sud-Ouest and la Ligue Nationale Contre le Cancer (Equipe labellisée 2005 and 2008). Work in the Gabrielli laboratory was supported by the National Health and Medical Research Council (NHMRC) Australia. R.B. is a recipient of a CJ Martin post-doctoral training fellowship from the NHMRC Australia.
References
- Araki M., Masutani C., Takemura M., Uchida A., Sugasawa K., Kondoh J., Ohkuma Y., Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Bornens M. Structure and duplication of the centrosome. J. Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Hergert P., Delouvee A., Euteneuer U., Formstecher E., Khodjakov A., Bornens M. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 2009;185:101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron A.T., Suman V.J., Nemeth E., Salisbury J.L. The pericentriolar lattice of PtK2 cells exhibits temperature and calcium-modulated behavior. J. Cell Sci. 1994;107:2993–3003. doi: 10.1242/jcs.107.11.2993. [DOI] [PubMed] [Google Scholar]
- Baum P., Furlong C., Byers B. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 1986;83:5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., Glover D.M. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Boutros R., Ducommun B. Asymmetric localization of the CDC25B phosphatase to the mother centrosome during interphase. Cell Cycle. 2008;7:401–406. doi: 10.4161/cc.7.3.5295. [DOI] [PubMed] [Google Scholar]
- Boutros R., Lobjois V., Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat. Rev. Cancer. 2007a;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- Boutros R., Lobjois V., Ducommun B. CDC25B involvement in the centrosome duplication cycle and in microtubule nucleation. Cancer Res. 2007b;67:11557–11564. doi: 10.1158/0008-5472.CAN-07-2415. [DOI] [PubMed] [Google Scholar]
- Charbonnier J.B., Christova P., Shosheva A., Stura E., Le Du M.H., Blouquit Y., Duchambon P., Miron S., Craescu C.T. Crystallization and preliminary X-ray diffraction data of the complex between human centrin 2 and a peptide from the protein XPC. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006;62:649–651. doi: 10.1107/S1744309106019415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Periasamy A. Characterization of two-photon excitation fluorescence lifetime imaging microscopy for protein localization. Microsc. Res. Tech. 2004;63:72–80. doi: 10.1002/jemt.10430. [DOI] [PubMed] [Google Scholar]
- Chen Z., Indjeian V.B., McManus M., Wang L., Dynlacht B.D. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., Muller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Doxsey S. Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol. 2001;2:688–698. doi: 10.1038/35089575. [DOI] [PubMed] [Google Scholar]
- Forrest A., Gabrielli B. Cdc25B activity is regulated by 14-3-3. Oncogene. 2001;20:4393–4401. doi: 10.1038/sj.onc.1204574. [DOI] [PubMed] [Google Scholar]
- Friedberg F. Centrin isoforms in mammals. Relation to calmodulin. Mol. Biol. Rep. 2006;33:243–252. doi: 10.1007/s11033-006-9004-z. [DOI] [PubMed] [Google Scholar]
- Gabrielli B.G., De Souza C.P., Tonks I.D., Clark J.M., Hayward N.K., Ellem K.A. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J. Cell Sci. 1996;109:1081–1093. doi: 10.1242/jcs.109.5.1081. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Li C., Thompson E.A., Maller J.L., Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Kasbek C., Yang C.H., Yusof A.M., Chapman H.M., Winey M., Fisk H.A. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol. Biol. Cell. 2007;18:4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J.V. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 2003;162:1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U.R., Nigg E.A. SUMO-dependent regulation of centrin-2. J. Cell Sci. 2009;122:3312–3321. doi: 10.1242/jcs.050245. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., Nigg E.A. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Klink V.P., Wolniak S.M. Centrin is necessary for the formation of the motile apparatus in spermatids of Marsilea. Mol. Biol. Cell. 2001;12:761–776. doi: 10.1091/mbc.12.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblenz B., Schoppmeier J., Grunow A., Lechtreck K.F. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J. Cell Sci. 2003;116:2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- Kramer A., Mailand N., Lukas C., Syljuasen R.G., Wilkinson C.J., Nigg E.A., Bartek J., Lukas J. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat. Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- Krishnan R.V., Masuda A., Centonze V.E., Herman B. Quantitative imaging of protein–protein interactions by multiphoton fluorescence lifetime imaging microscopy using a streak camera. J. Biomed. Opt. 2003;8:362–367. doi: 10.1117/1.1577574. [DOI] [PubMed] [Google Scholar]
- Lutz W., Lingle W.L., McCormick D., Greenwood T.M., Salisbury J.L. Phosphorylation of centrin during the cell cycle and its role in centriole separation preceding centrosome duplication. J. Biol. Chem. 2001;276:20774–20780. doi: 10.1074/jbc.M101324200. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanz J., Yang A., Blouquit Y., Duchambon P., Assairi L., Craescu C.T. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 2006;273:4504–4515. doi: 10.1111/j.1742-4658.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Lukas J., Fry A.M., Bartek J., Nigg E.A. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Nishi R., Okuda Y., Watanabe E., Mori T., Iwai S., Masutani C., Sugasawa K., Hanaoka F. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 2005;25:5664–5674. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A., Moudjou M., Paintrand M., Salisbury J.L., Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Paoletti A., Bordes N., Haddad R., Schwartz C.L., Chang F., Bornens M. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell. 2003;14:2793–2808. doi: 10.1091/mbc.E02-10-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser S.L., Straatman K.R., Fry A.M. Molecular dissection of the centrosome overduplication pathway in S-phase arrested cells. Mol. Cell. Biol. 2009;29:1760–1773. doi: 10.1128/MCB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendes K.K., Rasala B.A., Forbes D.J. Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol. Cell. Biol. 2008;28:1755–1769. doi: 10.1128/MCB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., Glover D.M. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 2007a;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D.M., Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007b;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- Ruiz F., Beisson J., Rossier J., Dupuis-Williams P. Basal body duplication in Paramecium requires gamma-tubulin. Curr. Biol. 1999;9:43–46. doi: 10.1016/s0960-9822(99)80045-1. [DOI] [PubMed] [Google Scholar]
- Ruiz F., Garreau de Loubresse N., Klotz C., Beisson J., Koll F. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr. Biol. 2005;15:2097–2106. doi: 10.1016/j.cub.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Salisbury J.L. A mechanistic view on the evolutionary origin for centrin-based control of centriole duplication. J. Cell Physiol. 2007;213:420–428. doi: 10.1002/jcp.21226. [DOI] [PubMed] [Google Scholar]
- Salisbury J.L., Suino K.M., Busby R., Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 2002;12:1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Boutros R., Froment C., Monsarrat B., Ducommun B., Dozier C. CHK1 phosphorylates CDC25B during the cell cycle in the absence of DNA damage. J. Cell Sci. 2006;119:4269–4275. doi: 10.1242/jcs.03200. [DOI] [PubMed] [Google Scholar]
- Sohn J., Kristjansdottir K., Safi A., Parker B., Kiburz B., Rudolph J. Remote hot spots mediate protein substrate recognition for the Cdc25 phosphatase. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16437–16441. doi: 10.1073/pnas.0407663101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A.J., Morgan G., Giddings T.H., Jr, White E.A., Marchione R., McDonald H.B., Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W.Y., Spektor A., Luciano D.J., Indjeian V.B., Chen Z., Salisbury J.L., Sanchez I., Dynlacht B.D. CP110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol. Biol. Cell. 2006;17:3423–3434. doi: 10.1091/mbc.E06-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Miron S., Mouawad L., Duchambon P., Blouquit Y., Craescu C.T. Flexibility and plasticity of human centrin 2 binding to the xeroderma pigmentosum group C protein (XPC) from nuclear excision repair. Biochemistry. 2006;45:3653–3663. doi: 10.1021/bi0524868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.