Abstract
MicroRNAs (miRNAs), which are noncoding RNAs that posttranscriptionally inhibit expression of target genes, have recently emerged as important regulators of many cellular functions such as cell differentiation. The epithelial di/tripeptide membrane transporter PepT1 is expressed in highly differentiated cells (the villous tip) but not in undifferentiated cells (the crypt) of the small intestine. Here, we investigated the regulation of PepT1 expression by miRNAs and its functional consequences. We observed a reverse correlation between the expression levels of PepT1 and mature miRNA-92b (miR-92b) during the differentiation of intestinal epithelial Caco2-BBE cells, suggesting a miR-92b-mediated regulation of PepT1 expression. We demonstrate that miR-92b suppressed PepT1 expression at both mRNA and protein levels, with subsequent reduced PepT1 transport activity, in Caco2-BBE cells by directly targeting the PepT1 3′-untranslated region. In addition, miR-92b suppresses bacterial peptide-induced proinflammatory responses in intestinal epithelial cells by inhibiting PepT1 expression. Altogether, our study provides for the first time evidence for the regulation of PepT1 expression at a posttranscriptional level by miRNAs in intestinal epithelial cells during pathophysiological states.
Keywords: Caco2-BBE, differentiation, intestinal inflammation, microRNAs
one important function of intestinal epithelial cells is the absorption of small peptides from the diet via an apical membrane peptide transporter (12). PepT1 is a member of the proton-oligopeptide cotransporter family SLC15, which mediates the transport of di/tripeptides from intestinal lumen into epithelial cells.
A differential pattern of PepT1 expression in the small intestine has been reported. Along the vertical axis of the small intestine, PepT1 expression is most abundant at the villous tip, where it associates with apical lipid raft membrane microdomains (21) and decreases toward the crypt (24). Along the longitudinal axis, the level of PepT1 expression decreases from the duodenum to the ileum (24). PepT1 is generally not expressed in the normal colon (17, 24); however, its expression has been observed in inflamed colons from patients with inflammatory bowel disease (IBD) (17, 32). Using cultured epithelial cells, we have demonstrated that PepT1 is expressed at mRNA and protein levels in the human intestinal epithelial Caco2-BBE cells but not in the human colonic HT29-Cl.19A cells (17). In addition to dietary oligopeptide transport, PepT1 also mediates transport of the bacterial proinflammatory peptides such as formyl-methionyl-leucyl-phenylalanine (fMLP) (4, 18), muramyl dipeptide (MDP) (30), and l-Ala-γ-d-Glu-meso-DAP (Tri-DAP) (9). These studies indicate an important role for this transporter in the gut under both physiological and pathological conditions.
Given the importance of PepT1, understanding the molecular mechanisms underlying the regulation of its expression is necessary. Previous studies have shown the regulation of PepT1 expression at a transcriptional level (10, 19, 20, 22, 26–28). In a pathological context, we recently reported that pathogenic bacteria induce PepT1 expression in colonocytes via the transcription factor Cdx2 (23). So far, regulation of PepT1 expression at a posttranscriptional level has not yet been explored.
MicroRNAs (miRNAs), which are small noncoding RNAs (21–23 nucleotides), have recently emerged as a new class of regulators of gene expression. miRNAs posttranscriptionally regulate gene expression by binding to the 3′-untranslated regions (UTRs) of their target mRNAs (16, 31). Although the role of most miRNAs remains elusive, they have been implicated in vital cellular functions as diverse as intestinal epithelial cells differentiation (11), proliferation, and apoptosis (7). In the present study, we investigated the posttranscriptional regulation of PepT1 in intestinal epithelial cells by miRNAs.
MATERIALS AND METHODS
Cell culture.
Caco2-BBE cells were grown in DMEM (Invitrogen) supplemented with 14 mM NaHCO3, 10% FBS, and penicillin-streptomycin (Invitrogen). Cells were kept at 37°C in a 5% CO2 atmosphere and 90% humidity, and the medium was changed every 2 days.
miRNAs, plasmids construction, transfection, luciferase, and GFP repression experiments.
Hsa-miR-92b (pre-miR precursor AM17100, product ID PM10102), anti-hsa-miR-92b (anti-miR inhibitor AM17000, product ID AM10102), pre-miR negative control (AM17110), and anti-miR negative control (AM17010) were obtained from Ambion. Caco2-BBE cells cultured on plastic plates were transfected with 40 nM miRNA precursors or 40 nM antisense miRNA for the indicated times, by using Lipofectamine 2000 (Invitrogen) and Opti-MEMI reduced serum medium (Invitrogen) according to the manufacturer's instructions. Cells were changed to fresh growth media after 6 h, and total RNA and total protein were extracted from the cells.
For the luciferase repression experiments, the 3′-UTR of hPepT1 was cloned into the SpeI/HindIII sites of the pMIR-REPORT Luciferase vector (Ambion) by using the following primers: forward 5′-GCA CTA GTA GGT CAG GAG GCA AGT GGA G-3′; reverse 5′-GCA AGC TTT AAG AAC AGA AAT TTA TTT CAC-3′. Caco2-BBE cells cultured on 24-well plastic plates were cotransfected with 1 μg of the hPepT1 3′-UTR-luciferase construct and 40 nM of miRNA precursor by using Lipofectamine 2000. Luciferase activity was measured using the Dual-Luciferase Reporter Assay system (Promega) and a Luminoskan Ascent luminometer (Thermo Electron). Values were normalized to lysate protein concentration.
For the green fluorescent protein (GFP) repression experiment, the 3′-UTR of hPepT1 was cloned into the XhoI/HindIII sites of the pEGFP-C1 vector (BD Biosciences Clontech) by using the following primers: forward 5′-GCC TCG AGA GGT CAG GAG GCA AGT GGA G-3′; reverse 5′-GCA AGC TTT AAG AAC AGA AAT TTA TTT CAC-3′. Caco2-BBE cells seeded on coverslips were cotransfected with 1 μg of the hPepT1 3′-UTR-GFP construct and 40 nm of the miRNA precursor by use of Lipofectamine 2000. After 2 days of transfection, GFP production was visualized by fluorescent microscopy using a Zeiss Axioskop2 plus microscope.
Measurement of cell resistance.
Resistance of Caco2-BBE cells was monitored using the electric cell-substrate impedance sensing (ECIS) 1600R device (Applied BioPhysics). Cells were seeded in ECIS 8W1E electrodes at a density of 20,000 cells/400 μl per electrode, and resistance was measured in real time at a frequency of 500 Hz and a voltage of 1 V (5).
Isolation of the epithelium from the crypts and villi.
Epithelial cells from crypts and villi were isolated from the small intestine of 8-wk-old FVB wild-type male mice as previously described by Flint et al. (14). Briefly, small intestine was inverted and cut into pieces of 1–2 cm. The small intestine pieces were washed in HBSS− + 0.5 mM DTT for 5 min at 4°C with constant stirring (step 1). The pieces were collected and then incubated in 150 ml of chelating buffer (27 mM trisodium citrate, 5 mM Na2HPO4, 96 mM NaCl, 8 mM KH2PO4, 1.5 mM KCl, 0.5 mM DTT, 55 mM d-sorbitol, 44 mM sucrose, pH 7.3) at 4°C for 20 min with constant stirring. The villi detached from the pieces were collected by centrifugation of the chelating buffer at 1,500 g for 5 min at 4°C (step 2). The remaining pieces were resuspended in 20 ml of fresh chelating buffer in a 50-ml tube. The tube was manually inverted 20 times, and the pieces were collected and transferred to 20 ml of fresh chelating buffer. Manual wash was repeated 10 times (step 3). The pieces were then incubated in 100 ml of chelating buffer at 4°C for 10 min with constant stirring (step 4). The crypts detached into the chelating buffer were collected by centrifugation at 1,500 g for 5 min at 4°C. Steps 3 and 4 were repeated to increase the quantity of the crypts collected. The purity of villi and crypts was examined by microscopy, and the samples were stored at −80°C until use. All procedures using mice were approved by the Institutional Animal Care and Use Committee at Emory University, IACUC 156-2008.
Protein extraction and Western blot analysis.
Proteins were extracted by use of RIPA buffer [150 mM NaCl, 0.5% sodium deoxycholate, 50 mM Tris·HCl, pH 8, 0.1% SDS, 0.1% Nonidet P-40, 2 mM Na3VO4, 10 mM NaF supplemented with protease inhibitors (Roche Diagnostics)]. The homogenates were centrifuged at 13,000 rpm for 20 min at 4°C. Proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad). Membranes were probed for 1 h at room temperature with anti-human PepT1 (17), anti-mouse PepT1 (4), anti-phospho-Erk1/2 and anti-Erk (Cell Signaling), anti-phospho-p38 and anti-p38 (Cell Signaling), anti-Cdx2 (Santa Cruz), anti-β-actin (Cell Signaling), or anti-GAPDH (Ambion). After washing, membranes were further incubated for 1 h at room temperature with appropriate horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences). Immunoreactive proteins were detected with the enhanced chemiluminescence detection system (Amersham Biosciences).
RNA extraction and quantitative real-time RT-PCR.
Total RNA was extracted by use of TRIzol reagent (Invitrogen) and reverse transcribed by use of the first-strand cDNA synthesis kit (Fermentas). Quantitative RT-PCR (qRT-PCR) was performed using SYBR Green qPCR Master Mix (Fermentas) on a Mastercycler Realplex4 (Eppendorf). The 18S and GAPDH expression levels were used as housekeeping genes, and fold induction was calculated by the Ct method as follows: ΔΔCt = (CtTarget − Cthousekeeping)group 1 − (CtTarget − Cthousekeeping)group 2; the final data were derived from 2−ΔΔCt. The primers used were as follows: hPepT1 sense 5′-CGT GCA CGT AGC ACT GTC CAT-3′, hPepT1 antisense 5′-GGC TTG ATT CCT CCT GTA CCA-3′; GAPDH sense 5′-GTC GGA GTC AAC GGA TTT GG-3′, GAPDH antisense 5′-AAG CTT CCC GTT CTC AGC CT-3′; 18S sense 5′-CCC CTC GAT GAC TTT AGC TGA GTG T-3′, 18S antisense 5′-CGC CGG TCC AAG AAT TTC ACC TCT-3′; mouse PepT1 sense 5′-CGT GCA AGT AGC ACT GTC CAT-3′, mouse PepT1 antisense 5′-GGC TTG ATT CCT CCT GTA CCA-3′; mouse Cdx2 sense 5′-ACC TTC TGG ACA AGG ACG TG-3′, mouse Cdx2 antisense 5′-GCG GAG GAC TGA CAA AGT TC-3′.
Quantification of mature miRNA expression.
Total RNA extracted by use of the RNeasy kit (Qiagen) was reverse transcribed by using the NCode miRNA first-strand cDNA synthesis kit (Invitrogen) to quantify mature miRNA expression according to the manufacturer's instruction. qRT-PCR was performed as described above by using the universal primer provided in the NCode miRNA first-strand cDNA synthesis kit and one of the following forward primers: hsa-miR-92b 5′-TAT TGC ACT CGT CCC GGC CTC C-3′; mmu-miR-20a 5′-TAA AGT GCT TAT AGT GCA GGT AG-3′.
Uptake experiments.
Caco2-BBE cells were transfected with miR-92b precursor by using the siPORT NeoFX transfection agent (Ambion) in growth media following the manufacturer's instructions, and uptake experiments were performed 48 h after transfection as previously described (8). Briefly, cells were washed, incubated with HBSS+-10 mM MES (pH 6.2) at 37°C, and then incubated for 15 min at room temperature with HBSS+-10 mM MES (pH 6.2) containing 20 nM [3H]KPV ± 20 mM Gly-Leu. Cells were washed three times in ice-cold PBS, and cell-associated radioactivity was determined by use of a β-counter (Beckman). The results, expressed as specific uptake of [3H]KPV mediated by hPepT1, are calculated as follows: (Uptake of [3H]KPV) − (Uptake of [3H]KPV + Gly-Leu).
Statistical analysis.
Values were expressed as means ± SE. Statistical analysis was performed using unpaired two-tailed Student's t-test by InStat v3.06 (GraphPad) software. P < 0.05 was considered statistically significant.
RESULTS
miRNAs regulate PepT1 expression during intestinal epithelial cell differentiation.
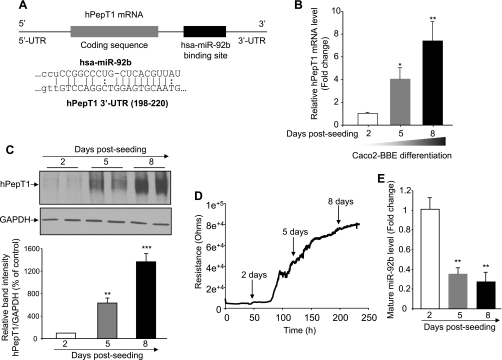
Since the expression of hPepT1 has been known to be increased during the differentiation of intestinal epithelial cells (1, 2), we hypothesized that miRNAs could be involved in the posttranscriptional regulation of hPepT1 expression during this process. Searching the MicroCosm Targets web site (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) revealed hsa-miR-92b (miR-92b) as a potential regulator of hPepT1 expression (Fig. 1A). To investigate the possibility that miR-92b could regulate hPepT1 expression during intestinal epithelial cell differentiation, the human intestinal epithelial Caco2-BBE cells were cultured for 2, 5, and 8 days, and expression levels of hPepT1 and miR-92b were assessed. qRT-PCR and Western blot analyses showed low levels of hPepT1 mRNA and protein expression at day 2 postseeding, when cells were not confluent and poorly differentiated (Fig. 1, B and C). hPepT1 expression was increased 5 days postseeding, when Caco2-BBE cells were subconfluent and partially differentiated, and reached an extremely high level when cells were completely confluent and well differentiated 8 days postseeding (Fig. 1, B and C). To confirm the differentiation status of Caco2-BBE cells during the conventional culture period, we measured the epithelial resistance of the cells in real time by the ECIS technique. Cells were seeded on the ECIS electrodes at the same density that was used for cell seeding on the culture plates (20,000 cells/electrode). Figure 1D shows an increase in cell resistance during the measurement, indicating that Caco2-BBE cells were increasingly differentiated and were mostly differentiated 8 days postseeding.
Fig. 1.
Expression level of human (h) PepT1 is increased, and that of microRNA-92b (miR-92b) is decreased during Caco2-BBE cell differentiation. A: schematic representation of the hPepT1 mRNA including the 3′-UTR with hsa-miR-92b binding site (top), and sequence alignment of hPepT1 3′-UTR and miR-92b target site predicted by MicroCosm Targets web site (bottom). Caco2-BBE cells were cultured on plastic plates for 2, 5, and 8 days, and hPepT1 mRNA and protein expression levels were analyzed by quantitative RT-PCR (qRT-PCR; B) and Western blot (C), respectively. Bar graphs in C show the relative intensity of blots (top) from 3 independent determinations with values represent means ± SE. D: resistance of Caco2-BBE cells (20,000 cells/400 μl/electrode) was measured in real-time at 500 Hz, 1 V using the electric cell-substrate impedance sensing (ECIS) device. E: mature miR-92b levels in Caco2-BBE cells at 2, 5, and 8 days postseeding were analyzed by qRT-PCR. Values represent means ± SE of n = 6/condition from 1 experiment repeated twice with similar results (B and E). *P < 0.05; **P < 0.005; ***P < 0.001 vs. day 2 (white bar).
The relative expression levels of miR-92b in Caco2-BBE cells during differentiation were then quantified by using the N-Code miRNA first-strand synthesis kit, which allows the detection of mature miRNAs, and qRT-PCR. Interestingly, we found that accompanying the increase in hPepT1 expression during Caco2-BBE cell differentiation is a decrease in mature miR-92b expression level (Fig. 1E). Together, these results suggest that miR-92b may have a role in regulating hPepT1 expression during intestinal epithelial cell differentiation.
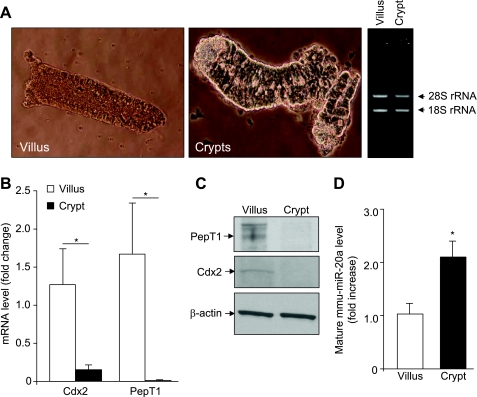
In an effort to verify the in vitro observations, we isolated villi and crypts from 8-wk-old FVB male mice, and the abundance of putative miRNAs involved in the regulation of mouse PepT1 toward the crypt-villus axis was quantified by qRT-PCR. Figure 2A shows that the isolated villi and crypts were pure and intact. Total RNA from villi or crypts was extracted and its integrity was verified by electrophoresis. As shown in Fig. 2A, 28S and 18S ribosomal RNA bands were very distinct and sharp, indicating that total RNA from both villi and crypts were intact. qRT-PCR and Western blot analyses showed that expression levels of PepT1 and the transcription factor Cdx2 were enriched in the villi but were not detected in the crypts (Fig. 2, B and C). Since it has been shown that PepT1 is a marker of differentiated intestinal epithelial cells (24) and Cdx2 levels are elevated in the villi (29), this result confirms the purity of the isolated crypt and villous fractions.
Fig. 2.
Expression patterns of PepT1 and mmu-miR-20a along the mouse crypt-villus axis are opposite. A: villi and crypts were isolated from jejunum of 8-wk-old FVB male mice. Pictures of the extracted villus and crypt fractions were taken with a Nikon Eclipse TS100 microscope at ×10 and ×40 magnifications, respectively. Total RNA from villus and crypt fractions were extracted, and their integrity was verified. PepT1 and Cdx2 expression levels were assessed by qRT-PCR (B) and Western blot (C). D: levels of mature form of mmu-miR-20a in the villus and crypt fractions were quantified by qRT-PCR. Values represent means ± SE of n = 6/group from 1 experiment repeated twice with similar results. *P < 0.05.
Screening the 3′-UTR of mouse (m) PepT1 against the public database (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) for possible complementation of miRNAs did not reveal mmu-miR-92b, a murine original equivalent of hsa-miR-92b, as a potential miRNA for mPepT1. Several predicted potential murine miRNA regulators of mPepT1 were selected, and their expression levels along the mouse intestinal crypt-villus axis were analyzed by quantitative RT-PCR. In agreement with the in vitro data, we found that expression of the mature mmu-miR-20a, a murine original candidate miRNA for mouse PepT1, was significantly decreased in the villi compared with that observed in the crypts, as assessed by qRT-PCR (Fig. 2D). Together, these data suggest that miRNAs may be involved in the regulation of PepT1 expression during intestinal epithelial cell differentiation.
MiR-92b inhibits hPepT1 expression in Caco2-BBE cells.
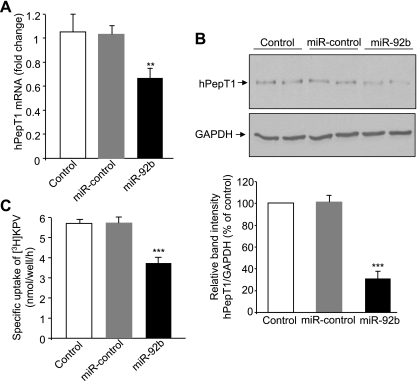
To examine whether miR-92b directly regulates hPepT1 expression, Caco2-BBE cells were transfected with miR-92b precursor, miR-control precursor or vehicle (control). We found that miR-92b effectively decreased hPepT1 mRNA and protein levels in Caco2-BBE cells after 48 h of transfection (Fig. 3, A and B). Furthermore, miR-92b-mediated downregulation of hPepT1 expression resulted in a ∼35% reduction of hPepT1 activity to transport KPV, a specific substrate of hPepT1 (8), into Caco2-BBE cells (Fig. 3C). In contrast, transfection of cells with miR-control or vehicle did not exhibit any significant effects on hPepT1 mRNA and protein expression levels or transport activity of hPepT1 (Fig. 3, A–C). These results demonstrate that miR-92b specifically downregulates hPepT1 expression in Caco2-BBE cells.
Fig. 3.
MiR-92b downregulates hPepT1 mRNA and protein expression in Caco2-BBE cells. Caco2-BBE cells were transfected with 40 nM miR-92b precursor or 40 nM miR-control precursor or vehicle (control) by using Lipofectamine 2000. hPepT1 expression 2 days posttransfection was assessed by qRT-PCR (A) and Western blot (B). Bar graphs in B show the relative intensity of blots (top) from 3 independent determinations with values represent means ± SE. C: hPepT1 transport activity 2 days posttransfection was expressed by measuring the specific uptake of KPV in Caco2-BBE cells. Values represent means ± SE of n = 6/condition from 1 experiment repeated twice with similar results (A and C). **P < 0.005; ***P < 0.001 vs. control.
MiR-92b directly targets the hPepT1 3′-UTR.
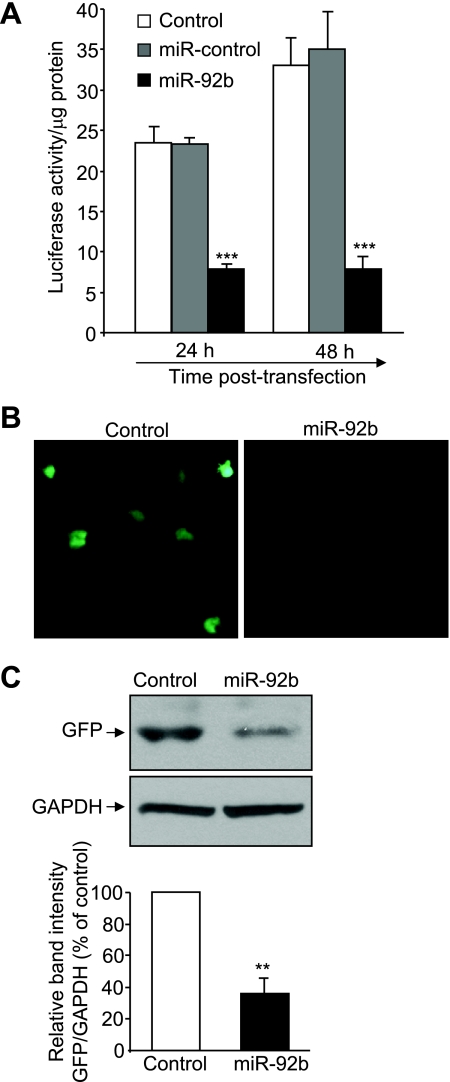
To examine whether miR-92b directly targets the 3′-UTR of hPepT1 mRNA, hPepT1 3′-UTR was cloned into a luciferase reporter gene (Luc-hPepT1 3′-UTR), and this construct was transfected into Caco2-BBE cells in the presence of miR-92b precursor or miR-control precursor. The targeting of miRNAs to hPepT1 3′-UTR was then assessed by measuring luciferase activity in these cells. As shown in Fig. 4A, miR-92b significantly reduced luciferase activity in Caco2-BBE cells by ∼67 and ∼78% after 24 and 48 h of transfection, respectively. In contrast, miR control did not inhibit luciferase activity. These results indicate that miR-92b binds to hPepT1 3′-UTR.
Fig. 4.
MiR-92b directly targets the 3′-UTR of hPepT1 mRNA. The hPepT1 mRNA 3′-UTR was cloned downstream of a luciferase reporter gene (hPepT1 3′-UTR-luc) or a green fluorescent protein (GFP)-coding sequence (hPepT1 3′-UTR-GFP). A: Caco2-BBE cells were transfected with the hPepT1 3′-UTR-luc construct in the presence or absence (control) of 40 nM miR-92b or 40 nM miR-control. Luciferase activity was measured at the indicated times and normalized to total protein concentration. Values represent means ± SE of n = 6/condition from 1 experiment repeated twice with similar results. B and C: Caco2-BBE cells were transfected with the hPepT1 3′-UTR-GFP construct in the presence or absence (control) of 40 nM miR-92b. GFP expression in cells 48 h posttransfection was assessed by immunofluorescent microscopy (B) or Western blot (C). Bar graphs in C show the relative intensity of blots (top) from 3 independent determinations with values represent means ± SE. **P < 0.005; ***P < 0.001 vs. control.
To confirm these results, the hPepT1 3′-UTR was cloned downstream of a GFP-coding sequence (GFP-hPepT1 3′-UTR), and the binding of miRNAs to this construct was assessed by determining GFP expression. We found that only miR-92b significantly decreased GFP expression as assessed by immunofluorescence microscopy (Fig. 4B) and Western blot analysis (Fig. 4C). Together, these data demonstrate that miR-92b downregulates hPepT1 expression by directly targeting the 3′-UTR of hPepT1 mRNA.
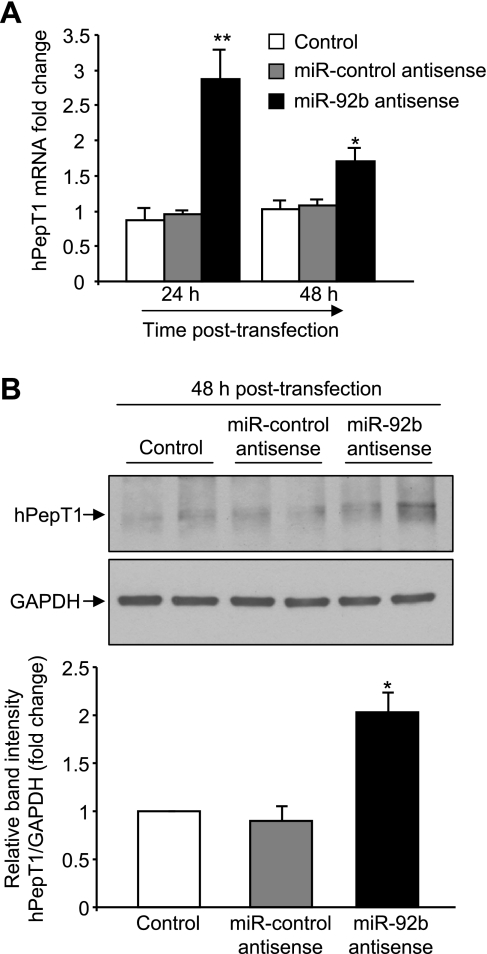
MiR-92b antisense increases hPepT1 expression in Caco2-BBE cells.
To confirm the miR-92b-mediated inhibition of hPepT1 expression, Caco2-BBE cells were transfected with an antisense of miR-92b or miR-control antisense and hPepT1 expression was assessed. qRT-PCR and Western blot revealed that only miR-92b antisense significantly increased hPepT1 mRNA and protein expression (Fig. 5, A and B). These results strongly support the finding that miR-92b represses hPepT1 expression.
Fig. 5.
MiR-92b antisense increases hPepT1 expression in Caco2-BBE cells. Caco2-BBE cells were transfected with 40 nM miR-92b antisense or 40 nM miR-control antisense or vehicle (control) by using Lipofectamine 2000 for the indicated times. hPepT1 mRNA and protein expression was assessed by qRT-PCR (A) and Western blot (B), respectively. Values represent means ± SE of n = 6/condition from 1 experiment repeated twice with similar results (A). Bar graphs in B show the relative intensity of blots (top) from 3 independent determinations with values represent means ± SE. *P < 0.05; **P < 0.005 vs. control.
MiR-92b suppresses bacterial peptide-induced proinflammatory response in Caco2-BBE cells by inhibiting hPepT1 expression.
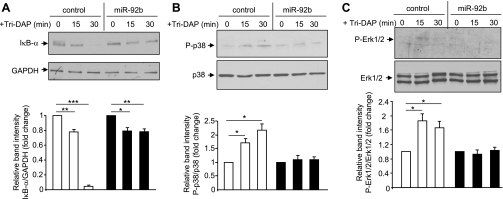
We recently showed that PepT1 transports the bacterial tripeptide Tri-DAP into intestinal epithelial cells, inducing inflammatory responses (9). We then sought to examine whether miR-92b could suppress Tri-DAP-induced intestinal inflammation by inhibiting hPepT1 expression. For that, Caco2-BBE cells pretransfected with miR-92b were stimulated with Tri-DAP for the indicated times, and NF-κB activation was assessed by Western blot analysis of degraded IκB-α levels. We found that miR-92b efficiently inhibited Tri-DAP-induced IκB-α degradation in Caco2-BBE cells (Fig. 6A).
Fig. 6.
MiR-92b decreases l-Ala-γ-d-Glu-meso-DAP (Tri-DAP)-induced proinflammatory responses in Caco2-BBE cells by inhibiting hPepT1 expression. Caco2-BBE cells pretransfected with 40 nM miR-92b or vehicle (control) were stimulated with Tri-DAP for the indicated times. Levels of IκB-α (A), phospho-p38 (B), and phospho-Erk1/2 (C) were analyzed by Western blot analysis. Bar graphs show the relative intensity of blots (top) from 3 independent determinations with values represent means ± SE. *P < 0.05; **P < 0.005; ***P < 0.001.
Since we previously showed that hPepT1 is required for Tri-DAP-induced activation of mitogen-activated protein kinases (9), we next examined the effect of miR-92b on this inflammatory response. Figure 6, B and C, shows that miR-92b significantly inhibited the increase in phospho-p38 and phospho-Erk1/2 levels in Caco2-BBE cells upon Tri-DAP stimulation. Together, these results shows that miR-92b suppresses Tri-DAP-induced proinflammatory responses in Caco2-BBE cells by inhibiting hPepT1 expression.
DISCUSSION
The small intestine displays a differential pattern of PepT1 expression. Along its vertical axis, PepT1 expression is most abundant at the villous tip and decreases toward the crypt (24). This suggests a regulation mechanism underlying PepT1 expression during the differentiation of intestinal epithelial cells. Here we provide evidence for the involvement of miRNAs as regulators in such process.
On the basis of our previously published miRNA profile in intestinal epithelial cells during differentiation (11), miR-92b was selected since its expression level was decreased during Caco2-BBE cell differentiation and therefore it is a potential regulator of hPepT1 expression in this process. We then demonstrated that expression level of miR-92b was decreased during the differentiation of Caco2-BBE cells. Most importantly, miR-92b directly binds to the 3′-UTR of hPepT1 mRNA and downregulates hPepT1 expression at both mRNA and protein levels in Caco2-BBE cells. These results indicate that miR-92b acts as a repressor of hPepT1 expression in intestinal epithelial cells. In this context, previous results have shown that miRNAs are involved in the control of cell differentiation in different tissues. For example, it has been shown that miR-194 is upregulated during intestinal epithelial cell differentiation (15), miR-273 is implicated in the neuronal differentiation of Caenorhabditis elegans (3), miR-181 is involved in human hematopoietic cell differentiation (6), miR-375 is important to the development of pancreatic inlets (25), miR-143 is thought to play a role in adipocyte differentiation (13), and a role for miR-7 in intestinal epithelial cell differentiation has been revealed (23). Importantly, we have recently reported specific miRNA expression profiles for different differentiation statuses of intestinal epithelial cells and shown that miRNAs could play a role in determining the unique physiological characteristics of intestinal epithelial cells (11). Although we showed a strong evidence for the effect of miR-92b on 3′-UTR of hPepT1, we cannot rule out the possibility that miR-92b could indirectly regulate hPepT1 expression by targeting an unidentified regulator of this transporter.
Importantly, we showed that miR-92b efficiently suppressed intestinal inflammation triggered by proinflammatory bacterial peptides by inhibiting expression of hPepT1. A role for hPepT1 in the intestinal inflammation has been suggested. hPepT1 expression is induced in inflamed colons from IBD patients (17) and in inflamed colonocytes infected with pathogenic bacteria (22). PepT1 is directly involved in intestinal inflammation by transporting bacterial proinflammatory peptides into intestinal epithelial cells (9, 30). Therefore, the capacity of miR-92b to suppress bacterial peptide-induced inflammatory responses in intestinal epithelia by acting as a repressor of hPepT1 expression is of importance. This suggests that modulating hPepT1 expression during intestinal inflammation using miR-92b could have promising therapeutic effects. In vivo studies using animal models would be interesting to test this possibility. It would be also of interest to examine the correlation between expression levels of PepT1 and miR-92b in biopsies from patients suffering from intestinal inflammation. Since one miRNA can potentially target hundreds of genes, we cannot rule out the possibility that miR-92b decreases bacterial peptide-induced inflammatory responses in intestinal epithelia by downregulating expression of other target genes rather than PepT1. To determine whether miR-92b effects are solely through PepT1, ectopical expression of PepT1 such that it cannot be targeted by miR-92b and examination of the miR92b-mediated effects on bacterial peptide-triggered inflammation could be helpful and need to be further studied.
Finally, since miR-92b has a role in the pathophysiological status of intestinal epithelia by regulating hPepT1 expression, it is of importance to investigate the regulation of miR-92b during such states. The mechanisms by which specific miRNAs are regulated remain largely unknown. Several transcription factors have been shown to be involved in miRNA regulation. For example, the hepatocyte nuclear factor-1α was recently shown to control the expression of miR-194 during Caco2 cell differentiation (15). It would be interesting to assess the regulation of miR-92b by the potential transcription factors during intestinal epithelial cell differentiation. Understanding the mechanism underlying regulation of miR-92b would help to more efficiently modulate hPepT1 expression as well as to better understand the role of miR-92b in physiological characteristics of intestinal epithelial cells in general.
In conclusion, we demonstrate that miR-92b downregulates hPepT1 expression, causing reduced hPepT1 transport activity in intestinal epithelial cells by targeting the 3′-UTR of hPepT1 mRNA. In addition, miR-92b suppresses bacterial peptide-induced proinflammatory responses in intestinal epithelial cells by inhibiting hPepT1 expression. To our knowledge, this is the first study providing evidence for the regulation of hPepT1 expression at a posttranscriptional level by miRNAs, enhancing our understanding of the mechanisms underlying the regulation of this intestinal oligopeptide transporter.
GRANTS
This work was supported by grants from the Department of Veterans Affairs and the National Institutes of Health of Diabetes and Digestive and Kidney Diseases [R24-DK-064399 center grant, R56DK084987 to D. Merlin, RO1-DK-55850 to S. V. Sitaraman] and a research fellowship award from the Crohn's and Colitis Foundation of America [to G. Dalmasso].
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1. Behrens I, Kissel T. Do cell culture conditions influence the carrier-mediated transport of peptides in Caco-2 cell monolayers? Eur J Pharm Sci 19: 433–442, 2003 [DOI] [PubMed] [Google Scholar]
- 2. Buyse M, Berlioz F, Guilmeau S, Tsocas A, Voisin T, Peranzi G, Merlin D, Laburthe M, Lewin MJ, Roze C, Bado A. PepT1-mediated epithelial transport of dipeptides and cephalexin is enhanced by luminal leptin in the small intestine. J Clin Invest 108: 1483–1494, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang S, Johnston RJ, Jr, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430: 785–789, 2004 [DOI] [PubMed] [Google Scholar]
- 4. Charrier L, Driss A, Yan Y, Nduati V, Klapproth JM, Sitaraman SV, Merlin D. hPepT1 mediates bacterial tripeptide fMLP uptake in human monocytes. Lab Invest 86: 490–503, 2006 [DOI] [PubMed] [Google Scholar]
- 5. Charrier L, Yan Y, Driss A, Laboisse CL, Sitaraman SV, Merlin D. ADAM-15 inhibits wound healing in human intestinal epithelial cell monolayers. Am J Physiol Gastrointest Liver Physiol 288: G346–G353, 2005 [DOI] [PubMed] [Google Scholar]
- 6. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86, 2004 [DOI] [PubMed] [Google Scholar]
- 7. Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 33: 1290–1297, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dalmasso G, Charrier-Hisamuddin L, Nguyen HT, Yan Y, Sitaraman S, Merlin D. PepT1-mediated tripeptide KPV uptake reduces intestinal inflammation. Gastroenterology 134: 166–178, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dalmasso G, Nguyen HT, Charrier-Hisamuddin L, Yan Y, Laroui H, Demoulin B, Sitaraman SV, Merlin D. PepT1 mediates transport of the proinflammatory bacterial tripeptide l-Ala-γ-d-Glu-meso-DAP in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 299: G687–G696, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dalmasso G, Nguyen HT, Yan Y, Charrier-Hisamuddin L, Sitaraman SV, Merlin D. Butyrate transcriptionally enhances peptide transporter PepT1 expression and activity. PLoS ONE 3: e2476, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dalmasso G, Nguyen HT, Yan Y, Laroui H, Srinivasan S, Sitaraman SV, Merlin D. MicroRNAs determine human intestinal epithelial cell fate. Differentiation 80: 147–154, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Daniel H. Molecular and integrative physiology of intestinal peptide transport. Annu Rev Physiol 66: 361–384, 2004 [DOI] [PubMed] [Google Scholar]
- 13. Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279: 52361–52365, 2004 [DOI] [PubMed] [Google Scholar]
- 14. Flint N, Cove FL, Evans GS. A low-temperature method for the isolation of small-intestinal epithelium along the crypt-villus axis. Biochem J 280: 331–334, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T, Watanabe M. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA 14: 1433–1442, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854, 1993 [DOI] [PubMed] [Google Scholar]
- 17. Merlin D, Si-Tahar M, Sitaraman SV, Eastburn K, Williams I, Liu X, Hediger MA, Madara JL. Colonic epithelial hPepT1 expression occurs in inflammatory bowel disease: transport of bacterial peptides influences expression of MHC class 1 molecules. Gastroenterology 120: 1666–1679, 2001 [DOI] [PubMed] [Google Scholar]
- 18. Merlin D, Steel A, Gewirtz AT, Si-Tahar M, Hediger MA, Madara JL. hPepT1-mediated epithelial transport of bacteria-derived chemotactic peptides enhances neutrophil-epithelial interactions. J Clin Invest 102: 2011–2018, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mutoh H, Satoh K, Kita H, Sakamoto H, Hayakawa H, Yamamoto H, Isoda N, Tamada K, Ido K, Sugano K. Cdx2 specifies the differentiation of morphological as well as functional absorptive enterocytes of the small intestine. Int J Dev Biol 49: 867–871, 2005 [DOI] [PubMed] [Google Scholar]
- 20. Nduati V, Yan Y, Dalmasso G, Driss A, Sitaraman S, Merlin D. Leptin transcriptionally enhances peptide transporter (hPepT1) expression and activity via the cAMP-response element-binding protein and Cdx2 transcription factors. J Biol Chem 282: 1359–1373, 2007 [DOI] [PubMed] [Google Scholar]
- 21. Nguyen HT, Charrier-Hisamuddin L, Dalmasso G, Hiol A, Sitaraman S, Merlin D. Association of PepT1 with lipid rafts differently modulates its transport activity in polarized and nonpolarized cells. Am J Physiol Gastrointest Liver Physiol 293: G1155–G1165, 2007 [DOI] [PubMed] [Google Scholar]
- 22. Nguyen HT, Dalmasso G, Powell KR, Yan Y, Bhatt S, Kalman D, Sitaraman SV, Merlin D. Pathogenic bacteria induce colonic PepT1 expression: an implication in host defense response. Gastroenterology 137: 1435–1447, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen HT, Dalmasso G, Yan Y, Laroui H, Dahan S, Mayer L, Sitaraman SV, Merlin D. MicroRNA-7 modulates CD98 expression during intestinal epithelial cell differentiation. J Biol Chem 285: 1479–1489, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ogihara H, Saito H, Shin BC, Terado T, Takenoshita S, Nagamachi Y, Inui K, Takata K. Immuno-localization of H+/peptide cotransporter in rat digestive tract. Biochem Biophys Res Commun 220: 848–852, 1996 [DOI] [PubMed] [Google Scholar]
- 25. Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230, 2004 [DOI] [PubMed] [Google Scholar]
- 26. Shimakura J, Terada T, Shimada Y, Katsura T, Inui K. The transcription factor Cdx2 regulates the intestine-specific expression of human peptide transporter 1 through functional interaction with Sp1. Biochem Pharmacol 71: 1581–1588, 2006 [DOI] [PubMed] [Google Scholar]
- 27. Shiraga T, Miyamoto K, Tanaka H, Yamamoto H, Taketani Y, Morita K, Tamai I, Tsuji A, Takeda E. Cellular and molecular mechanisms of dietary regulation on rat intestinal H+/peptide transporter PepT1. Gastroenterology 116: 354–362, 1999 [DOI] [PubMed] [Google Scholar]
- 28. Silberg DG, Sullivan J, Kang E, Swain GP, Moffett J, Sund NJ, Sackett SD, Kaestner KH. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology 122: 689–696, 2002 [DOI] [PubMed] [Google Scholar]
- 29. Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology 119: 961–971, 2000 [DOI] [PubMed] [Google Scholar]
- 30. Vavricka SR, Musch MW, Chang JE, Nakagawa Y, Phanvijhitsiri K, Waypa TS, Merlin D, Schneewind O, Chang EB. hPepT1 transports muramyl dipeptide, activating NF-kappaB and stimulating IL-8 secretion in human colonic Caco2/bbe cells. Gastroenterology 127: 1401–1409, 2004 [DOI] [PubMed] [Google Scholar]
- 31. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862, 1993 [DOI] [PubMed] [Google Scholar]
- 32. Wojtal KA, Eloranta JJ, Hruz P, Gutmann H, Drewe J, Staumann A, Beglinger C, Fried M, Kullak-Ublick GA, Vavricka SR. Changes in mRNA expression levels of solute carrier transporters in inflammatory bowel disease patients. Drug Metab Dispos 37: 1871–1877, 2009 [DOI] [PubMed] [Google Scholar]