Abstract
This protocol details the data quality assessment and control steps that are typically carried out during case-control association studies. The steps described involve the identification and removal of DNA samples and markers that introduce bias to the study. These critical steps are paramount to the success of a case-control study and are necessary before statistically testing for association. We describe how to use PLINK, a tool for handling SNP data, to carry out assessments of failure rate per-individual and per-SNP and to assess the degree of relatedness between individuals. We also detail other quality control procedures, including the use of SMARTPCA for the identification of ancestral outliers. These platforms were selected because they are user-friendly, widely used, and computationally efficient. Steps needed to detect and establish a disease association using case-control data are not discussed, as these are provided in a further protocol in the series. Issues concerning the study design and marker selection in case-control studies have been discussed in our earlier protocols. The protocol should take approximately 8 hours to complete.
Introduction
Biases in study design and errors in genotype calling have the potential to introduce systematic biases into genetic case-control association studies, leading to an increase in the number of false-positive and false-negative associations (see Box 1 for a glossary of terms). Many such errors can be avoided through careful collection of case and control groups and vigilant laboratory practices. A protocol for the successful ascertainment of unbiased case-control groups, focussing on sampling individuals from the same underlying population, is provided in an earlier publication in this series1. In this current protocol we assume that these guidelines regarding sample ascertainment have been followed. However, even when case-control association study design has been conducted appropriately, a thorough assessment of data quality – including testing whether ‘same-population sampling’ was successful - should still be undertaken. Such assessments allow the identification of substandard markers and samples, which should be removed prior to further analysis in order to reduce the number of false-positive and false-negative associations.
Box 1. Glossary.
- Cochran-Armitage trend test
Statistical test for analysis of categorical data when categories are ordered. It is used to test for association in a 2 × k contingency table. In genetic associations studies, because the underlying genetic model is unknown the additive version of this test is most commonly used.
- Confounding
A type of bias in statistical analysis causing spurious or distorted findings caused by a correlation between an extraneous variable (the confounding variable) and both the dependent/exposure variable (e.g. the genotype at a given locus) and the independent/outcome variable (e.g. case-control status).
- Failure rate
The proportion of missing genotypes. Genotypes are classified as missing if the genotype-calling algorithm cannot infer the genotype with sufficient confidence. Can be calculated across each individual and/or SNP.
- False-negative
Occurs when a true disease-associated variant is not associated with disease in a given study.
- False-positive
Occurs when a variant not in truth associated with disease status is significantly associated with disease in a given study.
- Genotype calling algorithm
A statistical algorithm that, per marker and per individual, converts intensity data from two allelic probes into a single genotype for analysis.
- Genotype call rate
The proportion of genotypes per marker with non-missing data.
- Genotype cluster plots
Per SNP graphical representations of the intensity data from two probes used during genotyping across all individuals, together with the final called genotype. Typically, across all individuals, the intensity of probe A is plotted against the intensity of probe B and the genotype for a given individual is represented by one of three different colors. Genotypes of the same class should be seen to cluster together and these clusters should be consistent across case and control groups.
- HapMap
An international project to create a haplotype map of the human genome. The publicly available data consists of ~3.2 million SNPs genotyped across four different samples sets of 60-90 individuals of African, Asian or European Ancestry (stage II). HapMap stage III consists of ~1.5 million SNP genotypes from a greater number of individuals and populations.
- Hardy-Weinberg equilibrium
Given a minor allele frequency of q the probabilities of the three possible genotypes (aa, Aa, AA) at a biallelic locus which is in Hardy-Weinberg equilibrium are ((1−q)2, 2q(1−q), q2). In a large, randomly mating, homogenous population these probabilities should be stable from generation to generation.
- Heterozygosity rate
The proportion of heterozygous genotypes for a given individual.
- Informative missingness
Occurs when the probability of a genotype being called missing is correlated with the true underlying genotype.
- Linkage Disequilibrium
Non-random association of alleles at two or more loci.
- Pair-wise identity by state
The proportion of loci where a given pair of individuals share the same alleles. Given by (IBS2 + 0.5×IBS1) / (N SNP pairs) where IBS2 and IBS1 are the number of loci where the two individuals have 2 alleles and 1 allele in common, respectively and N SNP pairs is the number of common, non-missing, SNPs.
- Population substructure
The presence of distinct groups of individuals with subtle differences in allele frequency such that genetic data can be used to cluster these individuals into separate groups.
- Principal components analysis
A mathematical procedure for calculating a number of orthogonal latent variables that summarize a data matrix containing many potentially correlated variables.
- r2
A measure of the linkage disequilibrium (genetic correlation) between two markers. An r2 of 1 indicates that the two markers are perfectly correlated and an r2 of 0 indicates that the two markers are completely independent.
This protocol deals with the quality control (QC) of genotype data from genome-wide and candidate gene case-control association studies. While the protocol applies to genotypes after they have been determined (‘called’) from probe intensity data, it is still important to understand how the genotype calling was conducted. Traditionally, for small-scale genotyping efforts, manual inspection of allele-probe intensities is carried out to call genotypes, and this is still the situation for many candidate gene/replication studies conducted at present. However, when undertaking genome-wide association (GWA) with so many markers, this is no longer practical. Genotype calling algorithms (implemented in proprietary software accompanying the genotyping platform, or employed externally in software such as Illuminus2 or Chiamo3) use mathematical clustering algorithms to analyse the raw intensity data and - for a given individual and a given marker locus - estimate the probability that their genotype is aa, Aa and AA. A threshold is then applied such that any genotype with a probability exceeding a certain cut-off is accepted and referred to as “called”; otherwise the genotype is not accepted and is referred to as “uncalled” or missing. The threshold applied can heavily impact on the genotype call rate and quality of the genotype data. If it is set too low and the separation of signal clouds is poor, erroneous genotypes can be assigned. However, calling only genotypes with high certainty can result in ‘informative missingness’ because failure to call may be dependent on genotype. For example, rare homozygous genotypes may, on average, have lower probabilities, thus introducing bias to allele frequencies based only on called genotypes4. Furthermore, a high calling threshold will produce a high proportion of unnecessarily uncalled (missing) genotypes thus reducing genomic coverage and power to detect association. The ultimate assessment of genotype quality is manual inspection of cluster plots and it is essential that after association testing these are inspected for any SNPs taken forward for replication regardless of QC intensity (to prevent wasteful replication efforts).
Genome-wide association
Because of the large number of marker loci tested for association in a genome-wide association (GWA) study, even a low rate of error/bias can be detrimental. If one million markers are tested for association and the proportion of poorly genotyped markers is 0.001 then - if the inaccurate calling results in a spurious association being detected - up to 1000 markers may be unnecessarily taken forward for replication due to false-positive association. In an attempt to remove these false positive associations one must undertake several QC steps to remove individuals or markers with particularly high error rates. If, as advised, many thousands of cases and controls have been genotyped to maximise power to detect association, the removal of a handful of individuals should have little effect on overall power. Furthermore, given the large number of markers genotyped in modern GWA studies, the removal of a (hopefully) small percentage of these should not greatly decrease the overall power of the study. That said, every marker removed from a study is potentially an overlooked disease association and thus the impact of removing one marker is potentially greater than the removal of one individual (though genotype imputation can be used to recover these markers5). In this protocol we advocate implementing QC on a ‘per-individual’ basis prior to conducting QC on a ‘per-marker’ basis to maximise the number of markers remaining in the study. This approach prevents markers being erroneously removed due to a subset of poorly genotyped individuals, but is susceptible to individuals being falsely removed on the basis of a poorly genotyped subset of markers. An alternative (and conservative) approach would be to complete both QC stages prior to removing any individuals or markers but data may be removed unnecessarily.
Per-individual QC
Per-individual QC of GWA data consists of at least four steps, 1) identification of individuals with discordant sex information, 2) identification of individuals with outlying missing genotype or heterozygosity rate, 3) identification of duplicated or related individuals, and 4) identification of individuals of divergent ancestry.
It is useful to begin by using genotype data from the X-chromosome to check for discordance with ascertained sex and thus highlight plating errors. Because males only have one copy of the X-chromosome they cannot be heterozygous for any marker not in the pseudo-autosomal region of the Y chromosome. Typically, when a genotype-calling algorithm detects a male heterozygote for an X-chromosome marker it calls that genotype as missing. Therefore, female DNA samples that are marked as male in the input files will have a lot of missing data because all of their heterozygous genotypes will be set to missing. Not all genotype calling algorithms automatically set heterozygous haploid genotypes to missing and by calling X chromosome markers blind to ascertained sex this functionality can be removed in those that do. Typically one expects male samples to have a homozygosity rate of 1 (though due to genotyping error there is variation around this) and females to have a homozygosity rate less than 0.2. Male DNA samples that are itmarked as female in the input files will have a higher than expected homozygosity rate and female samples marked as male will have a lower than expected heterozygosity rate. Therefore, the best way to detect individuals where discrepancies exist between the genotype information and the ascertained sex is to calculate the homozygosity rate across all X-chromosome SNPs for each individual in the sample and compare these to the expected rate. The sex of a case or control is typically only of relevance when this data is to be used during analysis, for example when carrying out a sex-stratified analysis, or when analysing the X chromosome. However, when samples with discordant sex information are detected it is important that these are investigated to ensure that another DNA sample has not been genotyped by mistake (because the wrong (sub)phenotype data may be connected to the genotypes). Unless the sample can be correctly identified using existing genotype data or it can be confirmed that sex was recorded incorrectly then individuals with discordant sex information should be removed from further analysis.
Large variations exist in DNA sample quality and these can have large effects on genotype call rate and genotype accuracy. Samples of low DNA quality or concentration often have below average call rates and genotype accuracy. The genotype failure rate and heterozygosity rate per individual are both measures of DNA sample quality. Typically, individuals with more than 3-7% missing genotypes have been removed3,6. Carefully scrutinizing the distribution of missing genotype rates across the entire sample set is the best way to ascertain the most appropriate threshold. Likewise, the distribution of mean heterozygosity (excluding the sex chromosomes) across all individuals should be inspected to identify individuals with an excessive or reduced proportion of heterozygote genotypes, which may be indicative of DNA sample contamination or inbreeding, respectively. Mean heterozygosity (which is given by (N–O)/N, where N is the number of non-missing genotypes and O is the observed number of homozygous genotypes for a given individual) will differ between populations and SNP genotyping panels. Due to the increased success rate and accuracy of modern high-throughput genotyping methodologies (including genotype calling algorithms), typically these measures jointly lead to only a small proportion of individuals being excluded from further analysis.
A basic feature of standard population-based case-control association studies is that all the samples are unrelated (i.e. the maximum relatedness between any pair of individuals is less than a second degree relative). If duplicates, first- or second- degree relatives are present, a bias may be introduced to the study because the genotypes within families will be over-represented, and thus the sample may no longer be a fair reflection of the allele frequencies in the entire population. In population-based case-control studies, all efforts should be made to limit the number of duplicate and related individuals in the design phase of a study (although the deliberate inclusion of duplicate samples can be used to determine genotyping error rate)7. To identify duplicate and related individuals, a metric (identity by state, IBS) is calculated for each pair of individuals based on the average proportion of alleles shared in common at genotyped SNPs (excluding the sex chromosomes). The method works best when only independent SNPs are included in the analysis. To achieve this, regions of extended linkage disequilibrium (LD) (such as the HLA) are entirely removed from the dataset8 and remaining regions are typically pruned so that no pair of SNPs within a given window (say, 50kb) is correlated (typically taken as r2>0.2). Following the calculation of IBS between all pairs of individuals, duplicates are denoted as those with an IBS of 1. The population mean IBS will vary depending on the allele frequency of genotyped markers within that population. Related individuals will share more alleles IBS than expected by chance, with the degree of additional sharing proportional to the degree of relatedness. The degree of recent shared ancestry for a pair of individuals (identity by descent, IBD) can be estimated using genome-wide IBS data (using software such as PLINK9. The expectation is that IBD = 1 for duplicates or monozygotic twins, IBD = 0.5 for first-degree relatives, IBD = 0.25 for second-degree relatives and IBD = 0.125 for third-degree relatives. Due to genotyping error, LD and population structure there is often some variation around these theoretical values and it is typical to remove one individual from each pair with an IBD > 0.1875, which is halfway between the expected IBD for third- and second-degree relatives. For these same reasons an IBD > 0.98 identifies duplicates.
Confounding can be a major source of bias in population-based case-control studies and is caused by underlying differences between the case and control subgroups other than those directly under study (typically, disease status), which correlate with the exposure variable. In the case of genetic studies, where the exposure of interest is genotype distribution, the main source of confounding is population stratification, in which genotypic differences between cases and controls are generated because of different population origins rather than any effect on disease risk10. For example, Campbell et al11 carried out association analysis on a panel of European American individuals discordant for height and detected significant association to LCT, a locus which has undergone strong selection in certain European populations and the frequency of variants within this gene differ greatly between populations. After matching cases and controls for population ancestry the evidence of association at this locus greatly decreased. While a well-designed population case-control study attempts to draw cases and controls from the same population, hidden fine-scale genetic substructure within that single population (or the inadvertent inclusion of individuals from another population) cannot be ruled out. The confounding occurs when the population substructure is not equally distributed between the case and control groups. In this scenario, a signal of association will arise for an ancestrally informative SNP, not because of an association with disease risk, but because of allele frequency differences between the founder populations that differentially comprise the cases and controls. Even a small degree of population stratification can adversely affect a GWA study due to the large sample sizes required to detect common variants underlying most complex diseases12. Therefore, after giving careful consideration to matching of cases and controls on population origin1, potential stratification must be examined and characterised during QC. Efforts should then be made to remove or reduce the effect of population stratification through the removal of individuals of divergent ancestry. Correction for fine-scale, or within-population, substructure can be attempted during association testing and this is detailed in a subsequent protocol in this series, together with methods for assessing the effect of confounding on genome-wide association test statistics.
The most common method for identifying (and subsequently removing) individuals with large-scale differences in ancestry is principal components analysis (PCA)13,14. An alternative yet related method, multidimensional scaling (implemented in PLINK), is available but requires a pair-wise IBD matrix to be constructed and is therefore more computationally complex. PCA is a multivariate statistical method used to produce a number of uncorrelated variables (or principal components) from a data matrix containing observations across a number of potentially correlated variables. The principal components are calculated so that the first principal component accounts for as much variation in the data as possible in a single component, followed then by the second component and so on. When using PCA to detect ancestry the observations are the individuals and the potentially correlated variables are the markers. A principal component model is built using pruned genome-wide genotype data from populations of known ancestry, for example to detect large-scale (continental level) ancestry one could use the HapMap genotype data from Europe (CEU), Asia (CHB+JPT) and Africa (YRI)15,16. Due to the large-scale genetic differences between these three ancestral groups the first two principal components are sufficient to separately cluster individuals from the three populations. The PCA model can then be applied to the GWA individuals to allow prediction of principal component scores for these samples, thus allowing them to be clustered in terms of ancestry alongside the HapMap samples. A common set of (approximately 50,000 independent markers must be used for the model building and prediction steps. Region of extended high LD (such as the HLA) should be removed prior to the analysis because these can overly influence the principal components model8. The method can also be used to cluster individuals based on fine-scale structure, though more principal components may be needed to fully capture this variation, and appropriate reference samples will be required.
Per-marker QC
Per-marker QC of GWA data consists of at least four steps, 1) identification of SNPs with an excessive missing genotype, 2) identification of SNPs demonstrating a significant deviation from Hardy-Weinberg equilibrium (HWE), 3) identification of SNPs with significantly different missing genotype rates between cases and controls and 4) the removal of all makers with a very low minor allele frequency.
The removal of sub optimal markers is key to the success of a GWA study, because they can present as false-positives and reduce the ability to identify true associations correlated with disease-risk. However, the criteria used to filter out low quality markers differ from study to study. Great care must be taken to only remove poorly characterised markers because every removed marker is potentially a missed disease variant. Classically, markers with a call rate less than 95% are removed from further study6,17, though some studies have chosen higher call-rate thresholds (99%) for markers of low frequency (minor allele frequency (MAF) <5%)3.
Most GWA studies choose to exclude markers that show extensive deviation from Hardy-Weinberg equilibrium (HWE) because this can be indicative of a genotyping or genotype calling error. However, deviations from Hardy-Weinberg equilibrium may also indicate selection, so a case sample can show deviations from HWE at loci associated with disease, and it would obviously be counter-productive to remove these loci from further investigation 18. Therefore, only control samples should be used when testing for deviations for HWE. The significance threshold for declaring SNPs to be in Hardy-Weinberg equilibrium has varied greatly between studies (p-value thresholds between 0.001 and 5.7 × 10−7 have been reported in the literature3,19). However, those studies which have set very low thresholds for HWE deviations have done so on proviso that all genotype cluster plots for SNPs showing some evidence of deviation from HWE (say, p<0.001) will be examined manually for quality. In practice this means that many SNPs with a HWE p-value less than 0.001 will be removed, though robustly genotyped SNPs below this threshold remain under study.
Testing for, and subsequently removing, SNPs with significant differences in missing genotype rate between cases and controls is another means of reducing confounding and removing poorly genotyped SNPs20. Calling case and control genotypes together, or using ‘fuzzy calls’21, greatly reduces this confounding but significant differences in genotype failure may still exist in the data and present as false-positive associations. In studies where cases and/or controls have been drawn from several different sources it is wise to test for significant differences in call rate, allele frequency and genotype frequency between these various groups to ensure that it is fair to treat the combined case or control set as one homogenous group.
The final step when conducting QC is to remove all SNPs with a very low MAF. Typically a MAF threshold of 1-2% is applied but studies with small sample size may need to set this threshold higher. The small size of the heterozygote and rare homozygote clusters makes these variants difficult to call using current genotype calling algorithms and they frequently present as false-positives in case-control association tests. Furthermore, even when well called, association signals seen at these rare SNPs are less robust because they are driven by the genotypes of only a few individuals. Given that power to detect association at rare variants is so low22, their removal does not overly impact on the study. However, even following the removal of rare variants and stringent individual and SNP QC, genotyping errors may still persist. Checking cluster plots manually is the best way to ensure genotype calls are robust and therefore it is essential that all SNPs associated with disease status be manually inspected for clustering errors prior to choosing SNPs for follow-up genotyping.
Candidate-gene association
Candidate gene association studies involve far fewer SNPs than GWA studies and therefore many of the GWA study QC procedures cannot be undertaken. One of the advantages of the GWA study approach is that more than 99% of the SNPs follow the null distribution of no association and can be used to detect evidence of confounding. This is not possible in a candidate gene approach because a) owing to the gene's candidacy there may be few SNPs falling under the null hypothesis of no association and b) far fewer SNPs are genotyped. With fewer genotyped SNPs, it is also more difficult to get accurate estimates of a) DNA quality through genotype failure rate and heterozygosity rate, b) population ancestry and c) familial relationships with others in the study. The detrimental effect these factors can have on a candidate gene association may be equal to that under the GWA scenario (although in a well-designed study of ethnically matched individuals, the prior probability of population stratification at a single locus is much lower than that of stratification at any locus across the genome) except here our ability to identify and remove erroneous individuals and SNPs is greatly reduced. This is perhaps another reason why candidate gene studies have typically not yielded many reproducible disease gene associations (in addition to the use of small samples size, poor coverage of genetic variation, and poor choice of candidates).
One should still attempt to identify and remove individuals with exceptionally low call rate. However, the threshold at which individuals are excluded will vary depending on the number of SNPs genotyped and will typically be higher than that used when carrying out a GWA study. For example, if a candidate gene study included 50 SNPs then removing individuals with more than 3% missing data would result in removing individuals missing more than only 2 SNPs. A more reasonable approach would be to remove those individuals missing 10 or more SNPs (a failure rate of 0.2).
QC of markers in candidate gene studies is more comparable to the GWA study approach as similar numbers of cases and controls should be involved. It is extremely important to examine the failure rate of the markers included in the candidate-gene study and exclude those with a high failure rate. When a SNP is identified with a high failure (>5%) an option is to return to the lab and attempt to re-genotype that SNP in the individuals with missing data. Given that SNPs included in a candidate gene study are chosen based on their ability to tag neighbouring SNPs23, the exclusion of a SNP due to elevated failure rate can seriously impair a candidate gene study. With this in mind, when a SNP is not genotyped with sufficient quality across individuals (and this can not be rectified by re-genotyping) it is advisable to return to the design stage and select another tag for the haplotype block in which the failed SNP resides. Detection of deviations from HWE in controls is still a relevant method for checking genotyping quality
Software
Standard statistical software (such as R24 or SPSS) can be used to conduct all of the analyses outlined above. However, for GWA studies, many researchers choose to use custom-built, freely available software such as PLINK9, GenABEL25, GS226 or snpMatrix (an R package which forms part of the bioconductor project (http://www.bioconductor.org/). These software can also be used for candidate gene association studies. The advantages of these compared to standard statistical software are a) they store the large genome-wide SNP data in memory efficient data structures, thus significantly improving computational efficiency and reducing disk usage and b) they more fully automate many of the necessary analyses. Although PLINK utilised in the present protocol, any of the other packages are equally suitable.
The next section describes protocols for QC of GWA and candidate gene data, respectively. The protocol is illustrated by the use of simulated datasets, which are available for download.
MATERIALS
EQUIPMENT
Genome-wide association
Data
- Genome-wide SNP data and software scripts
Software
Candidate gene association
Data
- Candidate-gene SNP data and software scripts
Software
Computer workstation with Unix/Linux operating systems
- PLINK software9 for genome-wide association analysis:
- Statistical software for data analysis and graphing such as:
- SPSS
PROCEDURE
Creation of BED files
1| The format in which genotype data are returned to investigators varies between genome-wide SNP platforms and genotyping centers. We assume that genotypes have been called by the genotyping centre and returned in the standard .ped and .map file formats (see Box 2). To obtain example genomewide .ped and .map files download the file raw-GWA-data.tgz (for candidate gene studies please see Box 3).
2| Type ‘tar xfvz raw-GWA-data.tgz’ at the shell prompt to unpack the gzipped .tar file and create the files raw-GWA-data.map and raw-GWA-data.ped.
-
3| Use Plink to create the BED, BIM and FAM files. type ‘plink --file raw-GWA-data --make-bed --out raw-GWA-data’ at the shell prompt.
▲ critical step BED files save the data in a more memory and time efficient manner (binary files) so facilitate the analysis of large-scale datasets9. PLINK creates a .log file (named raw-GWA-data.log) which details (amongst other information) the implemented commands, the number of cases and controls in the input files, any excluded data and the genotyping rate in the remaining data. This file is very useful for checking the software is successfully completing commands.
▲ critical step If genotypes are not available in PED file format GS226,27 and Plink both have functionality to read several other file formats and convert these into PED files (or even directly into BED files).
BOX2. PED and MAP files.
A PED file is a white-space (space or tab) delimited file where each line represents one individuals and the first six columns are mandatory and in the order ‘Family ID’, ‘Individual ID’, ‘Paternal ID’, ‘Maternal ID’, ‘Sex (1 = male, 2 = female, 0 = missing)’ and ‘Phenotype (1 = unaffected, 2 = affected, 0 = missing)’. The subsequent columns denote genotypes which can be any character (e.g. 1,2,3,4 or A,C,G,T). 0 denotes a missing genotype. Each SNP must have two alleles (i.e. both alleles are either present or absent). The order of SNPs in the PED file is given in the MAP file, where each line denotes a single marker and the four white-space separated columns are ‘Chromosome (1-22, X, Y or 0 for unplaced)’, ‘Marker name (typically a rs number)’, ‘Genetic distance in Morgans (this can be fixed to 0)’ and ‘Base-pair position (bp units)’.
Box 3. Candidate gene study.
Creation of PED files
1| The format in which genotype data are returned to investigators depends on where the genotyping was conducted and on which platform. We assume that genotypes have been called by the genotyping centre and returned in the standard .ped and .map file formats (see Box 1). Download the file ‘raw-PPARG-data.tgz’.
2| Type ‘tar xfvz raw-PPARG-data.tgz’ at the shell prompt to unpack the gzipped .tar file and create the files ‘raw-PPARG-data.map’ and ‘raw-PPARG-data.ped’.
Test markers for different genotype call rates between cases and contols
3| (At the Unix prompt type ‘plink --bfile raw-PPARG-data --test-missing --out raw-PPARG-data’ to test all markers for differences in call rate between cases and controls. The output of this test can be found in raw-PPARG-data.missing.
4| At the unix prompt type ‘perl run-diffmiss-qc.pl clean-inds-GWA-data’ to create a file called ‘fail-diffmiss-qc.txt’ which contains all SNPs with a significantly different (P<0.00001) missing data rate between cases and controls.
Run additional QC steps and remove failing markers and samples
5| At the unix prompt type ‘plink --file raw-PPARG-data --exclude fail-diffmiss-qc.txt –mind 0.1 –-maf 0.01 --geno 0.05 --hwe 0.00001 --recode --out clean-PPARG-data’ . . In addition to markers failing previous QC steps, those with a MAF<0.01 and a HWE P-value<0.00001 (in controls) are also removed. Samples with a genotype failure rate greater than 0.1 are also removed.
Identification of individuals with discordant sex information
4| At the Unix prompt type ‘plink --bfile raw-GWA-data --check-sex --out raw-GWA-data’ to calculate the mean homozygosity rate across X chromosome markers for each individual in the study.
5| Produce a list of individuals with discordant sex data by typing ‘grep PROBLEM raw-GWA-data.sexcheck > raw-GWA-data.sexprobs’ and open the file to obtain the family ids (column 1) and individuals id (column 2) for these individuals. Column 3 gives the ascertained sex and column 4 denotes the sex according to the genotype data. When the homozygosity rate is more than 0.2 but less than 0.8 the genotype data is inconclusive regarding the sex of an individual and these are marked in column 4 with a 0.
6| Report the IDs of individuals with discordant sex information to those who conducted sex phenotyping. Where the discrepancy cannot be resolved, add the family ID (FID) and individual ID (IID) of the samples to a file called ‘fail-sexcheck-qc.txt’ (one individual per line, tab delimited).
Identification of individuals with elevated missing data rates or outlying heterozygosity rate
7| At the shell prompt type ‘plink --bfile raw-GWA-data --missing --out raw-GWA-data’ to create the files raw-GWA-data.imiss and raw-GWA-data.lmiss. The fourth column in the imiss file (N_MISS) gives the number of missing SNPs and the sixth column (F_MISS) gives the proportion of missing SNPs per individual.
8| At te shell prompt type ‘plink --bfile raw-GWA-data --het --out raw-GWA-data’ to create the file raw-GWA-data.het where the third column gives the observed number of homozygous genotypes [O(Hom)] and the fifth column gives the number of non-missing genotypes [N(NM)], per individual.
9| Calclate the observed heterozygosity rate per individual using the formula (N(NM) - O(Hom))/N(NM). Create a graph where the observed heterozygosity rate per individual is plotted on the x-axis and the proportion of missing SNPs per individuals is plotted on the y-axis. This can be done using standard software such as Excel or statistical packages such as SPSS. A script for calculating the heterozygosity rate and producing the graph using R has been supplied (imiss-vs-het.Rscript). Type ‘R CMD BATCH imiss-vs-het.Rscript’ at the unix prompt to run this script and create the graph (raw-GWA-data.imiss-vs-het.pdf).
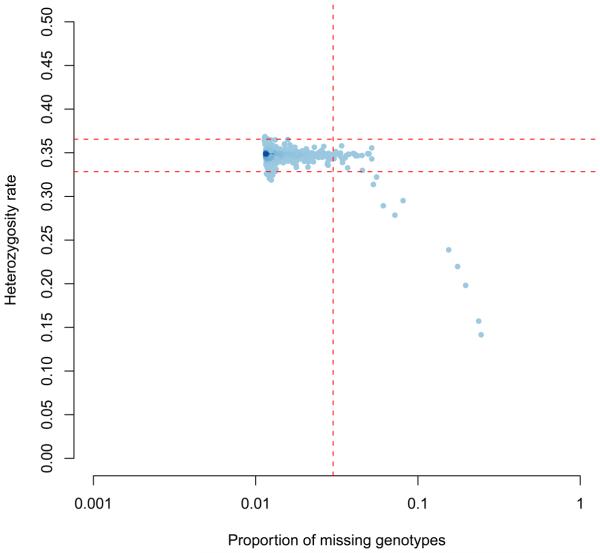
10| Exmine the plot to decide reasonable thresholds at which to exclude individuals based on elevated missing or extreme heterozygosity. We chose to exclude all individuals with a genotype failure rate ≥ 0.03 (Fig 1, vertical dashed line) and/or heterozygosity rate ± 3 standard deviations from the mean (Fig 1, horizontal dashed lines). Add the family ID and individual ID of the 30 samples failing this QC to a file named ‘fail-imisshet-qc.txt’.
Figure 1.
Genotype failure rate vs. heterozygosity across all individuals the study. Shading indicates sample density and dashed lines denote QC thresholds.
Identification of duplicated or related individuals
- 11| To reduce the computational complexity the number of SNPs used to create the IBS matrix can be reduced by pruning the dataset so that no pair of SNPs (within a given number of base-pairs) has an r2 greater that a given threshold (typically 0.2). Given that our current dataset was simulated ignoring LD this step is not applicable, but a list of SNPs for inclusion in this step can be downloaded from http://www.well.ox.ac.uk/~carl/gwa/nature-protocols/raw-GWA-data.prune.in.
- ▲ critical step In real datasets where LD is present the data can be pruned by typing at the shell prompt ‘plink --file raw-GWA-data --exclude high-LD-regions.txt --range --indep-pairwise 50 5 0.2 --out raw-GWA-data’ to create the file raw-GWA-data.prune.in, the list of SNPs to be kept in the analysis. This also excludes SNPs from extended regions of high LD listed in high-LD-regions.txt.
- 12| Type ‘plink --bfile raw-GWA-data –-extract raw-GWA-data.prune.in –-genome --out raw-GWA-data’ at the shell prompt to generate pair-wise IBS for all pairs of individuals in the study based on the reduced marker set.
- ▲ critical step Because this step can take much time it is advised to prefix above command with the Unix command nohup to allow the command to continue running on the machine after user log out. Placing an ampersand (&) at the end of the command will free the Unix terminal for further use.
13| Type ‘perl run-IBD-QC.pl raw-GWA-data’ at the unix prompt to identify all pairs of individuals with an IBD > 0.185. The code looks at the individual call rates stored in raw-GWA-data.imiss and output the ids of the individual with the lowest call-rate to ‘fail-IBD-QC.txt’ for subsequent removal
Identification of individuals of divergent ancestry
14| This step is conducted by merging study genotypes to HapMap phase 3 data from four ethnic populations. The genotypes from the two studies must match for strand so not to introduce error. Because not all SNPs are required for this analysis the more difficult to align A→T and C→G SNPs can be omitted. To create a new bed file excluding from the GWA data those SNPs which do not feature in the genotype data of the four original HapMap populations (HM3 data) type ‘plink --bfile raw-GWA-data –-extract hapmap3r2_CEU.CHB.JPT.YRI.no-at-cg-snps.txt --make-bed --out raw-GWA-data.hapmap-snps’ at the unix prompt.
- 15| Merge the raw-GWA-data.hapmap-snps files with the HapMap data and extract the pruned SNP set by typing ‘plink --bfile raw-GWA-data. hapmap-snps --bmerge hapmap3r2_CEU.CHB.JPT.YRI.founders.no-at-cg-snps.bed hapmap3r2_CEU.CHB.JPT.YRI.founders.no-at-cg-snps.bim hapmap3r2_CEU.CHB.JPT.YRI.founders.no-at-cg-snps.fam --extract raw-GWA-data.prune.in --make-bed --out raw-GWA-data.hapmap3r2.pruned’.
- ▲ critical step It is likely that the merge will not complete and PLINK will terminate with the message ‘ERROR: Stopping due to mis-matching SNPs -- check +/− strand?’. Read raw-GWA-data.hapmap3r2.log to see this message. Because all A→T and C→G SNPs have been removed prior to undertaking this analysis all SNPs which are discordant for strand between the two data sets are listed in raw-GWA-data.hapmap3r2.pruned.missnp. To align the strands across the data sets and successfully complete the merge simply repeat step (i) including the command ‘--flip raw-GWA-data.hapmap3r2.pruned.missnp’ and then repeat (ii).
16| create a copy of the bim and fam files by typing ‘cp raw-GWA-data.hapmap3r2.pruned.bim raw-GWA-data.hapmap3r2.pruned.pedsnp’ followed by ‘cp raw-GWA-data.hapmap3r2.pruned.fam raw-GWA-data.hapmap3r2.pruned.pedind’ at the Unix prompt.
17| conduct a principal components analysis on the merged data by typing ‘perl smartpca.perl -i raw-GWA-data.hapmap3r2.pruned.bed -a raw-GWA-data.hapmap3r2.pruned.pedsnp-b raw-GWA-data.hapmap3r2.pruned.pedind -o raw-GWA-data.hapmap3r2.pruned.pca -p raw-GWA-data.hapmap3r2.pruned.plot -e raw-GWA-data.hapmap3r2.pruned.eval -l raw-GWA-data.hapmap3r2.pruned.log -k 2 -t 2 -w pca-populations.txt’.
18| create a scatter diagram of the first two principal components, including all individuals in the file raw-GWA-data.hapmap3r2.pruned.pca.evec (the first and second principal components are columns 2 and 3 respectively). Use the data in column 4 to color the points according to sample origin. An R script for creating this plot (plot-pca-results.Rscript) has been provided (though any standard graphing software can be used).
- 19| Derive PC1 and PC2 thresholds so that only individuals who match the given ancestral population are included. For populations of European descent this will be either the CEU or TSI HapMap3 individuals. Here, we chose to exclude all individuals with a 2nd principal component score less than 0.072. Write the FID and IID of these individuals to a file called ‘fail-ancestry-QC.txt’.
- ▲ critical step Choosing which thresholds to apply (and thus which individuals to remove) is not a straightforward process. The key is to remove those individuals with greatly divergent ancestry as these samples introduce the most bias to the study. Identification of more fine-scale ancestry can be conducted by using less divergent reference samples (for example, within Europe stratification could be identified using the CEU, TSI (Italian), GBR (British), FIN (Finnish) and IBS (Iberian) samples from the 1000 genomes project (http://www.1000genomes.org)). Robust identification of fine-scale population structure often requires the construction of many (2-10) principal components.
Remove all individuals failing QC
20| At the unix prompt type ‘cat fail-* | sort -k1 | uniq > fail-qc-inds.txt’ to concatenate all the files listing individuals failing the previous QC steps into single file.
21| The file fail-qc-inds.txt should now contain a list of unique individuals failing the previous QC steps. To remove these from the dataset type ‘plink --bfile raw-GWA-data --remove fail-qc-inds.txt --make-bed --out clean-inds-GWA-data’ at the unix prompt.
Identify all markers with an excessive missing data rate
22| To calculate the missing genotype rate for each marker type ‘plink --bfile clean-inds-GWA-data --missing --out clean-inds-GWA-data’. The results of this analysis can be found in clean-inds-GWA-data.lmiss.
23| Plot a histogram of the missing genotype rate to identify a threshold for extreme genotype failure rate. This can be done using the data in column five of the clean-inds-GWA-data.lmiss file and any standard statistical/graphing software package. A script for creating this histogram in R has been provided (lmiss-hist.Rscript). We chose to a call-rate threshold of 3% (these SNPs will be removed later in the protocol).
Test markers for different genotype call rates between cases and contols
24| At the Unix prompt type ‘plink --bfile clean-inds-GWA-data --test-missing --out clean-inds-GWA-data’ to test all markers for differences in call rate between cases and controls. The output of this test can be found in clean-inds-GWA-data.missing.
25| At the unix prompt type ‘perl run-diffmiss-qc.pl clean-inds-GWA-data’ to create a file called ‘fail-diffmiss-qc.txt’ which contains all SNPs with a significantly different (P<0.00001) missing data rate between cases and controls.
Remove all markers failing QC
26| To remove poor SNPs from further analysis and create a clean GWA data file type ‘plink --bfile clean-inds-GWA-data --exclude fail-diffmiss-qc.txt –-maf 0.01 --geno 0.05 --hwe 0.00001 --make-bed --out clean-GWA-data’ at the Unix prompt. In addition to markers failing previous QC steps, those with a MAF<0.01 and a HWE P-value<0.00001 (in controls) are also removed.
● Timing
Steps 1-3, Creation of bed files: ~20 minutes
Steps 4-21, Individuals level QC: ~7 hours
Steps 22-26, Marker level QC: ~30 minutes
Inexperienced analysts will typically require more time. Given the computational nature of this protocol, timings will also vary with computational resources.
TROUBLESHOOTING
For help on the programs and websites used in this protocol, refer to the relevant websites:
ANTICIPATED RESULTS
A. Genome-wide association studies
Identify individuals with elevated missing data rates or outlying heterozygosity rate
Examining a plot of genotype failure rate vs. heterozygosity across all individuals in a study allows one to identify samples introducing bias to a study (Figure1). We chose to exclude all individuals with a genotype failure rate ≥ 0.03 (Fig 1, vertical dashed line) and/or heterozygosity rate ± 3 standard deviations from the mean (Fig 1, horizontal dashed lines).
Identify individuals of divergent ancestry
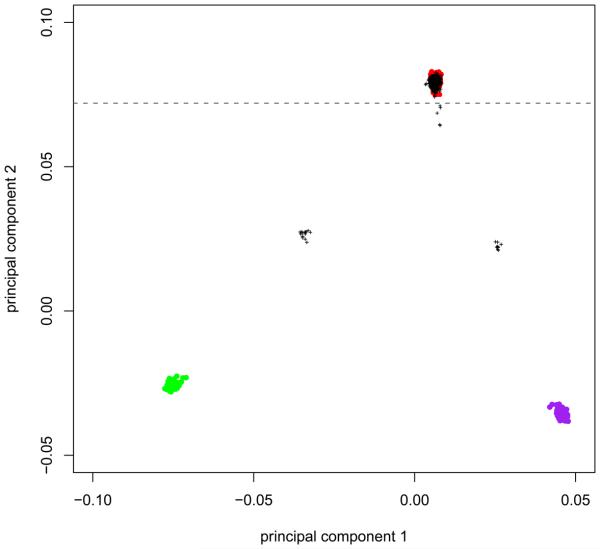
Principal component models were built using the CEU, CHB+JPT and YRI HapMap3 samples and then used to predict principal component scores for the cases and controls (of supposed European ancestry) using smartpca.pl (Figure 2). We chose to exclude 30 individuals that clustered away from the HapMap European samples (CEU). Depending on the population from which cases and controls are selected, the GWA study samples may not cluster precisely with population reference samples. Population stratification can still be reduced by removing individuals that lie away from the main cluster of GWA study samples (even if these do not cluster over a reference population). Alternatively, one can use more appropriate population reference samples in the PCA analysis.
Figure 2.
Ancestry clustering based on genome-wide association data. HapMap3 reference samples: CEU (red), CHB+JPT (purple) and YRI (green). GWA samples: black crosses. 11 cases and 19 controls with a 2nd principal component score less than 0.072 (grey dashed line) were marked for removal.
Identify all markers with an excessive missing data rate
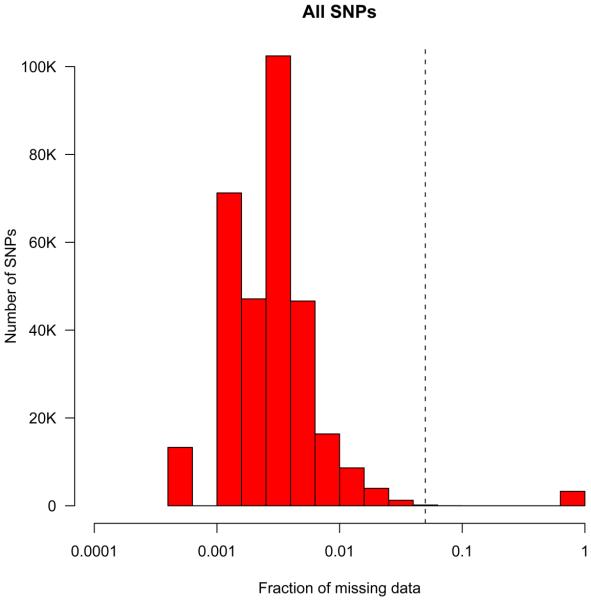
Scrutinizing the distribution of SNP call rate is one of the best ways to assess the success of the genotyping experiment (Figure 3). SNP with an excessive amount of missing genotypes should be removed to reduce false-positives.
Figure 3.
Histogram of missing data rate across all individuals passing ‘per-individual’ QC. The dashed vertical line represents the threshold (3%) at which SNPs were removed from further analysis due to an excess failure rate.
B. Candidate gene study
Test markers for different genotype call rates between cases and controls
For the PPARG SNPs in the file provided, no SNPs failed this QC check.
Run additional QC steps and remove failing markers and samples
The clean-PPARG-data files should contain 1971 cases and 1989 controls genotyped across 25 SNPs with a genotype call rate of 0.986.
Acknowledgements
CAA is funded by the Wellcome Trust (WT91745/Z/10/Z). APM is supported by a Wellcome Trust Senior Research Fellowship. KTZ is supported by a Wellcome Trust Research Career Development Fellowship.
Footnotes
The authors declare that they have no competing financial interests
REFERENCES
- 1.Zondervan KT, Cardon LR. Designing candidate gene and genome-wide case-control association studies. Nat Protoc. 2007;2:2492. doi: 10.1038/nprot.2007.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teo YY, et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 5.Marchini J, Howie B, Myers SR, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 6.Silverberg MS, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: causes, consequences and solutions. Nat Rev Genet. 2005;6:847. doi: 10.1038/nrg1707. [DOI] [PubMed] [Google Scholar]
- 8.Price AL, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 11.Campbell CD, et al. Demonstrating stratification in a European American population. Nat Genet. 2005;37:868. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 12.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1616. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 13.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.Fisher SA, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:967. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyre D, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 20.Moskvina V, Craddock N, Holmans P, Owen MJ, O'Donovan MC. Effects of differential genotyping error rate on the type I error probability of case-control studies. Hum Hered. 2006;61:55. doi: 10.1159/000092553. [DOI] [PubMed] [Google Scholar]
- 21.Plagnol V, Cooper JD, Todd JA, Clayton DG. A method to address differential bias in genotyping in large-scale association studies. PLoS Genet. 2007;3:e74. doi: 10.1371/journal.pgen.0030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34:188. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersson FH, et al. Marker selection for genetic case-control association studies. Nat Protoc. 2009;4:743. doi: 10.1038/nprot.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Development Core Team R: A language and environment for statistical computing. 2005 [Google Scholar]
- 25.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 26.Pettersson F, Morris AP, Barnes MR, Cardon LR. Goldsurfer2 (Gs2): a comprehensive tool for the analysis and visualization of genome wide association studies. BMC Bioinformatics. 2008;9:138. doi: 10.1186/1471-2105-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersson F, Jonsson O, Cardon LR. GOLDsurfer: three dimensional display of linkage disequilibrium. Bioinformatics. 2004;20:3241. doi: 10.1093/bioinformatics/bth341. [DOI] [PubMed] [Google Scholar]