Abstract
Transgenic plants and associated bacteria constitute a new generation of genetically modified organisms for efficient and environmental-friendly treatment of soil and water contaminated with polychlorinated biphenyls (PCBs). This review focuses on recent advances in phytoremediation for the treatment of PCBs, including the development of transgenic plants and associated bacteria. Phytoremediation, or the use of higher plants for rehabilitation of soil and groundwater, is a promising strategy for cost-effective treatment of sites contaminated by toxic compounds, including toxic PCBs. Plants can help mitigate environmental pollution by PCBs through a range of mechanisms: besides uptake from soil (phytoextraction), plants are capable of enzymatic transformation of PCBs (phytotransformation); by releasing a variety of secondary metabolites, plants also enhance the microbial activity in the root zone, improving biodegradation of PCBs (rhizoremediation). However, because of their hydrophobicity and chemical stability, PCBs are only slowly taken up and degraded by plants and associated bacteria, resulting in incomplete treatment and potential release of toxic metabolites into the environment. Moreover, naturally occurring plant-associated bacteria may not possess the enzymatic machinery necessary for PCB degradation. In order to overcome these limitations, bacterial genes involved in the metabolism of PCBs, such as biphenyl dioxygenases, have been introduced into higher plants, following a strategy similar to the development of transgenic crops. Similarly, bacteria have then been genetically modified that exhibit improved biodegradation capabilities and are able to maintain stable relationships with plants. Transgenic plants and associated bacteria bring hope for a broader and more efficient application of phytoremediation for the treatment of PCBs.
Introduction
Phytoremediation is an emerging technology that uses plants and associated bacteria for the treatment of soil and groundwater contaminated by toxic pollutants (1). The concept of using plants for remediation of organic pollutants emerged a few decades ago with the recognition that plants were capable of metabolizing toxic compounds, such as 1,1,1-trichloro-2,2-bis-(4'-chlorophenyl) ethane (DDT) and benzo[a]pyrene (2,3). Since then, phytoremediation acquired the status of a proven technology for the remediation of soil and groundwater contaminated by a variety of organic compounds, including pesticides, chlorinated solvents, explosives, polyaromatic hydrocarbons (PAHs), dioxins, and polychlorinated biphenyls (PCBs) (1,4–7). It is estimated that the budget invested in phytoremediation programs jumped from 50 million dollars in 1999 to 300 million dollars in 2007 (8).
Even though phytoremediation has been shown to efficiently reduce chemical hazards associated with various classes of organic and inorganic pollutants, it also suffers serious limitations that prevent large-scale field applications (1,7). As autotrophic organisms, plants usually lack the catabolic enzymes necessary to achieve full metabolism of recalcitrant organic compounds, often resulting in slow removal and incomplete degradation (9). Inherent limitations of plants for the metabolism of recalcitrant xenobiotic compounds led to the idea of modifying plants genetically by the introduction of bacterial or mammalian genes involved in the degradation of toxic chemicals, following a strategy that has been applied for decades with transgenic crops (10–12). Similarly, even though rhizoremediation plays a key role in the transformation of organic pollutants, naturally occurring plant-associated bacteria may not harbor the enzymatic machinery necessary for the efficient degradation of PCBs. In order to overcome this limitation, genetically modified bacteria have been constructed that exhibit improved biodegradation capabilities and are able to maintain stable relationships with plants. Transgenic plants and associated bacteria for phytoremediation could therefore constitute the new generation of genetically modified (GM) organisms (13).
Although phytoremediation technology has been extensively reviewed in the literature (4,6,9,13–24), only very few reviews have been published that focus specifically on PCBs (15,25,26). The present article summarizes new progress in phytoremediation of PCBs, including the use of transgenic plants and plant-associated bacteria.
Phytoremediation: Cleaning up Pollution with Plants
Living organisms are commonly exposed to a variety of natural (allelochemicals) or manmade toxic chemicals (xenobiotics). As a consequence, they have developed complex detoxification mechanisms to prevent harmful effects from exposure to these compounds (27–29). Bioremediation exploits the natural capability of living organisms to degrade toxic chemicals. Traditional remediation of PCB-polluted sites requires soil excavation and transport, prior to off-site treatment by solvent extraction, thermal alkaline dechlorination, incineration, or landfilling (15). These techniques are costly, damaging for the environment, and, in many cases, practically infeasible due to the range of the contamination (19). There is therefore a considerable interest in developing cost-effective alternatives based on microorganisms or plants. Bioremediation techniques, although requiring more time, are usually considered to represent between 10 and 50% of the cost of physical and chemical methods (7). Because of its potential for the sustainable mitigation of environmental pollution, bioremediation has been listed among the 'top ten technologies for improving human health' (30).
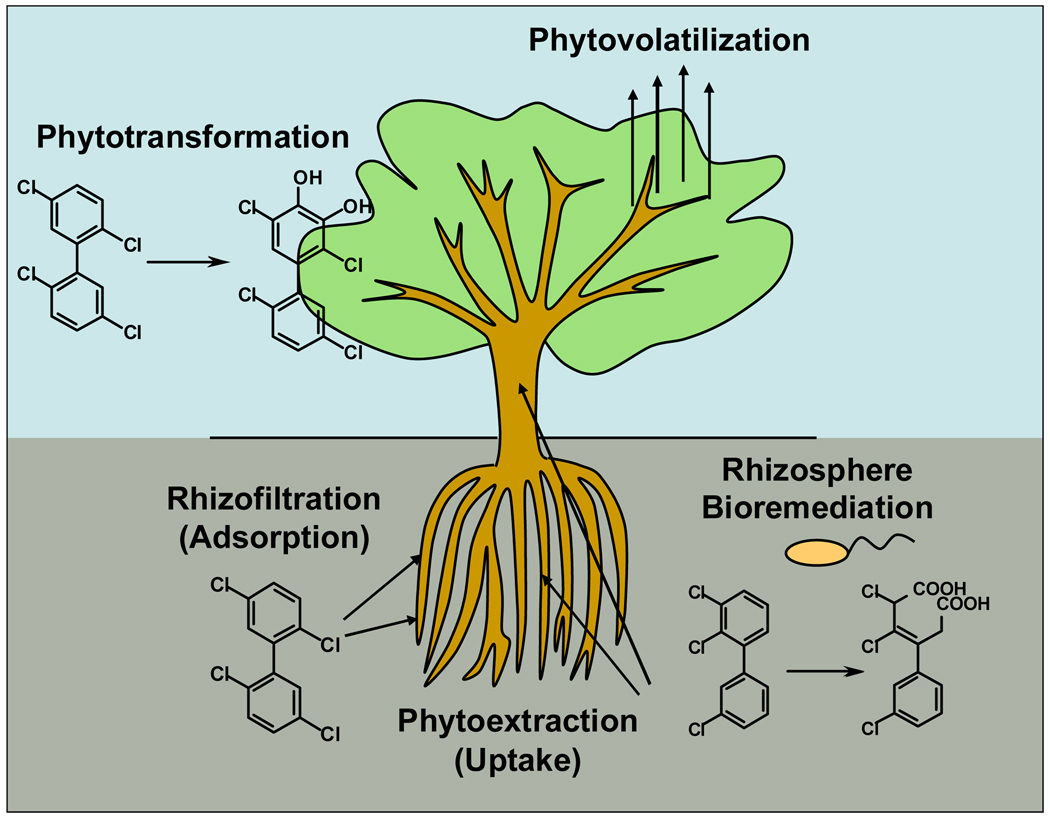
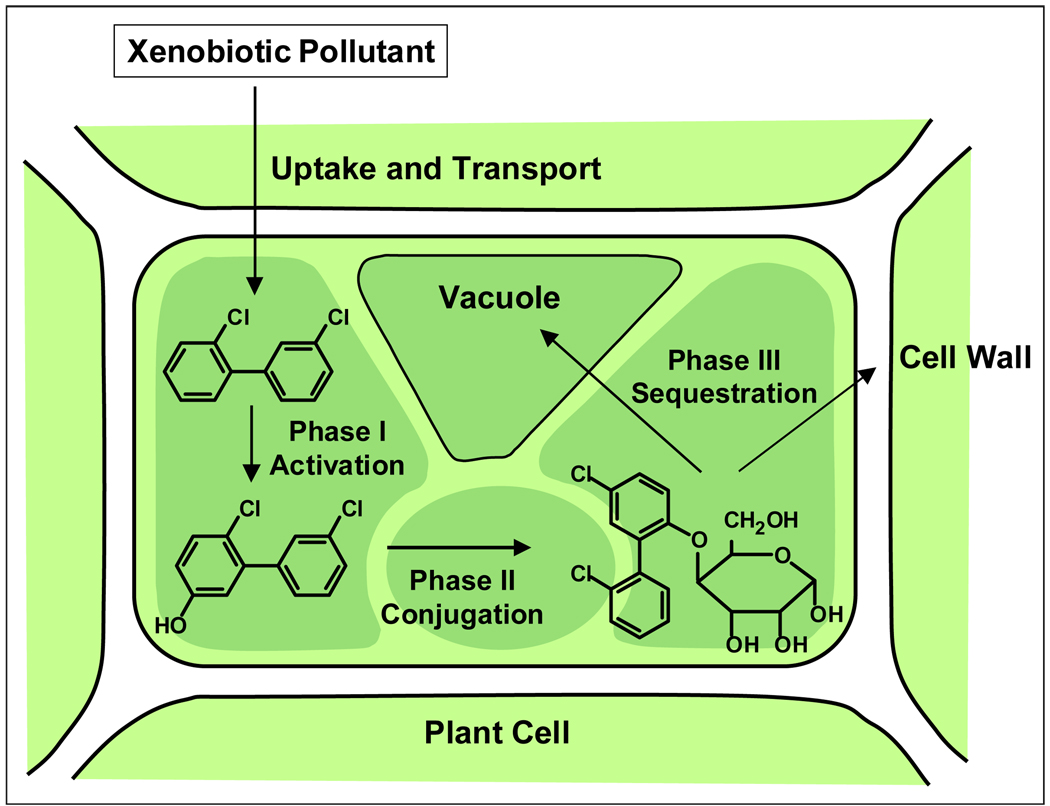
Phytoremediation encompasses a range of processes beyond direct plant uptake and metabolism, and it is best described as plant-mediated bioremediation (1,4,7,31,32). While definitions and terminology vary, the different phytoremediation processes can be summarized as in Figure 1: pollutants in soil and groundwater are taken up inside plant tissues (phytoextraction) or adsorbed to the roots (rhizofiltration); pollutants inside plant tissues are transformed by plant enzymes (phytotransformation) and/or volatilize into the atmosphere (phytovolatilization); and pollutants in soil are degraded by microbes in the root zone (rhizoremediation) or incorporated to soil material (phytostabilization) (1,6,10,32). Based on the observation that plants can metabolize pesticides, Sandermann (33) introduced the green liver concept, suggesting a detoxification sequence similar to what occurs in the liver of mammals (Figure 2) (3,33,34).
Figure 1.
Phytoremediation of organic pollutants, such as PCBs, may involve several processes: pollutants in soil and groundwater can be taken up inside plant tissues (phytoextraction) or adsorbed to the roots (rhizofiltration); pollutants inside plant tissues can be transformed by plant enzymes (phytotransformation) or can volatilize into the atmosphere (phytovolatilization); pollutants in soil can be degraded by microbes in the root zone (rhizoremediation) (1,6,9). Adapted from Van Aken (104).
Figure 2.
The three phases of the green liver model. Hypothetical pathway representing the metabolism of 2,3'-dichlorobiphenyl in plant tissues: Phase I, activation of the PCB by hydroxylation; Phase II, conjugation with a plant molecule (sugar); Phase III, sequestration of the conjugate into the vacuole or cell wall. Adapted from Van Aken (104).
Phytoremediation offers several advantages over other remediation strategies: low cost because of the absence of energy-consuming equipment and limited maintenance, no or limited negative impact on the environment because of the in situ nature of the process, and large public acceptance as an attractive green technology (19). In addition, phytoremediation offers potential beneficial side-effects, such as erosion control, site restoration, carbon sequestration, and feedstock for biofuel production (10,35). As autotrophic organisms, plants use sunlight and carbon dioxide as energy and carbon sources. From an environmental standpoint, plants can be seen as 'natural, solar-powered, pump-and-treat systems' for cleaning up contaminated soils (9).
However, the technology also suffers several limitations: phytoremediation is restrained to shallow contamination of 'moderately hydrophobic' compounds susceptible to be efficiently absorbed by the roots (36,37). More importantly, remediation by plants is often slow and incomplete: as a corollary to their autotrophic metabolism, plants usually lack the biochemical pathways necessary to achieve total mineralization of recalcitrant pollutants, such as PAHs and PCBs (7). Phytoremediation can therefore lead to undesirable effects, such as the accumulation of toxic metabolites that may be released to the soil, enter the food chain, or volatilize into in the atmosphere (6,9,14,38,39). In addition, planted trees need several years to reach mature size and, in temperate regions, plants have limited activity during the dormant season (7). Additional constraints to phytoremediation are not of technical order, but are the current regulations, competition with other methods, and proprietary rights (40). An important barrier to the development of transgenic plants for bioremediation is associated with the potential risk of horizontal gene transfer to related wild or cultivated plants (41). There is a critical need for integrated risk assessment of transgenic bioremediation technologies that should lead to community education and reevaluation of current regulations (42). Additional research is needed for the development of molecular risk mitigation strategies. It is likely that the next generation of transgenic organisms for phytoremediation will involve systems preventing such a transfer, for instance by the introduction of transgenes into chloroplastic DNA or the use of conditional lethality genes (43).
Even though cleaning up pollution with plants appears to be an ideal remediation technology that has been proven to be effective by extensive laboratory and greenhouse research, a contrasting small number of field applications has been successfully conducted. Although this contradictory observation is related to a combination of factors largely shared by most bioremediation systems, phytoremediation is likely victim of its own attractiveness, leading the technology to be oversold. By its nature, phytoremediation is assorted with specific limitations and failure to clearly identify them may lead to ineffectiveness of the remediation process.
PCBs: Chemistry, Sources, Transport, and Toxicity
PCBs are xenobiotic chlorinated aromatic compounds that are characterized by high physical and chemical stability and categorized as persistent organic pollutants (POPs) (15). Because of their thermal stability and high dielectric constant, PCBs have been used for a variety of industrial applications, including lubricants, dielectric fluids, and plasticizers. PCBs were manufactured widely during a half century (from 1929 to the 1970s) and an estimated 1.5 million tons of PCBs have been produced worldwide. Because of their toxicity and persistence in the environment, PCBs have been banned in most countries in 1979.
Local manufacture, usage, spill, and improper disposal of PCBs have led to extensive environmental contamination. Because of their high volatility and stability, PCBs have been largely dispersed by atmospheric transport. The octanol-water partition coefficient (log Kow) allows to predict the mobility of PCBs in the environment: higher-chlorinated PCBs, with log Kow above 6, are associated with particulate matter in the atmosphere, soils, and sediments; lower-chlorinated congeners exist in gaseous phase and can be transported over longer distance. As a consequence, soils generally contain a higher proportion of higher-chlorinated congeners, while air is dominated by lower-chlorinated fractions (44). Because higher-chlorinated PCBs are susceptible to microbial anaerobic dechlorination, anoxic sediments are often enriched in lower-chlorinated congeners. Today PCBs are still emitted from several sources, such as leaks of existing equipment (e.g., electrical capacitors and ballasts), volatilization from dredging sediments, and sewage sludge soil application. PCBs have been detected in virtually every compartment of the ecosystem, including air, water, soil, sediment, and living organisms. PCBs are highly hydrophobic, leading to their bioaccumulation in living organisms (biomagnification). In humans, PCBs are commonly detected in breast milk and blood, with concentrations increasing with age. Plants constitute the major route of entry of PCBs in the food chain (15).
Toxicity of PCBs has been known since the 1930s (45): although acute toxicity for adult humans is rather low, chronic exposure to PCBs induces serious neurobehavioral, immunological, reproductive, and endocrine disorders in children (46,47). According to the Department of Health and Human Services (DHHS), U.S. Environmental Protection Agency (EPA), and International Agency for Research on Cancer (IARC), PCBs are suspected to be carcinogenic in animals and humans (45,48,49). PCBs are listed as EPA Priority Pollutants (http://oaspub.epa.gov/) and are ranked at the fifth position in the 2007 CERCLA (Comprehensive Environmental Response, Compensation, and Liability Act) Priority List of Hazardous Substances (http://www.atsdr.cdc.gov/).
Microbial Degradation of PCBs
The chemical stability of PCBs renders them quite recalcitrant to microbial biodegradation (50). The presence of more chlorine atoms increases the chemical stability and decreases water-solubility of PCBs, making higher-chlorinated congeners more recalcitrant to biodegradation. In addition, metabolism of PCBs is often energetically unfavorable, requiring additional carbon source to support their biodegradation (co-metabolism).
Although they are classified as POPs, microbial biodegradation of PCBs is well documented (45,48,51–53). Two major microbial metabolic routes are known: anaerobic and aerobic pathways, depending of the degree of chlorination of the PCB congener, the redox conditions, and the type of microorganism involved (48).
Anaerobic Dechlorination of PCBs
Generally speaking, PCB congeners with four or more chlorine atoms undergo anaerobic reductive dechlorination, an energy-yielding process where PCBs serve as electron acceptor for the oxidation of organic carbon. Chlorine atoms are preferentially removed from the meta- and para-positions on the biphenyl structure, leaving lesser-chlorinated ortho-substituted congeners (54). Microorganisms that reductively dechlorinate PCBs are widespread in contaminated sediments and involve species related to Dehalococcoides (55–59). PCB dechlorination has been mostly attributed to complex bacterial consortia and little is known about metabolic pathways, molecular bases, and enzymes implicated in the process. Only few bacterial species able to dechlorinate PCBs in pure culture have been identified and their range of activity is limited to a few congeners (45,56). Sequencing the genome of Dehalococcoides ethenogenes 195, a well-characterized tetrachlorethene degrader, revealed the presence of several reductive dehalogenase genes potentially implicated in PCB transformation (45). However, to date, no enzyme involved in PCB anaerobic dechlorination has been isolated or characterized.
Aerobic Biodegradation of PCBs
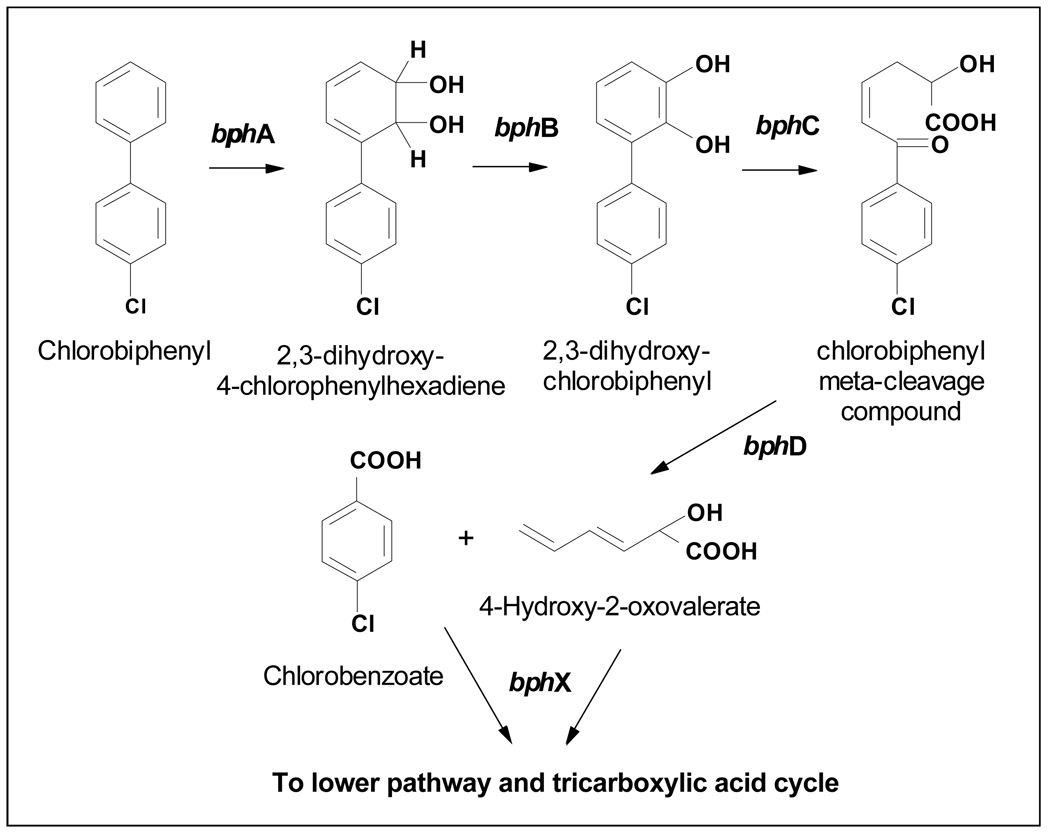
Lower-chlorinated PCB congeners – possibly produced by anaerobic dechlorination – undergo co-metabolic aerobic oxidation mediated by dioxygenases, resulting in ring opening and potentially complete mineralization of the molecule (50,53,60). Several bacterial strains are capable of oxidative degradation of PCBs, including mainly members of the genus Pseudomonas, Burkholderia, Comamonas, Rhodococcus, and Bacillus. The number of chlorine atoms per molecule and placement of chlorine atoms are important factors for the aerobic biodegradation via oxidative enzymes (53,60,61). Generally, PCB congeners with three or fewer chlorine atoms per molecule are easily degraded, and the ones with five or more are quite recalcitrant (requiring reductive dechlorination prior to aerobic mineralization). However, one of the most efficient PCB degraders characterized, Burkholderia xenovorans strain LB400, was shown to metabolize a hexachlorobiphenyl congener (51). Aerobic biodegradation of PCBs typically involves two clusters of genes, the first one responsible for transformation into chlorobenzoates and chlorinated aliphatic acids (biphenyl upper pathway), and the second one for further mineralization of chlorobenzoates and aliphatic acids (biphenyl lower pathway) (45,50,51). The upper pathway, which is similar for all described aerobic PCB degraders, involves seven genes grouped into one operon (biphenyl dioxygenase, bph) (Figure 3): A multi-component dioxygenase (bphA, bphE, bphF, and bphG) initiates hydroxylation of two adjacent biphenyl carbons to form an arene cis-diol. In the second step, a cis-2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase (bphB) further oxidizes the biphenyl ring to produce a cis-dihydroxychlorobiphenyl. In the third step, a second dioxygenase, 2,3-dihydroxybiphenyl 1,2-dioxygenase (bphC), opens the ring in ortho- or meta-position. The four step of the upper pathway involves a hydrolase (bphD) that cleaves the resulting molecule into chlorobenzoate and 2-hydroxypenta-2,4-dienoate (48).
Figure 3.
Bacterial aerobic degradation of lower-chlorinated PCBs is catalyzed by biphenyl dioxygenase (bph) gene cluster (upper pathway). Adapted from Furukawa et al. (61).
Phytoremediation of PCBs
The first reports on the potential of plants for bioremediation of PCBs were published in the late 1970s – early 1980s: Reinholtz and Volpe (62) (aquatic plants), Weber and Mrozek (uptake and translocation) (63), Schwartz and Lehmann (64) (detection of PCBs in plant tissues), and Bacci and Gaggi (65) (translocation and volatilization of PCBs from soil). Since then, significant advances have been made, showing the potential of plants and associated microbes for PCB metabolism. Processes recognized to be involved in phytoremediation of PCBs include rhizoremediation, phytoextraction, and phytotransformation.
Rhizoremediation of PCBs
Due to their high hydrophobicity, PCBs bind strongly to soil particles and are only poorly taken up inside plant tissues. Therefore, microbes in the rhizosphere play a dominant role in their biodegradation (13). Many reports have shown a significant increase of PCB attenuation in soil planted with a variety of plants, as compared with non-vegetated soils (15,16,19,66). There are many processes by which vegetation can stimulate microbial activity in soil and enhance biodegradation of recalcitrant PCBs:
Plant roots release organic compounds, such as sugar, amino acids, and organic acids, that can be used as electron donors to support aerobic co-metabolism or anaerobic dehalogenation of chlorinated compounds. In some instances, microbial aerobic metabolism will consume oxygen, resulting in anaerobic conditions favorable to PCB dehalogenation (16).
Plants secrete extracellular enzymes that can initiate transformation of PCBs and facilitate further microbial metabolism (67).
Plants release inducers that enhance microbial degradation. Plant phenolic exudates were shown to enhance the activity of the PCB degrader, B. xenovorans LB400 (68).
Plants increase soil permeability and oxygen diffusion in the rhizosphere, which potentially enhances microbial oxidative transformation by oxygenases (16,69,70).
Plant roots are also known to secrete diverse microbial growth factors (15).
Plant roots release organic acids and molecules that can act as surfactants, therefore mobilizing PCBs and rendering them more susceptible to be absorbed inside plant tissues (15).
Several publications have shown the positive effect of root exudates, including phenolic compounds, flavonoids, and terpenes, on microbial activity in soil and on biodegradation of PCBs (67,71–73). Vegetation was reported to significantly increase PCB removal from soil, as compared to non-planted soil, both due to higher microbial degradation of PCBs in the root zone and uptake inside plant tissues. Epuri and Sorensen (74) showed a higher mineralization of hexachlorobiphenyl in Aroclor 1260-contaminated soil planted with tall fescue (Festuca arundinacea), as compared with unplanted soil. Singer et al. (75) studied the interactive effects of different treatments on the degradation of Aroclor 1242 in soil, including bioaugmentation with PCB-degrading bacteria, biostimulation with inducers and surfactants, and vegetation with Brassica nigra. The authors observed a significantly higher PCB degradation in vegetated soil, as compared to non-planted controls, and concluded that plants enhanced PCB degradation by increasing oxygen diffusion in soil, amendment infiltration, and microbial enrichment. In a phytoremediation experiment using several plant species (alfalfa, flatpea, sericea lespedeza, deertongue, reed canarygrass, switchgrass, and tall fescue) for the bioremediation of PCB-contaminated soil, Aroclor 1248 was shown to be removed to a greater extent from all vegetated pots (38% or less PCB recovery), as compared with non-planted pots (82% PCB recovery) (76). In addition, plants increased enzymatic activity in soil that was shown to correlate with the levels of PCB biodegradation. Recently, Smith et al. (77) conducted greenhouse experiments on PCB-contaminated sediments following different treatments, including addition of organic amendment (mixture of straw and starch adjusted to a C:N ratio of 10:1) and vegetation with low and high-transpiring plants (including Scirpus fluviatilis, Tripsacum dactyloides, Carex aquatalis, and Spartina pectinata). The authors observed highest PCB removal following the addition of amendment with low-transpiring plant or no plant treatment, concluding that organic amendment resulted in oxygen consumption necessary to achieve anaerobic dechlorination of PCBs.
Molecular biology tools have also been used to locate PCB degraders in the roots of plants growing in PCB-contaminated soil: Pseudomonas fluorescens strain was constructed that expressed a green fluorescent protein (GFP) under the control of the meta-pathway Pm promoter from P. putida known to be induced by 3-chlorobenzoate, a product of 3-monochlorobiphenyl metabolism. When added to alfalfa roots (Medicago sativa) growing on 3-monochlorobiphenyl-contmainated soil, engineered bacteria indicated the presence of degrading microcolonies on the root surface and in crevices between root epidermal cells (78). Similarly, Hogan et al. (79) developed a real-time PCR assay based on SYBR Green and fluorescence resonance energy transfer (FRET) probes allowing the sensitive detection of transgenic P. fluorescens expressing bph operon from the PCB degrader, B. xenovorans LB400.
Uptake of PCBs inside Plant Tissues
In order to predict uptake of organic pollutants by plants, Briggs et al. (36) and Burken and Schnoor (37) developed experimental relationships based on log Kow. Based on their models, only 'moderately hydrophobic' compounds (0.5 < log Kow < 4.5) would be significantly taken up and translocated inside plant tissues. The efficiency of plant uptake of PCBs – with log Kow ranging from 4.5 (2-monochlorobiphenyl) to 8.2 (decachlorobiphenyl) – is expected to decrease fast with the degree of chlorination. Studying phytoextraction of Aroclor 1260-contaminated soil from three sites in Canada by nine plant species, Zeeb et al. (80) detected variable concentrations of PCBs in root tissues, and, to a lesser level, in shoot tissues. The authors observed higher concentration of tetrachloro- to hexachlorobiphenyls in shoots, although heptachloro- and nonachlorobiphenyls were also present in detectable amounts. These results suggest that, despite the predictions based on log Kow, higher-chlorinated congeners would be susceptible to be taken up inside plant tissues. On the other hand, using hydroponic hybrid poplars, Liu and Schnoor (81) observed that selected mono- to tetrachlorinated PCBs adsorbed to plant roots, but only lower-chlorinated PCBs were translocated to aerial parts (mono-, di-, and trichlorinated PCBs to upper stems and mono- and dichlorinated PCBs to shoots). In a field trial on Aroclor-contaminated soil, Aslund et al. (82) showed an increase of PCB concentration in the stems and leaves of pumpkin plants with time of exposure, while root concentration remained unchanged. The authors suggested that PCB transfer in plants occurs primarily via uptake and translocation, while other potential mechanisms, such as volatilization and deposition, have negligible contribution.
Plant Metabolism of PCBs
Plant metabolism of xenobiotic compounds is conceptually represented as a three-phase process known as the green liver model (Figure 2) (33,34): Phase I, the initial activation, consists of oxidation of PCBs to produce various hydroxylated products, characterized by a higher solubility and reactivity. Phase II involves conjugation of Phase I-activated compounds with molecules of plant origin (e.g., glutathione or aminoacids) forming adducts less toxic and more soluble than parent PCBs. Phase III involves sequestration of the conjugates in plant organelles (e.g., vacuole) or incorporation into plant structures (e.g., cell wall) (3,33,34).
Although plants were shown to contribute to PCB attenuation in soil since the 1970s, it was not before the 1990s that the capability of plants to metabolize PCBs was demonstrated. In pioneer work studying the transformation of 19 PCB congeners in plant cell cultures of Rosa spp. (Paul's Scarlet rose), Lee and Fletcher (83) observed that 11 individual congeners had been metabolized by more than 10%. Wilken et al. (84) studied the metabolism of 10 PCB congeners in 12 plant species and detected various mono- and dihydroxylated metabolites. Mackova et al. (85) used in vitro cell cultures of a variety of plants species (Armoracia rusticana, Solanum aviculare, Atropa belladonna, and Solanum nigrum) to characterize the metabolism of a commercial mixture (Delor 103) and observed that PCB transformation capability greatly differed from strain to strain. Using in vitro hairy root culture of S. nigrum (black nightshade), Kucerova et al. (86) showed that plant cells were capable of oxidizing mono- and dichlorinated PCBs into mono- and dihydroxylated biphenyls. Different laboratory experiments conducted with plant cell cultures showed that all mono- and dichlorobiphenyls were slightly hydroxylated, with the exception of 4,4'-dichlorobiphenyl, hypothesized to be sterically protected from enzymatic attack (86–88). Further studies using plant cell cultures showed that more persistent dichloro-, trichloro-, and tetrachlorobiphenyl congeners could also be metabolized by plant cells: Harms et al. (88) demonstrated that 3,3',4,4'-tetrachlorobiphenyl could be oxidized to several mono-hydroxylated intermediates by plant cell cultures of Rosa spp. and Lactuca sativa (lettuce). Following a similar pathway, 2,2',5,5'-tetrachlorobiphenyl was transformed to 3,4-dihydroxy-2,2',5,5'-tetrachlorobiphenyl. In experiments using black nightshade hairy root cultures exposed to several dichlorinated, trichlorinated, tetrachlorinated, and pentachlorinated PCB congeners, Rezek et al. (89) observed the formation of hydroxylated PCB metabolites from dichloro- and trichlorobiphenyl congeners, while tetrachloro- and pentachlorobiphenyl congeners were not metabolized.
In summary, plant metabolism of PCBs varies according to the plant species and degree of chlorination and substitution pattern. Initial steps in plant metabolism of PCBs involve oxidation of the biphenyl core, which is discouraged by the presence of electron-withdrawing chlorine atoms. Plant metabolism of PCBs appears therefore limited to tetra-chlorinated and lower congeners. In some instance, lower-chlorinated congeners are more recalcitrant than higher ones, suggesting the importance of substitution pattern. For instance, in the study cited above, Lee and Fletcher (83) observed that 4,4'-dichlorobiphenyl was not hydroxylated while 2,4,4'-trichlorobiphenyl was.
Plant Enzymes Involved in PCB Transformation
Several studies suggest that different oxygenases may be implicated in the initial metabolism of PCBs in plants (Phase I of the green liver model), including cytochrome P-450 monooxygenases (88) and peroxidases (85,87,90). Studying the metabolism of PCBs in rose cell cultures, Lee and Fletcher (83) observed a decrease of PCB metabolism by cytochrome P-450 inhibitors, while peroxidase inhibitors did not produce significant effect, suggesting the intervention of cytochrome P-450s. On the other hand, Koller et al. (91) reported extensive transformation and dechlorination of dichloro- and tetrachlorobiphenyl by commercial horseradish peroxidase (HRP). Also, using various in vitro plant cell cultures, Chroma et al. (87,92,93) observed a correlation between PCB transformation and various catabolic enzymes including peroxidases, Remazol Brillant Blue R (RBBR) oxidases, and cytochrome P-450s, suggesting the implication of these three enzymes in PCB metabolism in plants.
Although very little is known about conjugative enzymes involved in PCB metabolism (Phase II of the green liver model), knowledge gained from the degradation of other nucleophilic xenobiotics suggests that various transferases, such as glutathione S-transferases (e.g., conjugation of glutathione with several pesticides) and glycosyltransferases (e.g., conjugation of glucose with chlorophenols and DDT) are likely to be involved in the conjugation and compartmentation of PCB adducts in plant tissues (33,94). Plant tolerance (Brassica juncea) to several chlorinated pollutants, such as atrazine, metolachlor, and 1-chloro-2,4-dinitrobenzene (CDNB), was enhanced by the overexpression of enzymes involved in glutathione synthesis, including γ-glutamylcysteine synthetase (ECS) and glutathione synthetase (GS), further suggesting the potential implication of glutathione in PCB metabolism (95).
Our knowledge of plant metabolism of xenobiotics is still fragmentary and other enzymes are likely to be involved in PCB transformation. For instance, Magee et al. (96) reported recently dechlorination of 2,2',4,4',5,5'-hexachlorobiphenyl by crude extract of nitrate reductase from Medicago sativa and a pure commercial nitrate reductase from Zea mays. Also, plant dehalogenation of chlorinated solvents has been reported following a mechanism similar to microbial anaerobic dechlorination (97,98). Although no plant dehalogenase has been identified or characterized, such a mechanism could potentially lead to PCB dechlorination in plant tissues.
Transgenic Plants for Phytoremediation of PCBs
Although PCBs have been shown to be removed by plants, only rather slow biodegradation rates have been achieved in field trials, potentially leading to accumulation and volatilization of toxic compounds. Genetic transformation of plants for enhanced phytoremediation capabilities is typically achieved by the introduction of external genes whose products are involved in various detoxification processes (17,99). Microbes and mammals are heterotrophic organisms that possess the enzymatic metabolic enzymes to achieve a near-complete mineralization of organic molecules. Microbial and mammalian catabolic genes can therefore be used to complement the metabolic capabilities of plants (9).
Transgenic plants have been produced for phytoremediation of both heavy metals and organic pollutants (9,99). Early examples include tobacco plants expressing a yeast metallothionein gene and showing a higher tolerance to cadmium (100), Arabidopsis thaliana overexpressing a zinc transporter protein and showing a two-fold higher accumulation of zinc in roots (101), and tobacco plants expressing a human cytochrome P-450 for enhanced metabolism of trichloroethylene (11). The use of transgenic plants for phytoremediation applications has been reviewed recently in several articles (9,17,18,24,26,99,102–105). Table 1 presents a non-exhaustive list of transgenic plants and bacteria constructed for phytoremediation of PCBs.
Table 1.
Summary of publications about transgenic plants and bacteria for the phytoremediation of PCBs
| Compound | Gene | Source | Host Organism | Reference |
|---|---|---|---|---|
| PCBs | Biphenyl dioxygenase, bph, located on transposon TnPCB | Pseudomonas sp. strain LB400 | Sugar beet seeds (cv. Rex) | (110) |
| PCBs | Biphenyl dioxygenase, bphC | Comamonas testosteroni B-356 | Tobacco (Nicotiana tabacum) | (106) |
| 2',3,4-Trichloro-biphenyl | Oxygenolytic ortho-dechlorination (ohb) gene |
Pseudomonas aeruginosa strain 142 |
Sinorhizobium meliloti colonizing Alfalfa (Medicago sativa) | (115) |
| 3-Chloro-biphenyl | Biphenyl dioxygenase, bph Green fluorescent protein, gfp | Burkholderia xenovorans LB400 | alfalfa roots (Medicago sativa,,var. Resis, DLF Trifolium) | (78) |
| PCBs | Biphenyl dioxygenase, bphC on mini-transposon Tn5 | Burkholderia xenovorans LB400 | Pseudomonas fluorescens F113 colonizing alfalfa roots | (79) |
| 2',3,4-Trichloro-biphenyl | PCB-degrading genes, ortho-halobenzoate 1,2-dioxygenase (ohb) genes |
Pseudomonas aeruginosa strain 142 |
Sinorhizobium meliloti colonizing Alfalfa (Medicago sativa) | (116) |
| Individual PCBs congeners | Biphenyl dioxygenase, bph | Burkholderia xenovorans LB400 |
Pseudomonas fluorescens F113 colonizing Alfalfa (Medicago sativa) rhizosphere |
(111) |
| PCB-contaminated soil | Biphenyl dioxygenase, bph | Burkholderia xenovorans LB400 | Pseudomonas fluorescens colonizing rhizosphere of Willow (Salix sp.) | (117) |
| 4-Chloro-biphenyl | Biphenyl dioxygenases, bphA, bphE, bphF, bphG | Burkholderia xenovorans LB400 | Nicotiana tabacum, Nicotiana benthamiana | (107) |
| Individual PCBs congeners | Biphenyl dioxygenase, bph |
Burkholderia xenovorans LB400 |
Pseudomonas fluorescens F113 colonizing Willow (Salix sp.) rhizosphere | (114) |
| 2,3-Dihydroxy-biphenyl | Biphenyl dioxygenase, bphC |
Pseudomonas testosteroni B-356 |
Tobacco (Nicotiana tabacum) |
(108) |
Plant cytochrome P-450-mediated metabolism of PCBs produces toxic epoxide intermediates and trans-diol metabolites not easily further biodegraded. Unlike cytochrome P-450s, bacterial biphenyl dioxygenases produce cis-diol intermediates susceptible to ring cleavage and complete mineralization (26). In an attempt to overcome this limitation, components of bacterial biphenyl dioxygenase operon, bph, were introduced into plants. In pioneer work, Francova et al. (106) genetically modified tobacco plants (Nicotiana tobacum) by insertion of the gene responsible for 2,3-dihydroxybiphenyl ring cleavage, bphC, from the PCB degrader Comamonas testosteroni. bphC gene was cloned into plasmid pB1 121 under the control of the strong CaMV 35S promoter and introduced into the 'natural genetic engineer' Agrobacterium tumefaciens. Successful transformation was confirmed by amplification of bphC using PCR. Although the engineered plants were not tested for their capability to metabolize PCBs, this work constitutes a milestone in the development of transgenic plants for the transformation of PCBs. In a similar study, Mohammadi et al. (107) inserted bph genes from B. xenovorans LB400, one of the most efficient PCB degrader, into tobacco plants. Three components of the bph operon necessary for dioxygenation of the biphenyl ring, bphAE, bphF, and bphG, were individually cloned and expressed in transgenic plants. The authors showed that purified enzymes from the plants were capable of oxidizing 4-chlorobiphenyl into 2,3-dihydro-2,3-dihydroxy-4'-chlorobiphenyl. Recently, Novakova et al. (108) constructed transgenic tobacco plants expressing bphC from the PCB degrader, P. testosteroni B-356. When grown in the presence of 2,3-dihydroxybiphenyl (0.5 mM), one transgenic line, H2, exhibited higher resistance to the toxic compound, as compared to wild-type plants. Although successful application of this revolutionary strategy will require more development, such as engineering-improved PCB-degrading enzymes and coordinated expression of different genes, these results suggest that transgenic plants expressing the complete bacterial PCB metabolic pathway could help overcome inherent limitations of phytoremediation (26).
Transgenic Plant-Associated Bacteria for Rhizoremediation of PCBs
Plants are known to increase both microbial numbers and activity in soil, which can result in an increase of biodegradation activity (69,109). However, endogenous or rhizosphere bacteria capable of maintaining a stable relationship with plants may not harbor the metabolic enzymes necessary for the efficient catabolism of persistent pollutants (110,111). In an attempt to improve rhizoremediation performances, several research groups have cloned key catabolic genes of known xenobiotic degraders into specific rhizosphere bacteria (16,19,29,49,66,112,113). In pioneer study, Brazil et al. (110) introduced the genetically engineered transposon, TnPCB, containing bph genes from the PCB degrader, B. xenovorans LB400, into P. fluorescens F113, a bacterium colonizing the roots of many plants. The recombinant bacterium, strain F113pcb, expressed heterologous bph genes, as it was confirmed by its ability to utilize biphenyl as sole carbon source. Rhizosphere competence of strain F113pcb was identical to wild-type P. fluorescens, as confirmed by colonization experiments of sugar beet seedling roots. This study demonstrated that rhizosphere-adapted microbes can be genetically engineered to metabolize recalcitrant xenobiotics without affecting their ecological competence. Following a similar approach, bph operon from B. xenovorans strain LB400, was inserted into strain F113 under the control of the strong promoter, nodbox 4, from Sinorhizobium meliloti (111). The constructed strain, F113::1180, expressed high level of biphenyl dioxygenase and was capable of metabolizing biphenyl and several monochlorinated to trichlorinated PCB congeners at much higher rate than strain F113pcb. In addition, the transgenic strain, F113::1180, was able to metabolize Delor 103 better than initial bph donor strain, B. xenovorans LB400. Recently, another group reported higher PCB metabolization rates with transgenic P. fluorescens F113::1180 and B. xenovorans LB400, as compared to strain F113pcb (114). Using mesocosm experiments with PCB-contaminated soil, the authors reported a good survival ability of F113 strains in willow plant rhizosphere, suggesting that association of transgenic rhizosphere bacteria with plants constitute a promising approach for the treatment of PCB-contaminated soils.
Nitrogen-fixing bacterium, S. meliloti, lives in symbiotic association with roots of the leguminous alfalfa plants, M. sativa, providing its host with reduced nitrogen and increasing soil fertility. In an attempt to increase rhizoremediation performance, S. melitoti was transformed by introduction of a PCB-degrading plasmid containing the oxygenolytic ortho-dechlorination gene, ohb (115). Transformant strains were able to grow on 100 mg L−1 2',3,4-trichlorobiphenyl and dechlorinate 100% of PCBs, as compared to 15% achieved by wild-type bacteria. In another study, S. meliloti was transformed by introduction of a PCB-degrading plasmid harboring the bph operon. Transgenic S. meliloti was shown to degrade 2',3,4-trichlorobiphenyl. Plant chamber tests revealed that alfalfa plants inoculated with transgenic bacteria were capable of two-fold higher dechlorination of 2',3,4-trichlorobiphenyl, as compared to control alfalfa inoculated with wild-type S. meliloti (116). More recently, in order to improve bioremediation of the commercial mixture, Delor 103, in contaminated soil, de Carcer et al. (117) inoculated the roots of willows (Salix viminalis × schwerinii) with two genetically modified (GM) P. fluorescens strains: class 1 GM strain modified with a single chromosomal insertion of bph operon and class 2 GM strain with insertion of bph operon under the control of the nod regulatory system of S. meliloti. After about six months, analysis of PCBs showed a statistically significant increase of degradation rate in rhizosphere soil inoculated with class GM 1 and GM 2 P. fluorescens strains, as compared to control soil inoculated with wild-type strain. In addition, the presence of transgenic bacteria did not affect the microbial community in bulk soil.
An interesting approach to enhance rhizoremediation of PCBs is based on the concept of rhizoengineering. Rhizoengineering consists of using transgenic plants or metabolic mutants exuding modified patterns of plant secondary metabolites, therefore promoting the growth of specific bacterial groups capable of xenobiotic biodegradation. Using a 'rhizosphere metabolomic' approach, Narasimhan et al. (118) identified a large majority of phenylpropanoids, including flavonoids, in plant exudates. Different near-isogenic lines of Arabidopsis mutants overproducing flavonoids were then used to promote root colonization by P. putida PML2. P. putida PML2 is a rhizospheric bacterium that has the capability of metabolizing both flavonoids and PCBs. Results obtained showed that Pseudomonas PML2 colonized the roots of Arabidopsis flavonoid-overproducing mutants at higher levels. In addition, Pseudomonas PML2 was able to reach significantly higher depletion of 2-monochloro- and 4-monochlorobipheynls (90%) when growing in association with flavonoid-expressing Arabidopsis, as compared to flavonoid null mutant. The authors concluded that this approach complements the use of transgenic plants for bioremediation applications.
Conclusions
Transgenic bacteria have been used for industrial production of pharmaceuticals and human proteins (e.g., insulin) and transgenic plants have been used for the expression of insect or pesticide resistance (e.g., Bt-maize). From an environmental standpoint, agricultural plants expressing genes involved in the biodegradation of pesticides are the first transgenic organisms used for phytoremediation applications. Recently, non-agricultural plants and associated bacteria have been developed to mitigate pollution of soil and groundwater by toxic agrochemicals and other xenobiotic pollutants, including PCBs (17,99).
To date, only genes involved in Phase I of the green liver model have been introduced into transgenic plants for PCB degradation. Further developments may involve the introduction of multiple transgenes involved in different phases of the green liver model, which would help overcome a major limitation inherent to phytoremediation, i.e., the threat that accumulated toxic compounds would volatilize or otherwise contaminate the food chain (6,38,39,99). As an illustration of transgenic plants expressing enzymes involved in Phase II of the green liver model, Indian mustard (Brassica juncea) was modified to overexpress enzymes involved in glutathione metabolism (ECS and GS), resulting in enhanced tolerance to atrazine, metolachlor, and CDNB (95).
Another interesting approach to enhance phytoremedation efficiency would consist of engineering plants to secrete microbial enzymes being released into the environment to achieve ex-planta bioremediation, such as transgenic tobacco expressing extracellular fungal peroxidases for the removal of pentachlorophenol (PCP) (119).
Although it has not been used for the treatment of PCBs, an alternative strategy may involve the genetic transformation of endophytic bacteria. Unlike rhizospheric bacteria, endophytic bacteria colonize the internal tissues of plants (35,120). Many endophytes have been shown to play a role in the metabolism of toxic xenobiotic pollutants, therefore potentially enhancing phytoremediation (121). Barac et al. (122) described the conjugative transformation of natural endophytes harboring a toluene-degradation plasmid (pTOM) for improved in planta degradation of toluene. As suggested in a recent review by Weyens et al. (105), metal-tolerant endophytes equipped with enzymes capable of biodegradation of organic compounds would allow phytoremediation of sites co-contaminated with mixture of toxic metals and organic pollutants.
Finally, an important barrier to the field application of transgenic trees for bioremediation is associated with the true or perceived risk of horizontal gene transfer to wild or cultivated plants. There is therefore a critical need for further risk-benefit analysis and risk mitigation strategies to ensure that transgenic biotechnologies would result in wider acceptance and application of phytoremediation (42,43).
Acknowledgements
The authors are grateful to the University of Iowa Superfund Basic Research Program (NIEHS; award number P42ES05605).
References
- 1.Salt DE, Smith RD, Raskin I. Phytoremediation. Annu. Rev. Plant Phys. 1998;49:643–668. doi: 10.1146/annurev.arplant.49.1.643. [DOI] [PubMed] [Google Scholar]
- 2.Castelfranco P, Foy C, Deutsch D. Non-enzymatic detoxification of 2-chloro-4,6-bis(ehtylamino)-S-triazine (simazine) by extract of Zea maize. Weeds. 1961;9:580–591. [Google Scholar]
- 3.Cole DJ. Oxidation of xenobiotics in plants. Prog. Pest. Biochem. Technol. 1983;3:199–253. [Google Scholar]
- 4.Macek T, Mackova M, Kas J. Exploitation of plants for the removal of organics in environmental remediation. Biotechnol. Adv. 2000;18:23–34. doi: 10.1016/s0734-9750(99)00034-8. [DOI] [PubMed] [Google Scholar]
- 5.Meagher RB. Phytoremediation of toxic elemental and organic pollutants. Curr. Opin. Plant Biol. 2000;3:153–162. doi: 10.1016/s1369-5266(99)00054-0. [DOI] [PubMed] [Google Scholar]
- 6.Pilon-Smits E. Phytoremediation. Annu. Rev. Plant Biol. 2005;56:15–39. doi: 10.1146/annurev.arplant.56.032604.144214. [DOI] [PubMed] [Google Scholar]
- 7.Schnoor JL, Licht LA, McCutcheton SC, Wolfe NL, Carreira LH. Phytoremediation of organic and nutrient contaminants. Environ. Sci. Technol. 1995;29:318A–323A. doi: 10.1021/es00007a747. [DOI] [PubMed] [Google Scholar]
- 8.Campos VM, Merino I, Casado R, Pacios LF, Gomez L. Phytoremediation of organic pollutants. Span. J. Agric. Res. 2008;6:38–47. [Google Scholar]
- 9.Eapen S, Singh S, D'Souza SF. Advances in development of transgenic plants for remediation of xenobiotic pollutants. Biotechnol. Adv. 2007;25:442–451. doi: 10.1016/j.biotechadv.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Dietz AC, Schnoor JL. Advances in phytoremediation. Environ. Health Persp. 2001;109:163–168. doi: 10.1289/ehp.01109s1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doty SL, Shang TQ, Wilson AM, Tangen J, Westergreen AD, Newman LA, Strand SE, Gordon MP. Enhanced metabolism of halogenated hydrocarbons in transgenic plants containing mammalian cytochrome P450 2E1. P. Natl. Acad. Sci. USA. 2000;97:6287–6291. doi: 10.1073/pnas.97.12.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French CE, Rosser SJ, Davies GJ, Nicklin S, Bruce NC. Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase. Nat. Biotechnol. 1999;17:491–494. doi: 10.1038/8673. [DOI] [PubMed] [Google Scholar]
- 13.Macek T, Kotrba P, Svatos A, Novakova M, Demnerova K, Mackova M. Novel roles for genetically modified plants in environmental protection. Trends Biotechnol. 2008;26:146–152. doi: 10.1016/j.tibtech.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Arthur EL, Rice PJ, Anderson TA, Baladi SM, Henderson KLD, Coats JR. Phytorernediation - An overview. Crit. Rev. Plant Sci. 2005;24:109–122. [Google Scholar]
- 15.Campanella BF, Bock C, Schroder P. Phytoremediation to increase the degradation of PCBs and PCDD/Fs - Potential and limitations. Environ. Sci. Pollut. R. 2002;9:73–85. doi: 10.1007/BF02987318. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhry Q, Blom-Zandstra M, Gupta S, Joner EJ. Utilising the synergy between plants and rhizosphere microorganisms to enhance breakdown of organic pollutants in the environment. Environ. Sci. Pollut. R. 2005;12:34–48. doi: 10.1065/espr2004.08.213. [DOI] [PubMed] [Google Scholar]
- 17.Cherian S, Oliveira MM. Transgenic plants in phytoremediation: Recent advances and new possibilities. Environ. Sci. Technol. 2005;39:9377–9390. doi: 10.1021/es051134l. [DOI] [PubMed] [Google Scholar]
- 18.Doty SL. Enhancing phytoremediation through the use of transgenics and endophytes. New Phytol. 2008;179:318–333. doi: 10.1111/j.1469-8137.2008.02446.x. [DOI] [PubMed] [Google Scholar]
- 19.Gerhardt KE, Huang XD, Glick BR, Greenberg BM. Phytoremediation and rhizoremediation of organic soil contaminants: Potential and challenges. Plant Sci. 2009;176:20–30. [Google Scholar]
- 20.James CA, Strand SE. Phytoremediation of small organic contaminants using transgenic plants. Curr. Opin. Biotech. 2009;20:237–241. doi: 10.1016/j.copbio.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawahigashi H. Transgenic plants for phytoremediation of herbicides. Curr. Opin. Biotech. 2009;20:225–230. doi: 10.1016/j.copbio.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Singh OV, Jain RK. Phytoremediation of toxic aromatic pollutants from soil. Appl. Microbiol. Biot. 2003;63:128–135. doi: 10.1007/s00253-003-1425-1. [DOI] [PubMed] [Google Scholar]
- 23.Susarla S, Medina VF, McCutcheon SC. Phytoremediation: An ecological solution to organic chemical contamination. Ecol. Eng. 2002;18:647–658. [Google Scholar]
- 24.Van Aken B. Transgenic plants for enhanced phytoremediation of toxic explosives. Curr. Opin. Biotech. 2009;20:231–236. doi: 10.1016/j.copbio.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Mackova M, et al. Phytoremediation of polychlorinated biphenyls. In: Mackova M, Dowling D, Macek T, editors. Phytoremediation and Rhizoremediation: Theoretical Background. Dordrecht: Springer; 2006. pp. 143–169. [Google Scholar]
- 26.Sylvestre M, Macek T, Mackova M. Transgenic plants to improve rhizoremediation of polychlorinated biphenyls (PCBs) Curr. Opin. Biotech. 2009;20:242–247. doi: 10.1016/j.copbio.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Singer AC. The chemical ecology of pollutants biodegradation. In: Mackova M, Dowling D, Macek T, editors. Phytoremediation and Rhizoremediation. Theoretical Background. Dordrecht: Springer; 2006. pp. 5–21. [Google Scholar]
- 28.Singer AC, Crowley DE, Thompson IP. Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol. 2003;21:123–130. doi: 10.1016/S0167-7799(02)00041-0. [DOI] [PubMed] [Google Scholar]
- 29.Singer AC, Thompson IP, Bailey MJ. The tritrophic trinity: A source of pollutant-degrading enzymes and its implications for phytoremediation. Curr. Opin. Microbiol. 2004;7:239–244. doi: 10.1016/j.mib.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Daar AS, Thorsteinsdottir H, Martin DK, Smith AC, Nast S, Singer PA. Top ten biotechnologies for improving health in developing countries. Nat. Genet. 2002;32:229–232. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- 31.Burken JG. Uptake and metabolism of organic compounds: Green-liver model. In: McCutcheon SC, Schnoor JL, editors. Phytoremediation. Control and Transport of Contaminants. Hoboken: John Wiley; 2003. pp. 59–84. [Google Scholar]
- 32.Nzengung VA, O'Niell WL, McCutcheon SC, Wolfe NL. Sequestration and transformation of water soluble halogenated organic compounds using aquatic plants, algae, and microbial mats. In: McCutcheon SC, Schnoor JL, editors. Phytoremediation. Transformation and control of Contaminants. Hoboken: John Wiley; 2003. pp. 499–528. [Google Scholar]
- 33.Sandermann H. Higher plant metabolism of xenobiotics: The 'green liver' concept. Pharmacogenetics. 1994;4:225–241. doi: 10.1097/00008571-199410000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Coleman JOD, Blake-Kalff MMA, Davies TGE. Detoxification of xenobiotics by plants: Chemical modification and vacuolar compartimentation. Trends Plant Sci. 1997;2:144–151. [Google Scholar]
- 35.Doty SL, James CA, Moore AL, Vajzovic A, Singleton GL, Ma C, Khan Z, Xin G, Kang JW, Park AY, Meilan R, Strauss SH, Wilkerson J, Farin F, Strand SE. Enhanced phytoremediation of volatile environmental pollutants with transgenic trees. P. Natl. Acad. Sci. USA. 2007;104:16816–16821. doi: 10.1073/pnas.0703276104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briggs GG, Bromilow RH, Evans AA. Relationship between lipophilicity and root uptake and translocation of non-ionised chemicals by barley. Pestic. Sci. 1982;13:495–504. [Google Scholar]
- 37.Burken JG, Schnoor JL. Predictive relationships for uptake of organic contaminants by hybrid poplar trees. Environ. Sci. Technol. 1998;32:3379–3385. [Google Scholar]
- 38.Newman LA, Strand SE, Choe N, Duffy J, Ekuan G, Ruszaj M, Shurtleff BB, Wilmoth J, Heilman P, Gordon MP. Uptake and biotransformation of trichloroethylene by hybrid poplars. Environ. Sci. Technol. 1997;31:1062–1067. doi: 10.1289/ehp.98106s41001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon JM, Van Aken B, Schnoor JL. Leaching of contaminated leaves following uptake and phytoremediation of RDX, HMX, and TNT by poplar. Int. J. Phytoremediat. 2006;8:81–94. doi: 10.1080/15226510500507128. [DOI] [PubMed] [Google Scholar]
- 40.Marmiroli N, McCutcheon SC. Making phytoremediation a successful technology. In: McCutcheon SC, Schnoor JL, editors. Phytoremediation. Transformation and Control of Contaminants. Hoboken: John Wiley; 2003. pp. 85–119. [Google Scholar]
- 41.Miller H. The EPA's war on bioremediation. Nat. Biotechnol. 1997;15:486. doi: 10.1038/nbt0697-486. [DOI] [PubMed] [Google Scholar]
- 42.Linacre NA, Whiting SN, Baker AJM, Angle JS, Ades PK. Transgenics and phytoremediation: The need for an integrated risk assessment, management, and communication strategy. Int. J. Phytoremediat. 2003;5:181–185. doi: 10.1080/713610179. [DOI] [PubMed] [Google Scholar]
- 43.Davison J. Risk mitigation of genetically modified bacteria and plants designed for bioremediation. J. Ind. Microbiol. Biot. 2005;32:639–650. doi: 10.1007/s10295-005-0242-1. [DOI] [PubMed] [Google Scholar]
- 44.Hornbuckle KC, Smith GL, Miller SM, Eadie BJ, Lansing M. Magnitude and origin of PCBs resuspended in open waters of Lake Michigan. Abstr. Pap. Am. Chem. S. 2004;228 017-ENVR. [Google Scholar]
- 45.Pieper DH, Seeger M. Bacterial metabolism of polychlorinated biphenyls. J. Mol. Microb. Biotech. 2008;15:121–138. doi: 10.1159/000121325. [DOI] [PubMed] [Google Scholar]
- 46.ATSDR. Agency for Toxic Substances and Disease Registry. US Department of Health and Human Services, Public Health Service; 2000. Toxicological profile for polychlorinated biphenyls (PCBs) [PubMed] [Google Scholar]
- 47.Mayes BA, McConnell EE, Neal BH, Brunner MJ, Hamilton SB, Sullivan TM, Peters AC, Ryan MJ, Toft JD, Singer AW, Brown JF, Menton RG, Moore JA. Comparative carcinogenicity in Sprague-Dawley rats of the polychlorinated biphenyl mixtures Aroclors 1016, 1242, 1254, and 1260. Toxicol. Sci. 1998;41:62–76. doi: 10.1093/toxsci/41.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borja J, Taleon DM, Auresenia J, Gallardo S. Polychlorinated biphenyls and their biodegradation. Process. Biochem. 2005;40:1999–2013. [Google Scholar]
- 49.Pieper DH. Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biot. 2005;67:170–191. doi: 10.1007/s00253-004-1810-4. [DOI] [PubMed] [Google Scholar]
- 50.Furukawa K, Fujihara H. Microbial degradation of polychlorinated biphenyls: Biochemical and molecular features. J. Biosci. Bioeng. 2008;105:433–449. doi: 10.1263/jbb.105.433. [DOI] [PubMed] [Google Scholar]
- 51.Field JA, Sierra-Alvarez R. Microbial transformation and degradation of polychlorinated biphenyls. Environ. Pollut. 2008;155:1–12. doi: 10.1016/j.envpol.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 52.Ohtsubo Y, Kudo T, Tsuda M, Nagata Y. Strategies for bioremediation of polychlorinated biphenyls. Appl. Microbiol. Biot. 2004;65:250–258. doi: 10.1007/s00253-004-1654-y. [DOI] [PubMed] [Google Scholar]
- 53.Vasilyeva GK, Strijakova ER. Bioremediation of soils and sediments contaminated by polychlorinated biphenyls. Microbiology. 2007;76:639–653. [Google Scholar]
- 54.Olson PE, Reardon KF, Pilon-Smits EAH. Ecology of rhizosphere bioremediation. In: McCutcheon SC, Schnoor JL, editors. Phytoremediation. Transformation and Control of Contaminants. Hoboken: John Wiley; 2003. pp. 317–353. [Google Scholar]
- 55.Abraham WR, Nogales B, Golyshin PN, Pieper DH, Timmis KN. Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr. Opin. Microbiol. 2002;5:246–253. doi: 10.1016/s1369-5274(02)00323-5. [DOI] [PubMed] [Google Scholar]
- 56.Bedard DL, Bailey JJ, Reiss BL, Jerzak GV. Development and characterization of stable sediment-free anaerobic bacterial enrichment cultures that dechlorinate Aroclor 1260. Appl. Environ. Microb. 2006;72:2460–2470. doi: 10.1128/AEM.72.4.2460-2470.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho YC, Sokol RC, Frohnhoefer RC, Rhee GY. Reductive dechlorination of polychlorinated biphenyls: Threshold concentration and dechlorination kinetics of individual congeners in Aroclor 1248. Environ. Sci. Technol. 2003;37:5651–5656. doi: 10.1021/es034600k. [DOI] [PubMed] [Google Scholar]
- 58.Cho YC, Sokol RC, Rhee GY. Kinetics of polychlorinated biphenyl dechlorination by Hudson River, New York, USA, sediment microorganisms. Environ. Toxicol. Chem. 2002;21:715–719. [PubMed] [Google Scholar]
- 59.Tiedje JM, Boyd SA. Anaerobic degradation of chlorinated aromatic hydrocarbons. Dev. Ind. Microbiol. Series. 1987;27:117–127. [Google Scholar]
- 60.Kohler HPE, Kohlerstaub D, Focht DD. Co-metabolism of polychlorinated-biphenyls - Enhanced transformation of Aroclor 1254 by growing bacterial cells. Appl. Environ. Microb. 1988;54:1940–1945. doi: 10.1128/aem.54.8.1940-1945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Furukawa K, Suenaga H, Goto M. Biphenyl dioxygenases: Functional versatilities and directed evolution. J. Bacteriol. 2004;186:5189–5196. doi: 10.1128/JB.186.16.5189-5196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reinholtz W, Volpe AA. Elimination of polychlorinated biphenyl pollutants from water by means of aquatic plants. Abstr. Pap. Am. Chem. S. 1977;173:41–41. [Google Scholar]
- 63.Weber JB, Mrozek E. Polychlorinated biphenyls - Phytotoxicity, absorption and translocation by plants, and inactivation by activated carbon. B. Environ. Contam. Tox. 1979;23:412–417. doi: 10.1007/BF01769980. [DOI] [PubMed] [Google Scholar]
- 64.Schwartz TR, Lehmann RG. Determination of polychlorinated-biphenyls in plant tissue. B. Environ. Contam. Tox. 1982;28:723–727. doi: 10.1007/BF01605643. [DOI] [PubMed] [Google Scholar]
- 65.Bacci E, Gaggi C. Polychlorinated-biphenyls in plant foliage translocation or volatilization from contaminated soils. B. Environ. Contam. Tox. 1985;35:673–681. doi: 10.1007/BF01636572. [DOI] [PubMed] [Google Scholar]
- 66.Wood TK, Shim H, Burken JG, Ryoo D, Bowers KC, Chauhan S. Rhizosphere competitiveness of trichloroethylene-degrading, poplar-colonizing recombinants. Abstr. Pap. Am. Chem. S. 2000;219:U167–U168. doi: 10.1128/aem.66.11.4673-4678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fletcher JS, Hegde RS. Release of phenols from perennial plant roots and their potential importance in bioremediation. Chemosphere. 1995;31:3009–3016. [Google Scholar]
- 68.Hegde RS, Fletcher JS. Influence of plant growth stage and season on the release of root phenolics by mulberry as related to development of phytoremediation technology. Chemosphere. 1996;32:2471–2479. [Google Scholar]
- 69.Anderson TA, Guthrie EA, Walton BT. Bioremediation in the rhizosphere. Environ. Sci.Technol. 1993;27:2630–2636. [Google Scholar]
- 70.Shimp JF, Tracy JC, Davis LC, Lee E, Huang W, Erickson LE, Schnoor JL. Beneficial effects of plants in the remediation of soil and groundwater contaminated with organic materials. Crit. Rev. Env. Sci. Tec. 1993;23:41–77. [Google Scholar]
- 71.Fletcher JS, Donnley PK, Hegde RS. Proceedings of the 14th Annual Symposium on Current Topics in Plant Biochemistry, Physiology and Molecular Biology. Columbia: University of Missouri; 1995. Plant assisted polychlorinated biphenyl (PCB) biodegradation; pp. 42–43. [Google Scholar]
- 72.Focht DD. Strategies for the improvement of aerobic metabolism of polychlorinated-biphenyls. Curr. Opin. Biotech. 1995;6:341–346. [Google Scholar]
- 73.Gilbert ES, Crowley DE. Plant compounds that induce polychlorinated biphenyl biodegradation by Arthrobacter sp. strain B1B. Appl. Environ. Microb. 1997;63:1933–1938. doi: 10.1128/aem.63.5.1933-1938.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Epuri V, Sorensen DL. Phytoremediation of Soil and Water Contaminants. ACS Symposium Series; 1997. Benzo(a)pyrene and hexachlorobiphenyl contaminated soil: Phytoremediation potential; pp. 200–222. [Google Scholar]
- 75.Singer AC, Smith D, Jury WA, Hathuc K, Crowley DE. Impact of the plant rhizosphere and augmentation on remediation of polychlorinated biphenyl contaminated soil. Environ. Toxicol. Chem. 2003;22:1998–2004. doi: 10.1897/02-471. [DOI] [PubMed] [Google Scholar]
- 76.Chekol T, Vough LR, Chaney RL. Phytoremediation of polychlorinated biphenyl-contaminated soils: The rhizosphere effect. Environ. Int. 2004;30:799–804. doi: 10.1016/j.envint.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 77.Smith KE, Schwab AR, Banks MK. Phytoremediation of polychlorinated biphenyl (PCB)-contaminated sediment: A greenhouse feasibility study. J. Environ. Qual. 2007;36:239–244. doi: 10.2134/jeq2006.0089. [DOI] [PubMed] [Google Scholar]
- 78.Boldt TS, Sorensen J, Karlson U, Molin S, Ramos C. Combined use of different GFP reporters for monitoring single-cell activities of a genetically modified PCB degrader in the rhizosphere of alfalfa. FEMS Microbiol. Ecol. 2004;48:139–148. doi: 10.1016/j.femsec.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 79.Hogan J, Sherlock O, Ryan D, Whelan C, Francesconi S, Rivilla R, Dowling DN. Fluorescence resonance energy transfer (FRET) based molecular detection of a genetically modified PCB degrader in soil. FEMS Microbiol. Lett. 2004;236:349–357. doi: 10.1016/j.femsle.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 80.Zeeb BA, Amphlett JS, Rutter A, Reimer KJ. Potential for phytoremediation of polychlorinated biphenyl-(PCB-)contaminated soil. Int. J. Phytoremediat. 2006;8:199–221. doi: 10.1080/15226510600846749. [DOI] [PubMed] [Google Scholar]
- 81.Liu JY, Schnoor JL. Uptake and translocation of lesser-chlorinated polychlorinated biphenyls (PCBs) in whole hybrid poplar plants after hydroponic exposure. Chemosphere. 2008;73:1608–1616. doi: 10.1016/j.chemosphere.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aslund MLW, Rutter A, Reimer KJ, Zeeb BA. The effects of repeated planting, planting density, and specific transfer pathways on PCB uptake by Cucurbita pepo grown in field conditions. Sci. Total Environ. 2008;405:14–25. doi: 10.1016/j.scitotenv.2008.07.066. [DOI] [PubMed] [Google Scholar]
- 83.Lee I, Fletcher JS. Involvement of mixed-function oxidase systems in polychlorinated biphenyl metabolism by plant-cells. Plant Cell Rep. 1992;11:97–100. doi: 10.1007/BF00235262. [DOI] [PubMed] [Google Scholar]
- 84.Wilken A, Bock C, Bokern M, Harms H. Metabolism of different PCB congeners in plant-cell cultures. Environ. Toxicol. Chem. 1995;14:2017–2022. [Google Scholar]
- 85.Mackova M, Macek T, Ocenaskova J, Burkhard J, Demnerova K, Pazlarova J. Biodegradation of polychlorinated biphenyls by plant cells. Int. Biodeter. Biodegr. 1997;39:317–325. [Google Scholar]
- 86.Kucerova P, Mackova M, Chroma L, Burkhard J, Triska J, Demnerova K, Macek T. Metabolism of polychlorinated biphenyls by Solanum nigrum hairy root clone SNC-9O and analysis of transformation products. Plant Soil. 2000;225:109–115. [Google Scholar]
- 87.Chroma L, Moeder M, Kucerova P, Macek T, Mackova M. Plant enzymes in metabolism of polychlorinated biphenyls. Fresen. Environ. Bull. 2003;12:291–295. [Google Scholar]
- 88.Harms H, Bokern M, Kolb M, Bock C. Transformation of organic contaminants by different plant systems. In: McCutcheon SC, Schnoor JL, editors. Phytoremediation. Transformation and Control of Contaminants. Hoboken: John Wiley; 2003. pp. 285–316. [Google Scholar]
- 89.Rezek J, Macek T, Mackova M, Triska J. Plant metabolites of polychlorinated biphenyls in hairy root culture of black nightshade Solanum nigrum SNC-90. Chemosphere. 2007;69:1221–1227. doi: 10.1016/j.chemosphere.2007.05.090. [DOI] [PubMed] [Google Scholar]
- 90.Mackova M, Chroma L, Kucerova P, Burkhard J, Demnerova K, Macek T. Some aspects of PCB metabolism by Horseradish cells. Int. J. Phytoremediat. 2001;3:401–414. [Google Scholar]
- 91.Koller G, Moder M, Czihal K. Peroxidative degradation of selected PCB: a mechanistic study. Chemosphere. 2000;41:1827–1834. doi: 10.1016/s0045-6535(00)00132-6. [DOI] [PubMed] [Google Scholar]
- 92.Chroma L, Macek T, Demnerova K, Mackova M. Decolorization of RBBR by plant cells and correlation with the transformation of PCBs. Chemosphere. 2002a;49:739–748. doi: 10.1016/s0045-6535(02)00397-1. [DOI] [PubMed] [Google Scholar]
- 93.Chroma L, Mackova M, Kucerova P, Derwiesche C, Burkhard J, Macek T. Enzymes in plant metabolism of PCBs and PAHs. Acta Biotechnol. 2002b;22:35–41. [Google Scholar]
- 94.Pflugmacher S, Schroder P, Sandermann H. Taxonomic distribution of plant glutathione S-transferases acting on xenobiotics. Phytochemistry. 2000;54:267–273. doi: 10.1016/s0031-9422(00)00116-3. [DOI] [PubMed] [Google Scholar]
- 95.Flocco CG, Lindblom SD, Smits EAHP. Overexpression of enzymes involved in glutathione synthesis enhances tolerance to organic pollutants in Brassica juncea. Int. J. Phytoremediat. 2004;6:289–304. doi: 10.1080/16226510490888811. [DOI] [PubMed] [Google Scholar]
- 96.Magee KD, Michael A, Ullah H, Dutta SK. Dechlorination of PCB in the presence of plant nitrate reductase. Environ. Toxicol. Phar. 2008;25:144–147. doi: 10.1016/j.etap.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 97.Nzengung VA, Jeffries PM. Sequestration, phytoreduction, and phytooxidation of halogenated organic chemicals by aquatic and terrestrial plants. Int. J. Phytoremediat. 2001;3:13–40. [Google Scholar]
- 98.Wolfe NL, Hoehamer CF. Enzymes used by plants and microorganisms to detoxify organic compounds. In: McCutcheon SC, Schnoor JL, editors. Phytoremediation. Control and Transport of Contaminants. Hoboken: John Wiley; 2003. pp. 499–528. [Google Scholar]
- 99.Eapen S, D'Souza SF. Prospects of genetic engineering of plants for phytoremediation of toxic metals. Biotechnol. Adv. 2005;23:97–114. doi: 10.1016/j.biotechadv.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 100.Misra S, Gedamu L. Heavy-metal tolerant transgenic Brassica napus L. and Nicotiana tabacum L. plants. Theor. Appl. Genet. 1989;78:161–168. doi: 10.1007/BF00288793. [DOI] [PubMed] [Google Scholar]
- 101.Van der Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, Verkleij JAC, Hooykaas PJJ. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 1999;119:1047–1055. doi: 10.1104/pp.119.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chrastilova Z, Mackova M, Novakova M, Macek T, Szekeres M. Transgenic plants for effective phytoremediation of persistent toxic organic pollutants present in the environment. J. Biotechnol. 2007;131:S38–S38. [Google Scholar]
- 103.Sylvestre M. Genetically modified organisms to remediate polychlorinated biphenyls. Where do we stand? Int. Biodeterior. Biodegrad. 2004;54:153–162. [Google Scholar]
- 104.Van Aken B. Transgenic plants for phytoremediation: Helping nature to clean up environmental pollution. Trends Biotechnol. 2008;26:225–227. doi: 10.1016/j.tibtech.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 105.Weyens N, van der Lelie D, Taghavi S, Vangronsveld J. Phytoremediation: Plant–endophyte partnerships take the challenge. Curr. Opin. Biotech. 2009;20:237–254. doi: 10.1016/j.copbio.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 106.Francova K, Sura M, Macek T, Szekeres M, Bancos S, Demnerova K, Sylvestre M, Mackova M. Preparation of plants containing bacterial enzyme for degradation of polychlorinated biphenyls. Fresen. Environ. Bull. 2003;12:309–313. [Google Scholar]
- 107.Mohammadi M, Chalavi V, Novakova-Sura M, Laliberte JF, Sylvestre M. Expression of bacterial biphenyl-chlorobiphenyl dioxygenase genes in tobacco plants. Biotechnol. Bioeng. 2007;97:496–505. doi: 10.1002/bit.21188. [DOI] [PubMed] [Google Scholar]
- 108.Novakova M, Mackova M, Chrastilova Z, Viktorova J, Szekeres M, Demnerova K, Macek T. Cloning the bacterial bphC gene into Nicotiana tabacum to improve the efficiency of PCB-phytoremediation. Biotechnol. Bioeng. 2009;102:29–37. doi: 10.1002/bit.22038. [DOI] [PubMed] [Google Scholar]
- 109.Limbert ESB, Betts WB. Influence of substrate chemistry and microbial metabolic diversity on the bioremediation of xenobiotic contamination. Genet. Eng. Biotechnol. 1996;16:159–180. [Google Scholar]
- 110.Brazil GM, Kenefick L, Callanan M, Haro A, Delorenzo V, Dowling DN, Ogara F. Construction of a rhizosphere Pseudomonad with potential to degrade polychlorinated-biphenyls and detection of bph gene-expression in the rhizosphere. Appl. Environ. Microb. 1995;61:1946–1952. doi: 10.1128/aem.61.5.1946-1952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Villacieros M, Whelan C, Mackova M, Molgaard J, Sanchez-Contreras M, Lloret J, de Carcer DA, Oruezabal RI, Bolanos L, Macek T, Karlson U, Dowling DN, Martin M, Rivilla R. Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression. Appl. Environ. Microbiol. 2005;71:2687–2694. doi: 10.1128/AEM.71.5.2687-2694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ang EL, Zhao HM, Obbard JP. Recent advances in the bioremediation of persistent organic pollutants via biomolecular engineering. Enzyme Microb. Tech. 2005;37:487–496. [Google Scholar]
- 113.Kucerova P, Mackova M, Macek T. Perspectives of phytoremediation in decontamination of organic pollutants and xenobiotics. Chem. Listy. 1999;93:19–26. [Google Scholar]
- 114.Rein A, Fernqvist MM, Mayer P, Trapp S, Bittens M, Karlson UG. Degradation of PCB congeners by bacterial strains. Appl. Microbiol. Biotechnol. 2007;77:469–481. doi: 10.1007/s00253-007-1175-6. [DOI] [PubMed] [Google Scholar]
- 115.Toure O, Chen YQ, Dutta SK. Sinorhizobium meliloti electrotransporant containing ortho-dechlorination gene shows enhanced PCB dechlorination. Fresen. Environ. Bull. 2003;12:320–322. [Google Scholar]
- 116.Chen YQ, Adam A, Toure O, Dutta SK. Molecular evidence of genetic modification of Sinorhizobium meliloti: Enhanced PCB bioremediation. J. Ind Microbiol. Biot. 2005;32:561–566. doi: 10.1007/s10295-005-0039-2. [DOI] [PubMed] [Google Scholar]
- 117.de Carcer DA, Martin M, Mackova M, Macek T, Karlson U, Rivilla R. The introduction of genetically modified microorganisms designed for rhizoremediation induces changes on native bacteria in the rhizosphere but not in the surrounding soil. ISME J. 2007;1:215–223. doi: 10.1038/sj.ismej.2007.27. [DOI] [PubMed] [Google Scholar]
- 118.Narasimhan K, Basheer C, Bajic VB, Swarup S. Enhancement of plant-microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol. 2003;132:146–153. doi: 10.1104/pp.102.016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iimura Y, Ikeda S, Sonoki T, Hayakawa T, Kajita S, Kimbara K, Tatsumi K, Katayama Y. Expression of a gene for Mn-peroxidase from Coriolus versicolor in transgenic tobacco generates potential tools for phytoremediation. Appl. Microbiol. Biot. 2002;59:246–251. doi: 10.1007/s00253-002-1008-6. [DOI] [PubMed] [Google Scholar]
- 120.Ryan RP, Ryan D, Dowling DN. Plant protection by the recombinant, root-colonizing Pseudomonas fluorescens F113rifPCB strain expressing arsenic resistance: improving rhizoremediation. Lett. Appl. Microbiol. 2007;45:668–674. doi: 10.1111/j.1472-765X.2007.02248.x. [DOI] [PubMed] [Google Scholar]
- 121.Van Aken B, Yoon JM, Just CL, Schnoor JL. Metabolism and mineralization of hexahydro-1,3,5-trinitro-1,3,5-triazine inside poplar tissues (Populus deltoides × nigra DN-34) Environ. Sci. Technol. 2004;38:4572–4579. doi: 10.1021/es049837a. [DOI] [PubMed] [Google Scholar]
- 122.Barac T, Taghavi S, Borremans B, Provoost A, Oeyen L, Colpaert JV, Vangronsveld J, van der Lelie D. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat. Biotechnol. 2004;22:583–588. doi: 10.1038/nbt960. [DOI] [PubMed] [Google Scholar]