Abstract
EndA is a membrane-attached surface-exposed DNA-entry nuclease previously known to be required for genetic transformation of Streptococcus pneumoniae. More recent studies have shown that the enzyme also plays an important role during the establishment of invasive infections by degrading extracellular chromatin in the form of neutrophil extracellular traps (NETs), enabling streptococci to overcome the innate immune system in mammals. As a virulence factor, EndA has become an interesting target for future drug design. Here we present the first mutational and biochemical analysis of recombinant forms of EndA produced either in a cell-free expression system or in Escherichia coli. We identify His160 and Asn191 to be essential for catalysis and Asn182 to be required for stability of EndA. The role of His160 as the putative general base in the catalytic mechanism is supported by chemical rescue of the H160A variant of EndA with imidazole added in excess. Our study paves the way for the identification and development of protein or low-molecular-weight inhibitors for EndA in future high-throughput screening assays.
INTRODUCTION
The surface-exposed DNA-entry nuclease EndA from Streptococcus pneumoniae is a membrane-bound enzyme required for DNA uptake during genetic transformation. The enzyme is responsible for processing double-stranded DNA into single-stranded fragments that in turn enter the microbial cell (1). Mutations within the endA gene effectively block DNA-uptake by S. pneumoniae due to the reduced nuclease activity (2). More recently, another important function of surface-exposed streptococcal EndA as a bacterial virulence factor was discovered. During infections, the enzyme is able to cleave the DNA scaffold of neutrophil extracellular traps (NETs) (3,4). NETs are anti-microbial chromatin barriers expelled by activated neutrophils and other cells of the innate immune system (5,6). By doing so, EndA enables streptococci to escape the innate immune response in mammals and supports spreading of the pathogenic organism, increasing the risk of a life-threatening invasive infection. Thus, EndA appears to be a potential target for the identification and/or design of specific nuclease inhibitors that could be turned into effective drugs fighting streptococcal infections.
A primary sequence analysis shows that some DNA-entry or competence-associated nucleases contain a conserved DRGH-motif and, additionally, exhibit the well-known H-N-H-motif indicating a structurally conserved active site (7). The H-N-H-motif is found in many other sequence-specific and nonspecific nucleases as well as in His–Cys box enzymes which can all be grouped together into the large ββα-Me finger nuclease superfamily (8). Within this family, the DRGH-motif containing nucleases obviously form two subgroups. The first subgroup consists of so-called sugar nonspecific enzymes (such as Anabaena nuclease, Serratia nuclease or mitochondrial Endonuclease G) found in all branches of life. As revealed by a BLAST-search for homologous proteins with Anabaena nuclease as a query, this first group currently comprises 1291 hits in 451 species including archaea, bacteria, fungi, metazoa and plants (9–11). In contrast to the first group, homologs of EndA were found with 629 hits in 207 species, but only in bacteria, with the notable exception of one hit in humans for the so-called dyad symmetry-binding protein (gi: 33339902).
In the present study, we report the molecular cloning and expression of recombinant EndA and demonstrate by site-directed mutagenesis and biochemical analyses that EndA is indeed a member of the DRGH-motif-containing nuclease family. Due to an apparent extreme toxicity of EndA we were not able to clone and express wild-type endA gene in Escherichia coli. However, the inactive variant EndA H160A could be cloned, expressed and purified successfully. Expression of wild-type EndA and several active-site mutants was achieved by using a cell-free expression system employing gene splicing generated by overlap extension PCR using the EndA H160A encoding plasmid DNA as template (12). The activity of EndA H160A expressed in E. coli could be restored in vitro by chemical rescue with excess imidazole that apparently replaces the histidine side chain absent in this variant. Our results provide a solid basis for future structural and functional studies of this important class of DNA-entry nucleases.
MATERIALS AND METHODS
Expression of pelB EndA H160A
The endA H160A gene was cloned in vector pET25b+ (Novagen). EndA H160A protein, fused at its N-terminal end to a pelB leader sequence for periplasmic localization, was expressed in E. coli BL21Gold(DE3) (Novagen). Bacteria were separated by centrifugation from the culture medium (LB broth). Proteins from cell lysates before and after induction, LB-broth before and after induction and from cell lysate after sonication as well as the soluble fraction after sonication and centrifugation were separated on a 12.5% SDS–PAGE gel.
Expression and purification of EndA H160A
His-GST double-tagged EndA H160A fused via a TEV protease cleavage site [EMBL Protein Expression and Purification Facility (pETM30)] was expressed in E. coli BL21 Star (DE3) cells (Invitrogen). Cells were grown in LB-broth containing kanamycin. Expression was induced at an OD600 of 0.5 by supplementing the medium with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (final concentration), and cells were harvested after a further 4 h. Soluble EndA H160A was purified according to the user manual for Protino® Ni-IDA 2000-packed columns with slight modifications. Columns were washed with 50 mM NaH2PO4/NaOH pH 8.0, 750 mM NaCl (containing high salt for nucleic acid removal). Eluted fractions were dialyzed directly against a TEV-protease reaction buffer (50 mM Tris–HCl pH 8.0, 1 mM DTT, 0.5 mM EDTA). His-GST was clipped off using His-tagged TEV-protease (MobiTec) with a concentration of 25 U/mg His-GST-EndA H160A overnight at 4°C. After dialyzing against 50 mM NaH2PO4/NaOH pH 8.0, 300 mM NaCl and 20 mM imidazole, the protein mixture was incubated with Ni2+-NTA agarose (Qiagen). Untagged EndA H160A was recovered from the flow through. In a final purification step the flow through was applied to a Superdex-75 HR10/30 column using 50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 0.01% Triton as eluant. Bovine serum albumin, ovalbumin, α-chymotrypsin, and cytochrome c were used as protein standards to determine the apparent molecular mass of EndA H160A. Fractions containing EndA H160A were collected and concentrated using CENTRIPLUS® (Centrifugal Filter Devices, MW cut-off: 10 000; MILLIPORE).
For small-angle X-ray scattering (SAXS) experiments, the Rosetta2 (DE3) expression strain (Novagen) was transformed with pET30M-EndA H160A. The cells were grown at 37°C in LB with 100 µg/ml kanamycin, 25 ug/ml chloramphenicol until A600 reached a value of 0.8–1.0. The temperature was then decreased to 18°C, and protein expression was induced by the addition of 0.4 mM IPTG. Protein expression continued overnight at 18°C. The cells were pelleted by centrifugation and lysed by sonication in 25 mM Tris–HCl pH 7.5, 1 M NaCl, in the presence of Complete Protease EDTA-free Inhibitors (Roche). The soluble fraction was bound in-batch to glutathione 4B resin (GE Healthcare) for 1 h at 4°C. Unbound protein was removed by repeated washes with 25 mM Tris–HCl pH 7.5, 1 M NaCl. EndA H160A was cleaved from the resin by overnight digestion with TEV protease at 4°C. The cleaved protein was concentrated to 50 mg/ml using Centricon Plus-70 concentrators (Millipore), then loaded onto a Superdex 200 16/60 column (GE Healthcare) equilibrated in 25 mM Tris–HCl pH 7.5, 75 mM NaCl. Peak fractions containing EndA H160A were pooled and concentrated to ∼50 mg/ml using an Amicon Ultra-15 concentrator (Millipore).
SAXS data acquisition and processing
SAXS data were collected at 25°C on the X9 beam line at the National Synchrotron Light Source (Brookhaven National Laboratory) or the 18ID beamline at the Advanced Photon Source (Argonne National Laboratory). The wavelength of the beam was 0.855 or 1.033 Å and the sample to detector distance was 2.0 or 2.36 m, respectively. SAXS data were collected on four concentrations of EndA H160A, ranging from 1 to 5 mg/ml in 25 mM Tris–HCl pH 7.5, 75 mM NaCl. Scattering data were circularly averaged and scaled to obtain a relative scattering intensity as a function of momentum transfer vector, q [(4πsinθ)/λ], after subtraction of buffer-scattering contributions.
SAXS data analysis and model construction
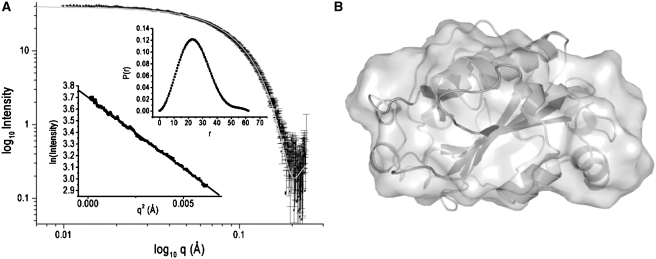
All scattering data were analyzed using the Primus software package; the GNOM45 software package was used for all P(r) and I0 analyses (13,14). The radius of gyration, Rg, and forward scattering, I0, were calculated from the second moment and the start of P(r), respectively, where Rg is the root mean square of all elemental volumes from the center-of-mass of the particle, weighted by their scattering densities, and I0 is directly proportional to the molar particle concentration, multiplied by the square of the scattering-particle molecular weight for particles with the same mean scattering density. Horse-heart cytochrome c was used as a standard reference protein for all I0 analysis. Guinier plots for EndA were linear over a q-range of 0.012–0.079 Å (Figure 2A).
Figure 2.
SAXS of EndA. (A) SAXS intensity I(q) data for EndA H160A (3.0 mg/ml); the insets show the P(r) pair-wise vector length distribution curve and Guinier fit of the I(q) scattering data. The gray line in the I(q) scattering plot is the calculated scattering curve of NucA generated with CRYSOL. (B) Ab-initio models derived from SAXS intensity data (surface representation) superimposed with a ribbon representation of the Anabaena NucA crystal structure (pdb-code 1zm8).
The three-dimensional shape of EndA was constructed from the SAXS data using the GASBOR22IQW program (q-range input for each analysis was from 0.01 to 0.04 Å−1), by calculating the distribution of linearly connected 1.9 Å spheres that best fit the scattering data (15). The calculation was repeated five times with different random starting points for the Monte Carlo optimization algorithm; no predefined shape or symmetry constraints were used. From these runs, the predicted structure with the lowest deviation of the calculated scattering profile from experimental data was used for interpretation (Figure 2B). To compare the SAXS-based models with the atomic structures, the SUPCOMB13 and CRYSOL programs were used (16,17).
Chemical rescue of the nuclease activity of EndA H160A
EndA H160A (20 nM) was incubated with a plasmid substrate (15 ng/µl, pBluescript SK(+), Stratagene) and a triple buffer containing 10 mM sodium acetate, 10 mM MES, 20 mM Tris–HCl, pH 8.0, 5 mM MgCl2 and 200 mM imidazole. After 2, 5, 10 and 20 min, aliquots were taken and the cleavage reaction stopped using a loading dye containing 250 mM EDTA. As a control, substrate plasmid was incubated for 20 min in the absence of EndA H160A. The reaction products were analyzed by agarose gel electrophoresis (0.8%). The same experiment was done at different pH values (pH 5.5, 6.5, 7.5, 8.5 and 9.5) in triple buffer whose pH was adjusted with HCl or NaOH, respectively. EndA H160A was diluted in the triple buffer at the pH used for the activity measurement. Each experiment was repeated three times. The ratio of open-circular to super-coiled DNA was calculated from the intensity of the bands using the TINA2.0 software package.
Sequence alignment
Sequences of the aligned DRGH-motif-containing nucleases were retrieved using the basic local alignment search tool BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) via the National Center for Biotechnology Information (NCBI) web server with the amino acid sequences of Anabaena nuclease (gi: 39041) or EndA (gi: 47374) as the query. Alignment of the primary structure of selected proteins was performed using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) via the web server of the European Bioinformatics Institute (EBI). According to the results obtained from our mutational and biochemical analyses the alignments were manually corrected using GeneDoc (http://www.nrbsc.org/gfx/genedoc/) or BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) software packages to match all amino acid residues with identical functions.
In vitro synthesis of EndA
DNA encoding EndA H160A cloned in plasmid pET25b+ (Novagen) was used as a template for overlap extension PCR based on the procedure described by Heckman and Pease to generate wild-type endA PCR product12. Using this as a template for further overlap extension PCR, all other variants of EndA (D157A, R158A, H160A, N179A, N182A and N191A) were generated. All constructs contain a T7 promoter sequence followed by a ribosomal binding site as described in the instruction manual for the PURExpress™ In vitro Protein Synthesis Kit (New England BioLabs). Protein synthesis was set up as a 25 µl reaction with 250-ng template DNA for each construct; incubation was for 2 h at 37°C.
In-gel activity assay
Aliquots of the unpurified protein-synthesis reaction mixture (3 µl) were analyzed on a 15% SDS–PAGE gel containing 30 µg/ml high-molecular-weight herring sperm DNA. After washing with water to remove SDS, the gel was incubated at ambient temperature in 40 mM Tris–HCl, pH 7.7, 2 mM MgCl2 to renature the nuclease. The gel was subsequently stained with ethidium bromide to detect in-gel DNA cleavage.
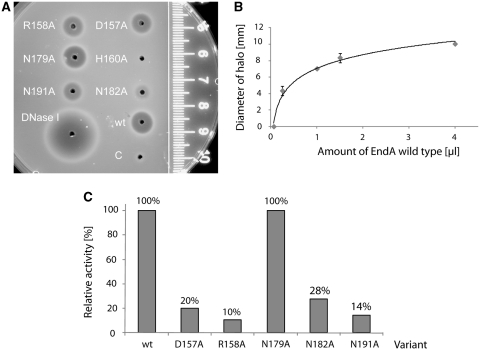
Single radiation enzyme diffusion assay
Petri dishes were prepared with 20 ml of 1% agarose solution in 40 mM Tris–HCl, pH 7.7, 2 mM MgCl2 containing 30 µg/ml high-molecular-weight herring sperm DNA. The agarose gel was pre-stained with ethidium bromide. Crude protein-synthesis reaction mixture (1 µl) and as control 1 µl DNaseI (Fermentas, 1 U/µl) with 1 µl of 40 mM Tris–HCl, pH 7.7, 2 mM MgCl2 were applied to wells with a diameter of 2 mm. Plates were incubated overnight at ambient temperature. The diameter (mm) of the halos was measured (18). For calibration, 0.062, 0.25, 1, 1.5 and 4 µl of the wild-type EndA protein synthesis reaction mixture were used. In the calibration curve, the diameter of the halos was plotted against the amounts of the wild-type EndA protein-synthesis reaction mixture. This curve was used to calculate the relative activity of the variants, whose concentration in the respective crude protein synthesis reaction mixture was estimated by a western blot analysis.
Western blot analysis
For analysis of the in vitro synthesis reaction, all EndA variants (crude protein-synthesis reaction) were separated on a 12.5% SDS–PAGE gel and electrophoretically transferred onto a PVDF transfer membrane (Amersham Hybond™ -P). Proteins were probed with a monoclonal mouse anti-FLAG M2-peroxidase conjugated antibody (SIGMA, dilution 1:500) and visualized using the Amersham™ ECL Plus Western Blotting Detection System. Quantitative western blot analysis was carried out using the TINA2.0 software package.
To analyze the various steps of the purification of EndA H160A, protein samples were separated on a 15% SDS–PAGE gel and electrophoretically transferred onto a PVDF transfer membrane (Amersham Hybond™ -P). Proteins were probed with a non-conjugated anti-GST-antibody (Pharmacia Biotech, dilution 1:2.000) and detected with anti-Sheep/Goat IgG-POD Fab-fragments (Boehringer Mannheim, dilution 1:2.500). The protein bands were visualized as described above.
Cleavage preferences of EndA
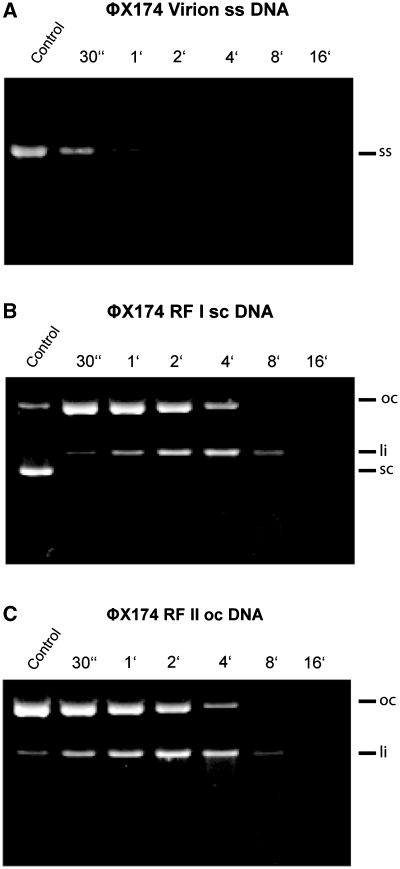
The wild-type EndA protein-synthesis reaction mixture was diluted 1:100 for cleavage of ΦX174 DNA (New England Biolabs): 8.4 nM single-stranded circular DNA 4.2 nM double-stranded, circular DNA and 4.2 nM double-stranded nicked DNA in 40 mM Tris–HCl, pH 7.7, 2 mM MgCl2 at ambient temperature. At the indicated time points, aliquots (10 µl) of each reaction mixture (100 µl) were analyzed on 0.8% agarose gels; the DNA was visualized by ethidium bromide staining.
RESULTS
Molecular cloning and expression of EndA in E. coli
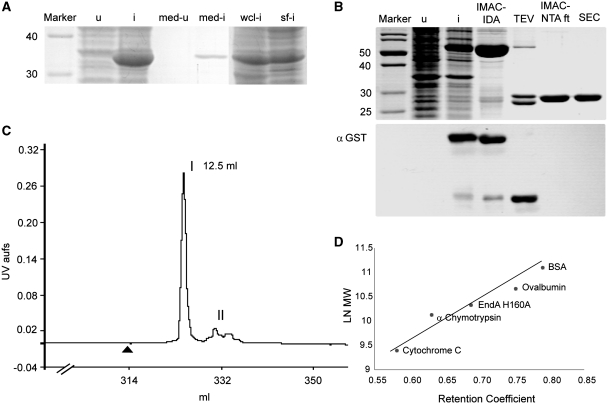
All efforts to clone and express EndA in E. coli using tightly repressed expression systems with or without secretion of the recombinant protein into the periplasmic space remained unsuccessful. In all cases, subsequent sequencing of recovered recombinant plasmids revealed mutations in the regions encoding the proposed active site and/or insertion of stop codons preventing the expression of full-length wild-type EndA protein (data not shown). We therefore generated the putatively inactive H160A variant with an exchange of the histidine residue from the DRGH-motif of EndA for alanine and verified its expression as a secreted protein due to fusion to the pelB leader sequence via cloning into pET-25(+) (Novagen) for periplasmic localization (Figure 1A). Soluble processed and secreted EndA H160A could be detected and recovered by Ni2+-NTA affinity chromatography from the culture medium as well as from the cell lysate of E. coli after induction of transformed cells. From SDS–PAGE analyses of this system it was obvious that the amount of unprocessed intracellular protein still containing the pelB leader sequence was relatively high even after induction with low amounts of IPTG (0.1 mM final concentration) and fermentation at reduced temperatures (22°C), explaining why the wild-type enzyme could not be expressed. Surprisingly, EndA H160A protein fused to pelB turned out to be highly soluble upon expression in E. coli in contrast to other nonspecific nucleases of the DRGH-motif containing type such as Anabaena nuclease or Serratia nuclease (Figure 1A) (19,20).
Figure 1.
(A) SDS–PAGE analysis of the expression of EndA H160A fused to the pelB leader sequence for periplasmic localization in E. coli. (u) cells before induction, suspended in loading buffer (i) cells after induction, suspended in loading buffer; (med-u) concentrated culture medium before induction, (med-i) concentrated culture medium after induction, (wcl-i) whole cell lysate after cell disruption by sonication and (sf-i) soluble fraction after centrifugation of the whole cell lysate. (B) Purification of His-GST double-tagged EndA H160A. Upper panel: SDS–PAGE analysis of the lysate of transformed BL21-Gold(DE3) cells before (u) and after (i) induction; (IMAC-IDA) His-GST-EndA H160A purified via immobilized metal ion affinity chromatography using a Ni2+-IDA-column; (TEV) cleavage of His-GST-EndA H160A with TEV protease; (IMAC-NTA ft) flow through after incubation with Ni2+-NTA-Agarose; (SEC) EndA-H160A after size-exclusion chromatography. Lower panel: western blot using anti-GST antibody to monitor the purification steps described above. (C) Purification of EndA H160A by size-exclusion chromatography: after clipping off the His-GST double tag and incubation with Ni-NTA-Agarose the collected flow through containing only untagged EndA H160A was applied to a Superdex 75 column and eluted at a flow rate of 1 ml/min. The black triangle indicates the time point of sample loading. Elution of proteins was monitored at 280 nm. Peak I represents EndA separated from a residual contaminant (II). (D) Retention coefficients (Superdex 75 column) of EndA and protein standards were determined and plotted against the natural logarithm of the molecular mass. The molecular weight of 25.5±4 kDa obtained for EndA, based on the fitted curve, corresponds reasonably well with the calculated molecular weight of 26.8 kDa indicating that EndA is a monomer under these conditions.
As intracellular EndA H160A was soluble and non-toxic to E. coli, we generated and expressed a His-GST double-tagged version of EndA H160A in E. coli using the pET30M expression vector (Figure 1B, upper panel). The recombinant fusion protein with a theoretical molecular mass of 55.6 kDa could be expressed and purified over a Ni2+-IDA-affinity chromatography column. After affinity purification, the double tag was clipped off by TEV-protease cleavage resulting in a non-tagged EndA H160A variant. His-tagged GST and TEV-protease were separated from EndA H160A using Ni2+-NTA-Agarose. The untagged nuclease could be recovered from the supernatant (Figure 1B, upper panel). A western blot analysis using anti-GST antibody was performed in order to verify nuclease recovery (Figure 1B, lower panel). In a final purification step, residual contaminations were removed from untagged EndA H160A by size-exclusion chromatography over a Superdex-75 HR10/30 column (Figure 1C). This analysis also revealed that EndA eluted as a monomeric protein with an apparent molecular mass of 25.5 ± 4 kDa (Mtheo = 26.8 kDa) as derived from three independent gel filtration runs (Figure 1D).
Analysis of EndA by SAXS
In order to determine its approximate size and shape, EndA was analyzed with SAXS (Figure 2A). Guinier plots were found to be linear over a q-range of 0.012–0.079 Å, consistent with the conclusion that the samples are monodisperse. Guinier analysis of the low-q regions yielded Rg values of 19.1 ± 0.3 Å averaged over four concentrations with a Dmax value of 55 Å; no significant variation of Rg was observed upon dilution. The calculated R value is similar to the value of 18.6 Å calculated from the crystal structure of the monomeric Anabaena nuclease A (pdb file: 1zm8) (21). Additionally, a I2 value of 1.9 is obtained by fitting the raw SAXS data to the NucA crystal structure using CRYSOL, suggesting that the two structures are similar in their global fold (16,21). Io analysis, based on the scattering of a standard protein (horse-heart cytochrome c), indicates that the molecular mass of EndA is 29.0 kDa, consistent with expectations for a monomer (Figure 2A). An ab initio shape was constructed from the I(q) scattering data using the program GASBOR22IQW (Figure 2B) (15). The resulting model has a globular shape with dimensions similar to those of NucA, further supporting the similarity of the shape and solution stoichiometry of the two proteins (21).
Chemical rescue of EndA H160A activity
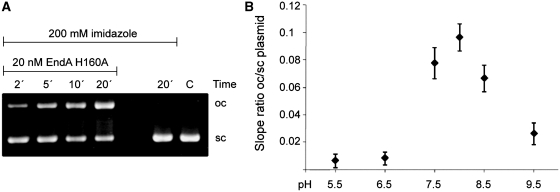
For EndA H160A, as expected, no nucleolytic activity was detectable in any activity assay we applied. However, since this variant was the only variant producible in E. coli, we tried to restore its activity by supplementing the reaction buffer with excess imidazole, which in principle could complement the deleted side chain that had been removed by substitution of the histidine residue by alanine (Figure 3) (22,23). Using a plasmid-activity assay the ratio of open circular to supercoiled DNA was calculated based on the intensity of the DNA bands obtained at selected time points at different pH values. Intriguingly, EndA H160A showed nucleolytic activity at pH values ranging from pH 7.5 to 8.5 at a concentration of 200 mM imidazole as detected by an increase with time of open, circular plasmid DNA (Figure 3B), demonstrating restoration of enzyme activity by supplementing the chemical group required for the initiation of catalysis. As a control, the plasmid DNA was incubated with 200 mM imidazole without enzyme for 20 min at the same pH values as used for the rescue experiments without any detectable effect on the plasmid DNA.
Figure 3.
Chemical rescue of the nuclease activity of the catalytically inactive EndA His160A variant by imidazole. (A) Aliquots of the reaction mixture containing 20 nM EndA H160A and 15 ng/µl pBSK plasmid in triple buffer at pH 8.0 were taken at the time points indicated and the reaction products analyzed by 0.8% agarose gel electrophoresis. The analysis shows an increase of open circular (oc) and a decrease of supercoiled (sc) plasmid DNA over time. Plasmid DNA incubated for 20 min in the absence of EndA H160A and plasmid (C) were loaded as controls. (B) The experiment was repeated at different pH-values (pH 5.5–9.5) and the ratio of open circular to super-coiled plasmid DNA plotted against time (n = 3). EndA H160A showed nucleolytic activity at slightly basic pH values ranging from pH 7.5 to 8.5.
Expression of wild-type EndA and variants using a cell-free transcription/translation system
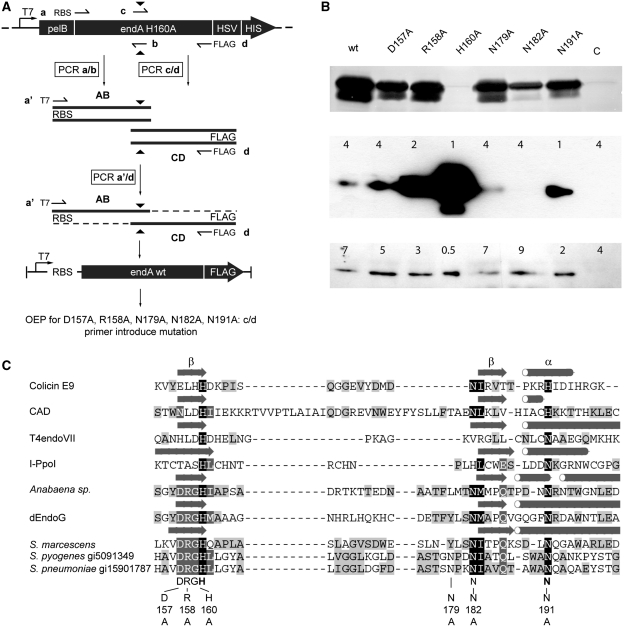
As it turned out to be impossible to express recombinant wild-type EndA protein in E. coli, we used a cell-free coupled transcription/translation system to produce wild-type EndA and several EndA variants. Wild-type EndA template DNA for cell-free expression was generated by overlap-extension PCR using the EndA H160A encoding plasmid to amplify two overlapping gene segments in which the codon for Ala160 was reverted to one coding for histidine (Figure 4A) (12). Codon replacement was mediated by two internal primers b and c containing the nucleotide substitution within the overlapping region of the segmented gene. The flanking primer a included a ribosomal binding site for extending the gene with a T7 promoter sequence by amplifying the hybridization products with the universal primer a’ also containing the RBS as complementary sequence for the second PCR. The resulting protein was FLAG-tagged at its C-terminus, due to flanking primer d containing the corresponding nucleotide sequence as an add-on. The amplified wild-type endA sequence was used in turn as a template for overlap-extension PCR to generate the templates for variants D157A, R158A, H160A, N179A, N182A and N191A. Variant H160A was amplified using primers a and d and in a second PCR extended with a T7-promoter sequence using primers a′ and d.
Figure 4.
(A) Generation of template DNA for in vitro protein synthesis via overlap extension PCR. Plasmid pET25+ harboring the gene coding for the inactive EndA H160A variant was used as a template for overlap extension PCR to generate the gene for wild-type EndA. The endA H160A gene was segmented via amplification in two separate PCR reactions with primer a containing a ribosomal binding site and primer b for segment AB, and primers c and d containing the sequence coding for the FLAG tag in segment CD. The full-length gene was generated by using primer a′ containing a T7 promoter sequence and primer d. Replacement of Ala160 by histidine was achieved by the two internal primer b and c introducing the nucleotide substitution (black triangle) within the overlap of the gene segments. The PCR product was the template for in vitro synthesis of wild-type EndA and for overlap extension PCR to produce the EndA variants D157A, R158A, N179A, N182A and N191A using primer c and d to introduce the mutations. The gene for the variant H160A was generated with primer a and d and in a second PCR extended with the T7 promoter sequence with primer a′, using d as reverse primer. (B) In-gel activity assay and western blot analysis of wild-type EndA and its variants. Upper panel: nuclease activity of the crude in vitro protein synthesis reaction mixture of wild-type EndA and variants D157A, R158A, H160A, N179A, N182A and N191A separated on an SDS–PAGE gel containing high-molecular-weight herring sperm DNA (ethidium bromide stained). The reaction mixture with the control template supplied with the In vitro Protein Synthesis Kit (New England BioLabs) was used as a negative control (C). Dark bands correspond to gel areas with digested DNA that are not stained with ethidium bromide. Middle and lower panel: western blot analysis of FLAG-tagged EndA and variants D157A, R158A, H160A, N179A, N182A, N191A using an anti-FLAG antibody. Numbers indicate the amounts (µl) of the respective crude synthesis reaction mixture loaded onto the gel. (C) Alignment of DNA-entry nucleases with other ββα-Me-finger enzymes (H-N-H/N- and DRGH-motif containing nucleases) of known structure. Indicated are amino acid residues selected for a substitution by alanine in S. pneumoniae EndA. Mutations at positions 157, 158 and 160 concern the conserved DRGH-motif. Histidine at position 160 also represents the first amino acid within the H-N-H/N-motif and plays a central role in catalysis. One of two asparagines at positions 179 and 182 could represent the first asparagine residue of the H-N-H/N-motif, with asparagine 191 being the second. The alignment was manually refined based on the structures of the listed enzymes and our mutational analysis (Colicin E9, pdb code 1emv; Caspase-activated DNase (CAD), pdb-code 1v0d; T4 endonuclease VII, pdb code 1e7d; I-PpoI, pdb-code 1a74; Anabaena nuclease (NucA), pdb code 1zm8; Endonuclease G (dEndoG) from D. melanogaster, pdb code 3ism; Serratia nuclease, pdb code 1smn). Secondary structure elements forming the core of the ββα-Me-finger active-site motif are indicated by arrows (β-sheets) and tubes (α-helices) as derived from the respective crystal structures. For the DNA-entry nucleases from S. pyogenes and S. pneumoniae (EndA) the gi numbers are indicated.
Guided by the sequence alignment of DRGH-motif containing proteins presented in Figure 4C we selected certain amino-acid residues for our mutational analysis of the presumptive active site of streptococcal EndA. Asp157, Arg158 and His160 from the conserved DRGH-motif, all localized in one of the two β-strands of the ββα-Me finger structure, were exchanged for alanine (7,24,25). His160 within the DRGH-motif corresponds to the first histidine residue in the H-N-H-motif and plays an essential role in catalysis acting as the general base (26,27). Furthermore, we also exchanged two asparagine residues (Asn179 and Asn182) for alanine. As candidates for the central asparagine residue in the H-N-H-motif, these residues might be involved in hydrogen-bond network formation and stabilization of the ββα-Me finger structure (28,29). According to our sequence alignment, another asparagine residue, Asn191, might assume the function of the metal ion ligand corresponding to the histidine residue at the last position of the H-N-H motif. In fact, DRGH-motif nucleases generally contain an H-N-N motif (7,24,25).
Even though we were able to detect, by Coomassie staining, protein synthesized with the control template supplied with our cell-free transcription/translation system, wild-type EndA and EndA variants could not be detected by Coomassie staining after separation on SDS–PAGE gels. However, a strong nuclease activity could be detected for all variants, except for EndA H160A and the control template supplied with the in vitro expression cocktail, using an in-gel activity assay after separation of synthesized proteins on SDS–PAGE gels containing high-molecular-weight herring sperm DNA, after refolding and ethidium bromide staining (Figure 4B, upper panel). It should be noted here, that the activity seen in the in-gel assay might not reflect the maximum activity of a given EndA variant, since all proteins were refolded in the same buffer but refolding efficiencies for the variants might vary for the wild-type and mutant proteins. All EndA variants could be detected by Western blot analysis employing an anti-FLAG antibody (Figure 4B, middle/lower panel). Intriguingly, the amounts of protein produced in the cell-free expression system apparently were inversely correlated to the activity of a given variant (for activity, see Figure 5). The variants were produced in different amounts although the template concentration used for the protein synthesis reaction was equal in all reactions. It is very likely that the synthesized nucleases in the coupled transcription/translation system are able to degrade the template DNA according to their specific activity. Thus, the amount of protein produced for a given variant increases in a feedback loop with the loss of activity of a given nuclease variant caused by the amino acid exchange. This is most obvious when comparing wild-type EndA with the inactive variant H160A, which was produced to the highest protein concentration measured via Western blot analysis (Figure 4B, middle and lower panel). Interestingly, the variant EndA-N182A is an exception to this rule. Although it only shows a moderate cleavage activity, this variant apparently was instable producing only a weak signal in the western blot when analyzed immediately after the synthesis reaction and a decreased signal when analyzed after a storage time of 2 weeks (data not shown).
Figure 5.
SRED assay: (A) Relative nuclease activity of the in vitro protein synthesis reaction mixture containing wild-type EndA, D157A, R158A, N179A, N182A and N191A tested with high-molecular-weight herring sperm DNA embedded in 1% agarose (prestained with ethidium bromide), 40 mM Tris–HCl, pH 7.7, 2 mM MgCl2. Variant H160A showed no measurable activity in this assay. The reaction mixture with the control template supplied with the in vitro Protein Synthesis Kit (New England BioLabs) was used as a negative control (C), DNaseI (Fermentas) as positive control. The ruler indicates the diameter of the halos. (B) Calibration curve for the estimation of relative nucleolytic activity from the size of the halos. The size of the halos (diameter in millimeters) was plotted against the amount (µl) of the protein synthesis reaction mixture containing wild-type EndA. (C) The relative nucleolytic activity of each EndA variant contained in the crude reaction mixture was determined from the diameter of the halos using the calibration curve in (B) and the concentration of the EndA variants in the crude reaction mixture determined by western blot analysis. The activity of wild-type EndA is set to 100%.
Determination of the activity of wild-type EndA and EndA variants
The nucleolytic activity of the crude protein-synthesis reaction mixture was quantified by measuring the size of the halo by a single-radiation enzyme diffusion (SRED) assay (Figure 5). For each enzyme variant, 1 µl of the synthesis reaction mixture was spotted onto an agar plate containing high-molecular-weight DNA. Whereas all EndA variants showed nuclease activity, the control template supplied for the in vitro expression cocktail showed none (Figure 5A). A quantity of 1 U of bovine pancreatic DNase I was used as a positive control. The relative concentration of each variant was determined by western blotting (Figure 4B, lower panel). To calibrate the system, different amounts of the wild-type EndA synthesis reaction mixture were used in a separate SRED assay and the size of the halos measured to obtain a calibration curve (Figure 5B). Since the concentrations of the variants relative to that of wild-type EndA was known from the western blot analysis, the relative activities of the variants could be determined (Figure 5C).
In a second type of activity assay, wild-type EndA was tested on single-stranded, double-stranded, supercoiled and nicked-phage DNAs, respectively, with a dilution of 1:100 of the crude protein-synthesis reaction mixture (Figure 6). No pronounced difference of wild-type EndA could be detected for the cleavage of single- or double-stranded DNA as all substrates were digested almost to the same extent, yet with a slight preference of the enzyme for single-stranded DNA. The protein synthesis mixtures obtained using either the template control supplied by the in vitro expression kit or the EndA H160A template, encoding the putatively inactivated enzyme, did not show any nucleolytic activity in this assay (data not shown).
Figure 6.
Substrate preference of wild-type EndA: Nucleolytic activity of wild-type EndA was tested on (A) ΦX174 DNA (single-stranded, circular), (B) RF I DNA (double-stranded, circular) and (C) RF II DNA (double-stranded, nicked). Wild-type EndA shows no substrate preference (ss—single stranded, sc—super coiled, oc—open circle, li—linear).
DISCUSSION
Streptococcus pneumoniae (pneumococci) are pathogenic Gram-positive bacteria causing serious infections of the respiratory system of mammalians and lethal meningitis (30). Other typical streptococcal diseases like streptococcal toxic shock syndrome (STSS), necrotizing fasciitis (NF) and tonsillitis are also triggered by group A streptococci (GAS; Streptococcus pyogenes) (31). This group of bacteria exhibit several serotypes expressing different extracellular virulence proteins, among them nucleases such as streptodornase (32). Several of these virulence factors are phage encoded, supporting recombination and spreading of newly arranged pathogenic genes presumably creating the high diversity of GAS strains (33). GAS strains express several types of streptodornases which support circumvention of the innate immune response of mammalians resulting in invasive infections (34,35). One mechanism for this was discovered recently as streptodornases are able to degrade the DNA scaffold of NETs (3). NETs are released by activated neutrophils and consist of expelled chromatin, elastase and other antimicrobial proteins and kill captured bacteria (5). This mechanism was also detected for S. pneumoniae (TIGR4) which express the competence associated nuclease EndA. This surface-bound nuclease plays a major role in the degradation of extracellular DNA to allow pneumococci to escape NETs and regain viability (4).
In the study presented here, we were unable to express active forms of EndA in E. coli even when using tightly regulated intracellular expression systems, anti-sense strategies or fusion of EndA to the pelB-leader sequence to secrete the recombinant wild-type enzyme into the periplasmic space. Due to the intrinsic toxicity of nucleases, cloning and recombinant expression of their genes in E. coli or other host organisms can be highly problematic unless, e.g. nuclease inhibitors are available for intracellular co-expression or the enzyme is produced as an inactive pro-enzyme that becomes activated by secretion into the extracellular space (20,36–38). EndA from S. pneumoniae is a secreted surface exposed endonuclease for which up to date no inhibitory protein is known (2). All cloning attempts for the EndA-gene in E. coli led to mutations in the region coding for the active site and/or insertion of stop codons in the open reading frame preventing expression of active full-length protein. EndA could only be cloned and expressed as an inactive variant in which the putative active-site histidine residue His160 was exchanged for alanine (H160A). This variant could be produced in high amounts either intracellularly as a His-GST double-tagged protein (Figure 1B) or upon secretion into the extracellular space of E. coli, which allowed recovery of the nuclease from the culture medium (Figure 1A). The DNA encoding the H160A variant was also used to correct for mutations that had accumulated in templates from previous cloning attempts in order to revert the coding sequence to the wild-type sequence according to gene bank entry (gi: 47374; data not shown).
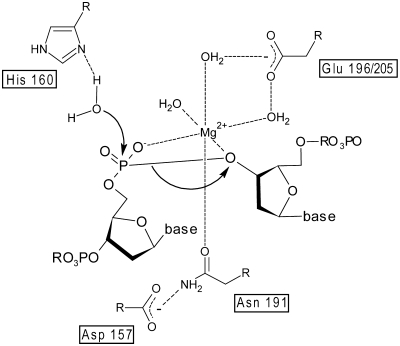
In order to perform a first mutational and biochemical analyses of EndA, we used overlap extension PCR to splice DNA templates coding for wild-type EndA and other desired variants (Figure 4A). Using this approach we were able to produce a series of EndA variants using a cell-free expression system. A subsequent analyses of in vitro translated wild-type EndA and EndA variants confirmed our expectations concerning amino acid residues constituting the active site of this DRGH-motif containing nuclease. According to our data and in agreement with the results obtained for so-called sugar-nonspecific nucleases such as Serratia nuclease, Anabaena nuclease or Endonuclease G, His160 from the DRGH-motif of EndA is crucial for catalysis, most likely by acting as a general base (Figure 7) (25,26,39). This assignment of the role of His160 as a general base in the catalytic mechanism of EndA is strongly supported by chemical rescue of the activity of the H160A variant with excess imidazole in the reaction buffer (Figure 3). At a slightly basic pH the imidazole is deprotonated and able to accept a proton from a water molecule nearby, thus very likely generating a nucleophile able to attack a phosphodiester bond. In the H160A variant, imidazole apparently occupies the space that is normally filled by the side chain of the active site histidine residue in the wild-type enzyme but absent in this variant. EndA H160A, therefore, is able to catalyze the cleavage of phosphodiester bonds in the presence of imidazole at a slightly basic pH. However, the activity observed for the rescued EndA H160A variant is approximately two orders of magnitude lower than that observed for the in vitro translated wild-type enzyme. In order to exploit the possibility of restoring activity by chemical rescue of inactive and thus non-toxic enzyme variants of EndA that can be produced in E. coli, we are currently generating other mutants with substitutions of the active-site histidine residue with the goal to produce variants with higher activity that can be used, e.g. in future high-throughput screening assays.
Figure 7.
Catalytic mechanism of wild-type EndA: the proposed catalytic mechanism of EndA is based on the established catalytic mechanism of the Serratia nuclease (26). His160 acts as the general base for the activation of a water molecule. The magnesium ion is coordinated by Asn191, which is held in place by Asp157, and the proS-oxygen as well as the 3′-oxygen of the phosphate diester. A glutamic acid residue residue nearby (Glu196 or Glu205 in EndA) is assumed to be indirectly involved in water-mediated magnesium ion binding.
As expected, replacement of amino acid residues from the conserved DRGH-motif of EndA at the first and second position by alanine (variants D157A and R158A) also diminishes nuclease activity, albeit to a lower extent (Figure 4). It is known from the abovementioned studies concerning Serratia nuclease, Anabaena nuclease and EndoG, that these residues are involved in proper positioning of the conserved asparagine residue, which is binding the divalent metal ion cofactor in the enzyme’s active site (Figure 7) (7,24). Furthermore, a so-called H-N-H-motif is characteristic for a large number of proteins covering a broad variety of functions such as growth inhibition of bacteria (bacteriocins), phage defense (restriction endonucleases) and recombination (homing endonucleases) (27). Sequence analyses show a conserved H-N-N-motif in EndA (Figure 4C). As two asparagines (N179, N182) were likely candidates for the second position within the H-N-N motif, we selected both of them for an alanine replacement. However, only variant N182A shows a marked drop in activity indicating that N182 very likely is part of the active site (Figure 5B) and probably involved in constraining the loop structure of the ββα-Me finger as shown for ColE7 at the corresponding position, thus contributing to the stability of the protein (28). This explains our finding that although variant N182A is less active than wild-type EndA, its concentration obtained in the cell free expression system is lower due to its general instability compared to variants with mutations directly affecting catalysis (Figure 5B). The amount of produced protein for all other variants in general is correlated with a decrease in activity. The less active a variant, the more protein can be produced in the cell-free expression assay, very likely due to degradation of the template DNA and/or mRNA by more active nuclease variants. The conserved aspargine residue, which corresponds to the last histidine residue in the H-N-H-motif is known to be the direct ligand responsible for metal ion binding as in the case of Serratia nuclease, Anabaena nuclease, and mitochondrial EndoG (7,25,29). Replacement of Asp191 by alanine in EndA leads to a decrease in activity suggesting that this residue is involved in catalysis, presumably by binding the divalent metal ion cofactor (Figures 5B and 7).
In the course of genetic transformation, Streptococci take up single-stranded linear DNA segments (40,41). Using different phage DNA substrates we could not detect a truly pronounced substrate preference for the isolated recombinant enzyme, although single-stranded virion DNA was cleaved slightly faster than double stranded DNA (Figure 6). EndA exerts a strong nicking activity towards supercoiled DNA arguing for a distributive haplotomic mechanism of action of the enzyme. This DNA cleavage mechanism is characteristic of monomeric as well as some dimeric nonspecific nucleases with independently acting subunits and supported by the fact that EndA behaves as a monomer in size-exclusion chromatography (Figure 1C). As single-stranded DNA serves as a substrate for EndA, the question of regulation of the enzyme activity on the membrane surface of streptococci arises. For the 17-kDa entry nuclease (NucA) from Bacillus subtilis, an inhibition of nuclease activity was observed in vitro by an 18-kDa protein (Nin) (42). Several Com proteins on the membrane of B. subtilis are involved in binding and transport of DNA into the cell similar as suggested for S. pneumoniae (43). It is, therefore, tempting to speculate that one of the Com proteins is interacting with EndA, thereby regulating its function and perhaps also being responsible for an inhibitory activity within the cell.
Employing inactivating mutations (H160A) or a cell-free expression system, we were able to produce recombinant forms of the streptococcal virulence factor EndA. We could demonstrate by an alignment-guided mutational analysis that EndA, which contains a DRGH- and an H-N-H(N)-motif, very likely follows a similar catalytic mechanism as described for other members of the ββα-Me finger nuclease superfamily, utilizing a divalent metal ion directly bound by Asn191 and a catalytically active histidine residue (His160) probably acting as the general base. Up to date, no mechanism for the regulation of this nuclease is known that, due to its involvement in bacterial pathogenesis, has become an interesting target for future drug design. Being able to produce and/or regenerate active forms of recombinant EndA and understanding its catalytic mechanism is a prerequisite for the future development of protein or low-molecular-weight inhibitors for this nuclease, responsible for the degradation of NETs and circumvention of the innate immune system during streptococcal infection.
FUNDING
German Research Foundation (DFG); the ‘Excellence Cluster Cardio-Pulmonary System’ (ECCPS), and the Dr -Herbert-Stolzenberg-Stiftung of the Justus-Liebig-University Giessen. Funding for open access charge: German Research Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the laboratory of Dr Lin Yang of the X9 beamline, at the National Synchrotron Light Source at Brookhaven National Laboratory and Dr Liang Guo of the 18-ID BioCat Beamline, at the Advanced Photon Source at the Argonne National Laboratory, for assistance with data collection. Use of the X9 beamline is supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No W-31-109-Eng-38.
REFERENCES
- 1.Lacks S, Neuberger M. Membrane location of a deoxyribonuclease implicated in the genetic transformation of Diplococcus pneumoniae. J. Bacteriol. 1975;124:1321–1329. doi: 10.1128/jb.124.3.1321-1329.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puyet A, Greenberg B, Lacks SA. Genetic and structural characterization of endA. A membrane-bound nuclease required for transformation of Streptococcus pneumoniae. J. Mol. Biol. 1990;213:727–738. doi: 10.1016/S0022-2836(05)80259-1. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 2006;16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 4.Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 2006;16:401–407. doi: 10.1016/j.cub.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 5.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 6.von Kockritz-Blickwede M, Goldmann O, Thulin P, Heinemann K, Norrby-Teglund A, Rohde M, Medina E. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood. 2008;111:3070–3080. doi: 10.1182/blood-2007-07-104018. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh M, Meiss G, Pingoud AM, London RE, Pedersen LC. The nuclease a-inhibitor complex is characterized by a novel metal ion bridge. J. Biol. Chem. 2007;282:5682–5690. doi: 10.1074/jbc.M605986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhlmann UC, Moore GR, James R, Kleanthous C, Hemmings AM. Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett. 1999;463:1–2. doi: 10.1016/s0014-5793(99)01499-4. [DOI] [PubMed] [Google Scholar]
- 9.Muro-Pastor AM, Flores E, Herrero A, Wolk CP. Identification, genetic analysis and characterization of a sugar-non-specific nuclease from the Cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 1992;6:3021–3030. doi: 10.1111/j.1365-2958.1992.tb01760.x. [DOI] [PubMed] [Google Scholar]
- 10.Miller MD, Benedik MJ, Sullivan MC, Shipley NS, Krause KL. Crystallization and preliminary crystallographic analysis of a novel nuclease from Serratia marcescens. J. Mol. Biol. 1991;222:27–30. doi: 10.1016/0022-2836(91)90734-n. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz-Carrillo A, Renaud J. Endonuclease G: a (dG)n X (dC)n-specific DNase from higher eukaryotes. EMBO J. 1987;6:401–407. doi: 10.1002/j.1460-2075.1987.tb04769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 13.Konarev PV, Sokolova VV, Sokolova AV, Koch MH, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 14.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 15.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svergun DI, Koch MH. CRYSOL – a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 17.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]
- 18.Nadano D, Yasuda T, Kishi K. Measurement of deoxyribonuclease I activity in human tissues and body fluids by a single radial enzyme-diffusion method. Clin. Chem. 1993;39:448–452. [PubMed] [Google Scholar]
- 19.Friedhoff P, Gimadutdinow O, Ruter T, Wende W, Urbanke C, Thole H, Pingoud A. A procedure for renaturation and purification of the extracellular Serratia marcescens nuclease from genetically engineered Escherichia coli. Protein Expr. Purif. 1994;5:37–43. doi: 10.1006/prep.1994.1005. [DOI] [PubMed] [Google Scholar]
- 20.Korn C, Meiss G, Gast F, Gimadutdinow O, Urbanke C, Pingoud A. Genetic engineering of Escherichia coli to produce a 1:1 complex of the anabaena sp. PCC 7120 nuclease NucA and its inhibitor NuiA. Gene. 2000;253:221–229. doi: 10.1016/s0378-1119(00)00267-5. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh M, Meiss G, Pingoud A, London RE, Pedersen LC. Structural insights into the mechanism of nuclease A, a betabeta alpha metal nuclease from Anabaena. J. Biol. Chem. 2005;280:27990–27997. doi: 10.1074/jbc.M501798200. [DOI] [PubMed] [Google Scholar]
- 22.Toney MD, Kirsch JF. Direct Bronsted analysis of the restoration of activity to a mutant enzyme by exogenous amines. Science. 1989;243:1485–1488. doi: 10.1126/science.2538921. [DOI] [PubMed] [Google Scholar]
- 23.Carter P, Wells JA. Engineering enzyme specificity by “substrate-assisted catalysis”. Science. 1987;237:394–399. doi: 10.1126/science.3299704. [DOI] [PubMed] [Google Scholar]
- 24.Miller MD, Cai J, Krause KL. The active site of Serratia endonuclease contains a conserved magnesium-water cluster. J. Mol. Biol. 1999;288:975–987. doi: 10.1006/jmbi.1999.2729. [DOI] [PubMed] [Google Scholar]
- 25.Loll B, Gebhardt M, Wahle E, Meinhart A. Crystal structure of the EndoG/EndoGI complex: mechanism of EndoG inhibition. Nucleic Acids Res. 2009;37:7312–7320. doi: 10.1093/nar/gkp770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meiss G, Gimadutdinow O, Haberland B, Pingoud A. Mechanism of DNA cleavage by the DNA/RNA-non-specific Anabaena sp. PCC 7120 endonuclease NucA and its inhibition by NuiA. J. Mol. Biol. 2000;297:521–534. doi: 10.1006/jmbi.2000.3586. [DOI] [PubMed] [Google Scholar]
- 27.Eastberg JH, Eklund J, Monnat R, Jr, Stoddard BL. Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism. Biochemistry. 2007;46:7215–7225. doi: 10.1021/bi700418d. [DOI] [PubMed] [Google Scholar]
- 28.Huang H, Yuan HS. The conserved asparagine in the HNH motif serves an important structural role in metal finger endonucleases. J. Mol. Biol. 2007;368:812–821. doi: 10.1016/j.jmb.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 29.Mate MJ, Kleanthous C. Structure-based analysis of the metal-dependent mechanism of H-N-H endonucleases. J. Biol. Chem. 2004;279:34763–34769. doi: 10.1074/jbc.M403719200. [DOI] [PubMed] [Google Scholar]
- 30.Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 2008;6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 31.Bronze MS, Dale JB. The reemergence of serious group A streptococcal infections and acute rheumatic fever. Am. J. Med. Sci. 1996;311:41–54. doi: 10.1097/00000441-199601000-00008. [DOI] [PubMed] [Google Scholar]
- 32.Ferreira BT, Benchetrit LC, De Castro AC, Batista TG, Barrucand L. Extracellular deoxyribonucleases of streptococci: a comparison of their occurrence and levels of production among beta-hemolytic strains of various serological groups. Zentralb. Bakteriol. 1992;277:493–503. doi: 10.1016/s0934-8840(11)80474-3. [DOI] [PubMed] [Google Scholar]
- 33.Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu MY, Smoot JC, Porcella SF, Parkins LD, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl Acad. Sci. USA. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Podbielski A, Zarges I, Flosdorff A, Weber-Heynemann J. Molecular characterization of a major serotype M49 group A streptococcal DNase gene (sdaD) Infect. Immun. 1996;64:5349–5356. doi: 10.1128/iai.64.12.5349-5356.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, et al. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl Acad. Sci. USA. 2005;102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ball TK, Suh Y, Benedik MJ. Disulfide bonds are required for Serratia marcescens nuclease activity. Nucleic Acids Res. 1992;20:4971–4974. doi: 10.1093/nar/20.19.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallis R, Reilly A, Barnes K, Abell C, Campbell DG, Moore GR, James R, Kleanthous C. Tandem overproduction and characterisation of the nuclease domain of colicin E9 and its cognate inhibitor protein Im9. Eur. J. Biochem. 1994;220:447–454. doi: 10.1111/j.1432-1033.1994.tb18642.x. [DOI] [PubMed] [Google Scholar]
- 38.Kleanthous C, Kuhlmann UC, Pommer AJ, Ferguson N, Radford SE, Moore GR, James R, Hemmings AM. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 39.Schafer P, Scholz SR, Gimadutdinow O, Cymerman IA, Bujnicki JM, Ruiz-Carrillo A, Pingoud A, Meiss G. Structural and functional characterization of mitochondrial EndoG, a sugar non-specific nuclease which plays an important role during apoptosis. J. Mol. Biol. 2004;338:217–228. doi: 10.1016/j.jmb.2004.02.069. [DOI] [PubMed] [Google Scholar]
- 40.Lacks S. Molecular fate of DNA in genetic transformation of Pneumococcus. J Mol. Biol. 1962;5:119–131. doi: 10.1016/s0022-2836(62)80067-9. [DOI] [PubMed] [Google Scholar]
- 41.Gabor M, Hotchkiss RD. Manifestation of linear organization in molecules of pneumococcal transforming DNA. Proc. Natl Acad. Sci. USA. 1966;56:1441–1448. doi: 10.1073/pnas.56.5.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vosman B, Kuiken G, Kooistra J, Venema G. Transformation in Bacillus subtilis: involvement of the 17-kilodalton DNA-entry nuclease and the competence-specific 18-kilodalton protein. J. Bacteriol. 1988;170:3703–3710. doi: 10.1128/jb.170.8.3703-3710.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Provvedi R, Chen I, Dubnau D. NucA is required for DNA cleavage during transformation of Bacillus subtilis. Mol. Microbiol. 2001;40:634–644. doi: 10.1046/j.1365-2958.2001.02406.x. [DOI] [PubMed] [Google Scholar]