Abstract
X-box binding protein 1 (XBP-1) is a key regulator required for cellular unfolded protein response (UPR) and plasma cell differentiation. In addition, involvement of XBP-1 in host cell–virus interaction and transcriptional regulation of viruses, such as human T-lymphotropic virus type 1 (HTLV-1), has been revealed recently. Two XBP-1 isoforms, XBP-1U and XBP-1S, which share an identical N-terminal domain, are present in cells. XBP-1S is a transcription activator while XBP-1U is the inactive isoform. Although the transactivation domain of XBP-1S has been identified within the XBP-1S-specific C-terminus, molecular mechanism of the transcriptional activation by XBP-1S still remains unknown. Here we report the interaction between p300/CBP-associated factor (PCAF) and XBP-1S through the C-terminal domain of XBP-1S. No binding between XBP-1U and PCAF is detected. In a cell-based reporter assay, overexpression of PCAF further stimulates the XBP-1S-mediated cellular and HTLV-1 transcription while knockdown of PCAF exhibits the opposite effect. Expression of endogenous XBP-1S cellular target genes, such as BiP and CHOP, is significantly inhibited when PCAF is knocked down. Furthermore, PCAF is recruited to the promoters of XBP-1S target genes in vivo, in a XBP-1S-dependent manner. Collectively, our results demonstrate that PCAF mediates the XBP-1S-dependent transcription through the interaction with XBP-1S.
INTRODUCTION
X-box binding protein 1 (XBP-1) belongs to the cyclic AMP response element binding protein/activating transcription factor (CREB/ATF) family of transcription factors. XBP-1 plays a major role in regulating unfolded protein response (UPR), which is triggered when endoplasmic reticulum (ER) is under stress (1). XBP-1 has two protein isoforms, XBP-1U and XBP-1S. Both isoforms share a common N-terminus containing a basic-region leucine zipper (bZIP) domain which is required for DNA binding. XBP-1U is the dominant isoform under non-stress conditions. Activation of UPR induces the endoribonuclease activity of inositol requiring enzyme 1, an ER transmembrane protein, which removes 26 nts from the open-reading frame of XBP-1 mRNA (2). This unconventional splicing occurs in cytoplasm and causes a frame shift at amino acid 165 of XBP-1, leading to the generation of XBP-1S by replacing the C-terminus of XBP-1U with a strong transactivation domain (2,3). XBP-1S is a transcription activator that up-regulates the expression of ER chaperones and other genes involved in membrane synthesis and the pathway of protein secretion (4,5). Overexpression of XBP-1S increases the secretory capacity of the cell and improves recombinant protein productivity in secretion-limited mammalian cells by expanding the surface area and volume of ER (5,6). It has been shown recently that high-level expression of recombinant secreted proteins in cells and environmental stresses during culture also induce the generation of XBP-1S (7). XBP-1S is also found to be essential in the terminal differentiation of the antibody producing plasma cells by enhancing the secretory machinery of the cell (8,9). The XBP-1-knockout B cells display impaired immunoglobulin secretion, which can be restored by ectopic expression of XBP-1S (8). Furthermore, the involvement of XBP-1 in tumorigenesis has been reported recently (10–12).
Recent studies show that cellular UPR can be induced by infection of various viruses, including Kaposi’s sarcoma-associated herpesvirus (13), West Nile virus (14), Japanese encephalitis virus (JEV) (15), hepatitis C virus (16,17), human cytomegalovirus (HCMV) (18,19), dengue virus serotype 2 (DEN-2) (20), severe acute respiratory syndrome coronavirus (21), coronavirus (22), Epstein-Barr virus (23) and Semliki Forest virus (24). Some viruses, such as JEV and DEN-2, use the ER of host cells as the primary site of glycoprotein synthesis, genomic RNA replication and virus particle maturation, and thus trigger ER stress as well as UPR (15,20). In the other case, some viral proteins, such as HCMV US11, traffic to the ER of host cells and induce UPR (18). The transactivator of human T-lymphotropic virus type 1 (HTLV-1), Tax, has been shown to be localized in the organelles associated with protein secretion including ER and Golgi complex (25), raising the possibility that HTLV-1 may affect cellular UPR as well. We previously discovered that XBP-1S stimulates basal and Tax-activated transcription of HTLV-1. Infection of HTLV-1 was found to induce UPR and up-regulation the expression of several UPR genes, including XBP-1. Furthermore, XBP-1 was identified as one of the Tax target genes in cells (26). Our results not only revealed a positive feedback loop between HTLV-1 and the host cells, but also suggested an important role for XBP-1 in transcriptional regulation of HTLV-1.
The localization of a transactivation domain within the C-terminus of XBP-1S helps to explain the transactivating ability of XBP-1S. However, the molecular mechanism of XBP-1S transactivation still remains to be determined. One possibility is that the C-terminus of XBP-1S may interact with a specific cellular co-activator, which is responsible for the up-regulation of XBP-1S target genes. Here, we identify a histone acetyltransferase (HAT), p300/CBP-associated factor (PCAF), as a XBP-1S-specific binding protein and demonstrate the functional significance of the PCAF-XBP-1S interaction in the XBP-1S-mediated transcription.
MATERIALS AND METHODS
Cells, short interfering RNAs, short hairpin RNAs and plasmids
HEK293, 293T and MCF7 cells were obtained from American Type Culture Collection. The short interfering RNAs (siRNAs) targeting PCAF (siPCAF-6: 5′-CGGAGTGTACTCCGCCTGCAA-3′ and siPCAF-7: 5′-CAGCAAATAATTGTCAGTCTA-3′) and p300 (sip300-7 siRNA: 5′-TTGGACTACCCTATCAAGTAA-3′ and sip300-10: 5′-CCCGGTGAACTCTCCTATAAT-3′) were purchased from Qiagen, and the short hairpin RNAs (shRNA) against PCAF (shPCAF: 5′-TAGATGAGGTGCTTTGAGCAGTTCTGAAA-3′) was obtained from Origene. Human XBP-1S and XBP-1U expression plasmids were previously described (26). The plasmids for expression of human PCAF and p300 were obtained from Open Biosystems. The plasmids containing a series of hemagglutinin (HA) tagged XBP-1 deletions were generous gifts from Dr Hiderou Yoshida (27). The firefly luciferase reporter plasmids, HTLV-Luc and BiP-Luc [including wild-type and ER stress response (ERSE) mutant BiP-Luc plasmids], were kindly provided by Dr Arnold Rabson and Dr Kazutoshi Mori, respectively (28,29).
Transient transfection and luciferase assays
Transient transfections of DNA plasmids into HEK293, 293T and MCF7 cells were performed using FuGENE 6 (Roche) according to the manufacturers’ instructions. To perform the cell-based overexpression assays, cells were grown to 50–80% confluence in 96-well plates and co-transfected with a luciferase reporter and an expression plasmid. Lipofectamine 2000 Reagent (Invitrogen) was utilized to co-transfect cells with DNA plasmids and siRNAs for the cell-based knockdown experiments. Firefly luciferase activities were measured 48 h post-transfection using the Bright-Glo assay system (Promega) and the activities were determined using an Infinite 200 multiplate reader (Tecan). HEK293 cells were used in the cell-based luciferase assays.
Co-immunoprecipitation and western blotting
293T cells were transiently co-transfected with indicated expression plasmids and the cell lysates were prepared 2 days post-transfection for co-immunoprecipitation (Co-IP). To get the high levels of ectopic expression, 293T, a highly transfectable derivative of HEK293, was chosen for the Co-IP study. The IP kit was purchased from Roche and Co-IP was performed according to the manufacturers’ instructions. The immunoprecipitated complexes were analyzed by western blotting. Western blotting was carried out according to the standard protocols. All the antibodies used in our study were obtained from Santa Cruz Biotechnology, except the anti-HA antibody (Sigma).
Quantitative reverse transcriptase-polymerase chain reaction
The UPR inducing compounds, tunicamycin (Tm) (Assay Designs) and thapsigargin (Tg) (Sigma), were dissolved in dimethyl sulfoxide (DMSO) to 10 mg/ml and 3 mM, respectively. All three cell lines, HEK293, 293T and MCF7, exhibited UPR after treating with Tm or Tg. Induction of the UPR genes in the treated cells were confirmed by quantitative reverse transcriptase-polymerase chain reaction (QRT–PCR) (data not shown). Among the cell lines used in this study, the endogenous XBP-1S target genes in MCF7 cells showed the highest sensitivity to the ectopic expression of XBP-1S (data not shown). Therefore, MCF7 cells were selected for the XBP-1S overexpression experiments followed by the examination of the transcriptional regulation of XBP-1S-dependent genes in vivo. Total RNAs of the transfected MCF7 cells or the Tm (10 µg/ml)/Tg (300 nM) treated HEK293 cells were isolated using RNeasy mini kit (Qiagen). One microgram of the total RNAs was converted into complementary DNA (cDNA) using ImPromTM-II Reverse Transcription System (Promega). Specific cDNAs were amplified using SYBR Green PCR Master Mix (Applied Biosystems). The primer pairs used in this study include: BiP (5′-GGTGAAAGACCCCTGACAAA-3′ and 5′-GTCAGGCGATTCTGGTCATT-3′), CHOP (5′-CTTCTCTGGCTTGGCTGACT-3′ and 5′-CCCTTGGTCTTCCTCCTCTT-3′), EDEM (5′-AGGTGCTGATAGGAGATGTGG-3′ and 5′-GGATTCTTGGTTGCCTGGTA-3′) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (5′-AACAGCCTCAAGATCATCAGC-3′ and 5′-GGATGATGTTCTGGAGGACC-3′). GAPDH was used as a control to normalize the cDNA inputs. Amplification and detection of the cDNAs were performed using ABI Prism 7000 Thermal-Cycler (Applied Biosystems).
Quantitative chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were carried out using EZ ChIP kit (Millipore) according to the manufacturer’s protocol with some modifications. HEK293 cells were treated with Tm (10 µg/ml) or Tg (300 nM) for 16 h prior to cross-linking. DNA fragments at around 200–1000 bp were achieved by sonication with Microson Ultrasonic Cell Disruptor (Misonix). For the IP, the indicated antibodies (i.e. anti-XBP-1 or anti-PCAF antibodies) were added to the sheared chromatin individually and incubated at 4°C overnight. The DNA/protein/antibody complex was then pulled down by protein G agarose and the DNA in the complex was purified using QIAquick PCR purification kit (Qiagen). Quantitative-PCR was performed to determine the relative amount of DNA that was immunoprecipitated by anti-XBP-1 or anti-PCAF antibodies in the presence of Tm or Tg. The primer pairs used to amplify the promoter regions of BiP and CHOP genes include: BiP (5′-GATGGGGCGGATGTTATCTA-3′ and 5′-CTCTCACACTCGCGAAACAC-3′) and CHOP (5′-GACACTACGTCGACCCCCTA-3′ and 5′-GGTTCCAGCTCTGATTTTGG-3′). Cells treated with DMSO were served as a negative control. For the overexpression study, MCF7 cells were co-transfected with a PCAF expression vector and one of the XBP-1 plasmids (XBP-1S or XBP-1U plasmids) 2 days prior to cross-linking. Cells co-transfected with a PCAF plasmid and an empty vector served as a negative control.
Statistical analysis
The data shown (including luciferase assays, QRT–PCR and quantitative ChIP) were analyzed using Student’s t-test at 5% significance level (P < 0.05).
RESULTS
XBP-1S interacts with PCAF through its C-terminal transactivation domain
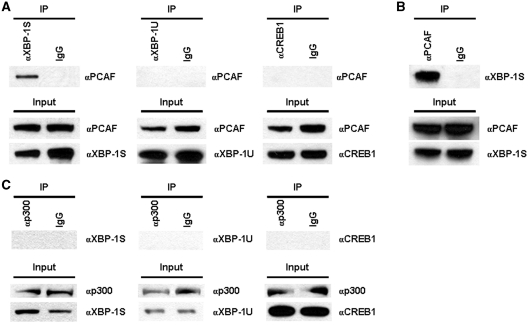
We previously demonstrated that XBP-1S, a member of CREB/ATF family proteins, stimulates basal and Tax-activated HTLV-1 transcription (26). It has been reported that two histone acetyltransferases (HATs), PCAF and p300, are required to activate HTLV-1 transcription through three 21-bp repeats known as Tax-responsive element (TRE) located with the HTLV-1 promoter (30). Each TRE contains a binding site for CREB/ATF proteins, suggesting a potential functional connection between HATs and XBP-1S. We first investigated the interaction between PCAF and two XBP-1 isoforms. Cells were transfected with an XBP-1S or XBP-1U expression plasmid followed by IP analyses (Figure 1A). The anti-XBP-1 antibody used in the assays can recognize both XBP-1 isoforms. The association between PCAF and another member of CREB/ATF protein family, CREB1, was also examined (Figure 1A). PCAF was found in the immunoprecipitated complexes of XBP-1S expressing cells, but not in XBP-1U or CREB1 expressing cells (Figure 1A). Reciprocal IP was carried out using an anti-PCAF antibody and XBP-1S was detected in the immunoprecipitated complexes, confirming the interaction between PCAF and XBP-1S (Figure 1B). Interaction between XBP-1S and another HAT, p300, was examined next. However, no association between p300, XBP-1S, XBP-1U and CREB1 was detected (Figure 1C), indicating a specific binding between PCAF and XBP-1S.
Figure 1.
PCAF associates with XBP-1S. (A) 293T cells were transfected with an expression plasmid to ectopically express XBP-1S, XBP-1U and CREB1, respectively. IP was performed using the cell lysates prepared from the transfected cells and the indicated antibodies. Normal IgG (IgG) was used as a negative control. The immunoprecipitated complexes and the protein inputs were analyzed by western blotting. (B) The cell lysates of XBP-1S expressing cells were used for IP with an anti-PCAF antibody. The presence of XBP-1S in the immunoprecipitates was determined by western blotting. (C) Cells were co-transfected with a p300 expression vector and an indicated plasmid (i.e. XBP-1S, XBP-1U, and CREB1 plasmids, respectively). IP was carried out using an anti-p300 antibody followed by western blotting.
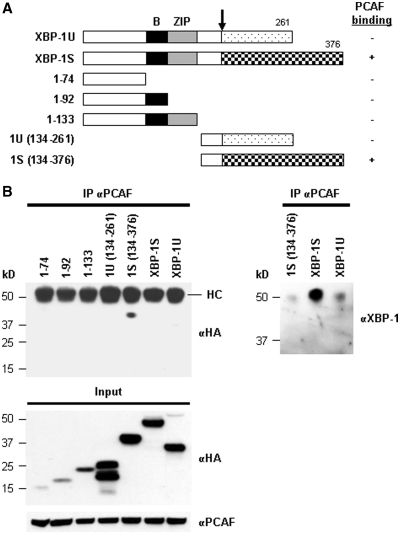
Domain study of XBP-1 was carried out using a series of HA-tagged XBP-1 truncations (Figure 2A). Cells were transfected with an individual XBP-1 truncation plasmid followed by IP using anti-PCAF antibodies. As shown in Figure 2B, only the XBP-1S-specific C-terminal region, which contained the transcriptional activation domain of XBP-1S, was found to associate with PCAF, but not the XBP-1U-specific C-terminus or any other regions of XBP-1. The heavy chains of anti-PCAF antibodies were also recognized by the secondary antibody used for the immunoblotting. Since the molecular weights of heavy chains and HA-tagged XBP-1S were similar (∼50 kDa), the blot could not reveal the presence of HA-XBP-1S in the immunoprecipitates. We did another western blot using an anti-XBP-1S antibody recognizing the common domain of XBP-1S and XBP-1U and confirmed the interaction between PCAF and HA-XBP-1S (Figure 2B, the anti-XBP-1 blot). It was noted that the interaction between PCAF and endogenous XBP-1S proteins were also detected in the HA-1S(134–376)- and HA-XBP-1U-transfected cells (Figure 2B). Collectively, the results demonstrate that PCAF binds to XBP-1S through the transcriptional activation domain of XBP-1S located in its C-terminal region.
Figure 2.
Domain study of XBP-1. (A) Diagram of XBP-1 truncations. All the constructs were HA-tagged. (B) 293T cells were transfected with the indicated plasmid to express an individual XBP-1 deletion. IP was performed using the anti-PCAF antibody followed by western blotting with anti-HA or anti-XBP-1 antibodies.
PCAF is required for XBP-1S-mediated activation of HTLV-1 and BiP transcription
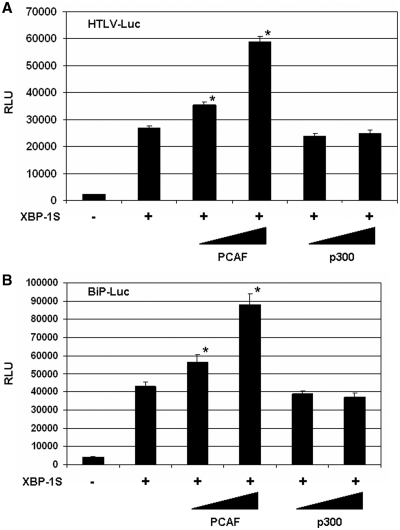
Functional significance of the PCAF-XBP-1S interaction was assessed in the XBP-1S-dependent transcription assays. XBP-1S is known to regulate the transcription of HTLV-1 and cellular gene BiP (4,26). The luciferase reporters, in which the expression of luciferase was driven by HTLV-1 and BiP promoters (i.e. HTLV-Luc and BiP-Luc, respectively), were utilized in the study. In the XBP-1S co-transfected cells, more than 10-fold increases in luciferase expression were observed in HTLV-1 and BiP promoters (Figure 3A and B). Further induction (more than 2-fold) of the XBP-1S-mediated activation of HTLV-1 and BiP promoters was detected in the PCAF-expressing cells (Figure 3A and B). However, overexpression of p300 had no significant effects on XBP-1S-dependent transcription (Figure 3A and B).
Figure 3.
Overexpression of PCAF stimulates XBP-1S-mediated transcription. HEK293 cells were transiently co-transfected with a luciferase reporter [(A) HTLV-Luc and (B) BiP-Luc] and indicated expression plasmids (i.e. XBP-1S, PCAF and p300). The amounts of PCAF and p300 plasmids were titrated at 3-fold increment. The total amounts of plasmids transfected were kept constant by adjusting the mock vector. *P < 0.05 versus control (i.e. cells transfected with a XBP-1S expression plasmid).
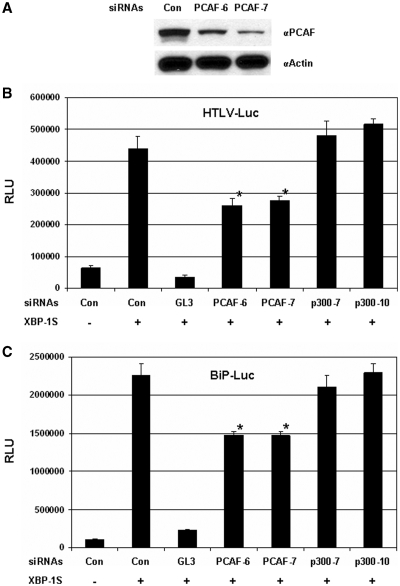
The impact of PCAF knockdown on the activation of HTLV-1 and BiP transcription by XBP-1S was studied next. The knockdown experiments were carried out using the siRNAs specifically targeting PCAF. The effectiveness of two PCAF siRNAs, PCAF-6 and PCAF-7, was confirmed by western blotting (Figure 4A). Two p300 siRNAs (i.e. p300-7 and p300-10) were utilized as controls since no association between p300 and XBP-1S was observed (Figure 1C). However, the protein levels of endogenous p300 in the cells were not high enough to be clearly revealed by western analyses. QRT–PCR was then used to confirm the actions of two p300 siRNAs. Forty to fifty percent decrease in p300 mRNA levels were detected in the cells transfected with the p300 siRNAs (data not shown).
Figure 4.
Knockdown of PCAF inhibits XBP-1S-mediated transcription. (A) Cell lysates of the PCAF siRNA-transfected HEK293 cells were analyzed by western blotting to determine the effectiveness of the siRNAs. Cells transfected with a non-specific siRNA were used as a negative control (i.e. Con.). For the luciferase-based assays, HEK293 cells were transiently co-transfected with a luciferase reporter [(B) HTLV-Luc and (C) BiP-Luc], a XBP-1S expression plasmid, and an indicated siRNA. The siRNAs used for the experiments included non-specific (i.e. Con.), luciferase (i.e. GL3), two PCAF (i.e. PCAF-6 and PCAF-7) and two p300 (i.e. p300-7 and p300-10) siRNAs. *P<0.05 versus control (i.e. cells co-transfected with a control non-specific siRNA and a XBP-1S expression plasmid).
Cells were co-transfected with the luciferase reporter (i.e. HTLV-Luc or BiP-Luc), a XBP-1S plasmid, and an indicated siRNA (Figure 4B and C). Compared to the transfection excluding the XBP-1S expression vector, 6- and 18-fold enhancement in the activation of HTLV and BiP promoters was observed (Figure 4B and C, the first two transfections). The GL3 siRNA, which specifically targeted the GL3 luciferase used in the HTLV-Luc and BiP-Luc reporters, was used as a positive control and caused 90% decreases in luciferase expression under the control of HTLV and BiP promoters (Figure 4B and C, the second and third transfections). The two PCAF siRNAs inhibited ∼40% luciferase expression driven by HTLV promoter, while no significant effects were caused by either p300 siRNA (Figure 4B). Similar observations were found in the BiP-Luc reporter assays (Figure 4C). Results obtained from the PCAF overexpression and knockdown reporter assays (Figures 3 and 4) demonstrate the functional involvement of PCAF in the genes regulated by XBP-1S.
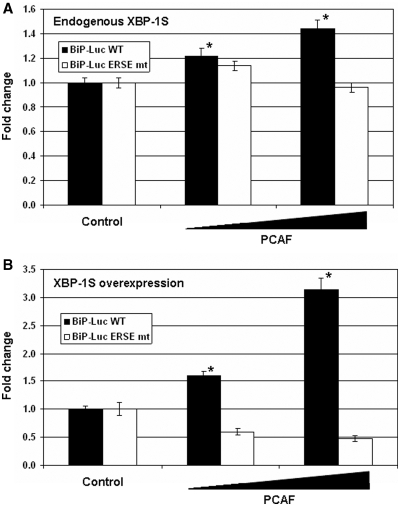
XBP-1S regulates the transcription of BiP by binding to the ERSE element located within the BiP promoter (2,31). We next wished to determine if the transcriptional activation of the BiP promoter by PCAF was mediated through ERSE as well. The wild-type and ERSE-mutant BiP-Luc reporter plasmids were utilized in the experiments. Extopic expression of PCAF significantly activated the Luc expression driven by the wild-type BiP promoter, while little or no effects were detected on the transcription driven by the ERSE-mutant BiP promoter (Figure 5A). Since the protein level of endogenous XBP-1S was low in the ER stress-free cells, only up to a 44% increase in BiP transcription was observed (Figure 5A). In the XBP-1S-overexpressing cells, PCAF exhibited stronger activation on the expression of luciferase driven by the wild-type BiP promoter (Figure 5B, up to 3-fold). However, no activating effects on the ERSE mutant BiP promoter were detected when both PCAF and XBP-1S were overexpressed (Figure 5B). Collectively, these results suggest that PCAF interacts with XBP-1S and mediates BiP transcription in an ERSE-dependent manner.
Figure 5.
Transcriptional activation of the BiP promoter by PCAF is mediated through ERSE. (A) HEK293 cells were transfected an indicated BiP-Luc reporter (i.e. wild-type or ERSE mutant) and a PCAF expression plasmid. Amounts of the PCAF plasmids were titrated at 3-fold increment. *P < 0.05 versus control (i.e. cells transfected with an empty plasmid and a wild-type BiP-Luc reporter). (B) HEK293 cells were co-transfected an indicated BiP-Luc reporter, a XBP-1S expression plasmid, and a PCAF expression plasmid (at 3-fold increment). *P < 0.05 versus control (i.e. cells transfected with an XBP-1S plasmid and a wild-type BiP-Luc reporter).
PCAF mediates the transcription of endogenous XBP-1S target genes
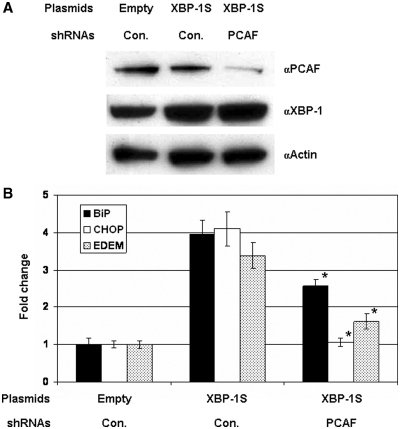
Requirement for PCAF in the mediation of endogenous XBP-1S target genes, including BiP, CHOP and EDEM (4), was investigated. We performed QRT–PCR assays to determine the impact of PCAF on the activation of XBP-1S target genes by knocking down the expression of PCAF. Compared to the DNA transfection, co-transfection of DNA plasmids and siRNAs was much more cytotoxic (data not shown). Therefore, a shRNA plasmid against PCAF was used to co-transfect cells along with an XBP-1 expression vector. Effectiveness of the PCAF shRNA was confirmed by western blotting (Figure 6A). Overexpression of XBP-1S resulted in 3- to 4-fold increases in the mRNA levels of BiP, CHOP, and EDEM (Figure 6B). Co-transfection of the PCAF shRNA in the XBP-1S-expressing cells led to 35, 74 and 52% inhibition of BiP, CHOP, and EDEM transcription, respectively (Figure 6B), demonstrating the involvement of PCAF in the XBP-1S-dependent transcription.
Figure 6.
Knockdown of PCAF inhibits the transcription of endogenous XBP-1S target genes. (A) MCF7 cells were co-transfected with the indicated plasmid (i.e. empty or XBP-1S expression vectors) and shRNA [i.e. non-specific (Con.) or PCAF]. Expression of PCAF and XBP-1S was analyzed by western blotting. (B) The mRNAs of the XBP-1S target genes, including BiP, CHOP and EDEM were quantified by QRT–PCR. Cells transfected with an empty vector and a non-specific shRNA served as a negative control. Knockdown of PCAF using a specific PCAF shRNA exhibited significant decreases in the expression of BiP CHOP, and EDEM, respectively (*P < 0.05 versus cells co-transfected with a control non-specific shRNA and a XBP-1S expression plasmid).
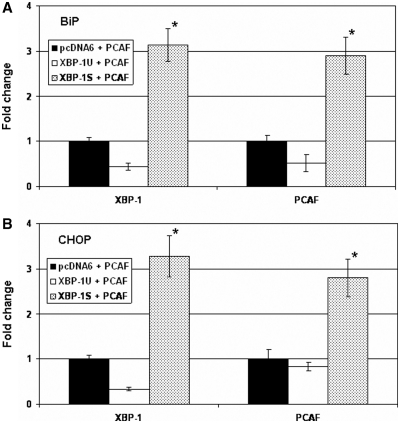
The in vivo recruitment of PCAF to the XBP-1S endogenous target genes was examined next. Cells were transfected with a PCAF expression plasmid and an indicated vector (i.e. empty, XBP-1U, and XBP-1S plasmids). Distribution of PCAF and XBP-1 on the promoters of BiP and CHOP was analyzed by quantitative ChIP. Fewer XBP-1 and PCAF proteins were located on BiP and CHOP genes when XBP-1U was overexpressed (Figure 7A and B). In the XBP-1S/PCAF co-transfected cells, more XBP-1S proteins were found to bind to the promoter region of BiP and CHOP genes (Figure 7A and B). It was expected since overexpression of XBP-1S activated the transcription of BiP and CHOP (Figure 6B). In addition, a 3-fold increase in PCAF binding to BiP and CHOP genes was detected in the XBP-1S/PCAF co-expressing cells (Figure 7A and B), providing the evidence that PCAF was recruited to BiP and CHOP promoters through the interaction with XBP-1S.
Figure 7.
XBP-1S recruits PCAF to the target genes of XBP-1S in vivo. MCF7 cells were co-transfected with a PCAF expression vector and an indicated plasmid [i.e. empty (pcDNA6), XBP-1U, or XBP-1S]. ChIP was carried out followed by quantitative PCR to quantify the binding of XBP-1S and PCAF to the promoters of BiP (A) and CHOP (B). Cells transfected with an empty vector and a PCAF plasmid was used as a negative control. *P < 0.05 versus negative controls.
Involvement of PCAF in the UPR-dependent activation of XBP-1S
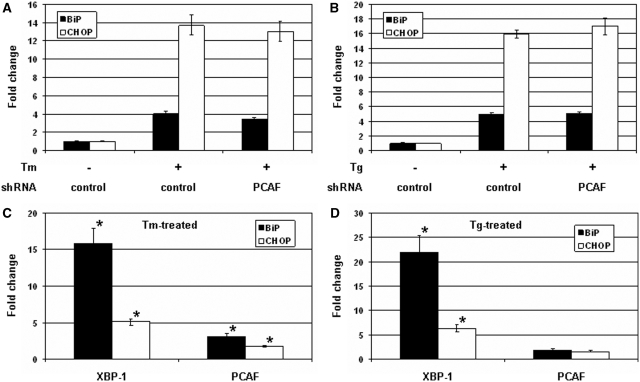
UPR induces the generation of XBP-1S which up-regulates its target genes required for secretory pathway, membrane synthesis, protein folding and ER-associated degradation (1). The involvement of PCAF for XBP-1S activation during UPR was studied by examining the expression BiP and CHOP genes. Cells were transfected with a control or PCAF shRNA followed by the treatment of Tm to induce UPR. The mRNAs isolated from the cells were analyzed by QRT–PCR. The mRNA levels of BiP and CHOP increased 4- and 14-fold, respectively, after Tm incubation (Figure 8A). Knockdown of PCAF only led to minor inhibition on the transcription of the two genes (Figure 8A). An identical set of assays was performed using Tg as the UPR inducing reagent. Little or no significant effects on BiP and CHOP mRNAs were detected in the PCAF shRNA-transfected cells (Figure 8B). We further carried out quantitative ChIP to examine the distribution of XBP-1S and PCAF on BiP and CHOP genes during UPR. Incubation of Tm resulted in 15- and 5-fold increases in XBP-1S binding to BiP and CHOP promoters, respectively, while only 3- and 1.7-fold increases in PCAF associating with the two genes (Figure 8C). In another set of experiments with Tg treatment, <2-fold increases in PCAF binding to endogenous BiP and CHOP genes were detected, while more than 20- (BiP) and 5-fold (CHOP) enhancement in XBP-1S binding (Figure 8D). Taken together the QRT–PCR and quantitative ChIP analyses suggest the limited involvement of PCAF in the mediation of XBP-1S target genes during UPR.
Figure 8.
Requirement of PCAF for the mediation of XBP-1S target genes under UPR. MCF7 cells were co-transfected with a non-specific (i.e. control) or PCAF shRNA, and incubated with 10 µg/ml Tm (A) or 300 nM Tg (B). Both Tm and Tg were dissolved in DMSO and the final concentration of DMSO in the culture was kept at 0.1%. Expression of endogenous BiP and CHOP genes was determined by QRT–PCR. Cells transfected with a control shRNA with 0.1% DMSO were served as a negative control. For quantitative ChIP assays, HEK293 cells were treated with 10 µg/ml Tm (C) or 300 nM Tg (D) and the bindings of XBP-1S and PCAF to the endogenous BiP and CHOP genes were analyzed by quantitative PCR. Cells incubated with 0.1% DMSO were used as a negative control. Fold changes were determined by comparing to the negative controls. *P < 0.05 versus negative controls.
Induction of UPR has no effects on the association between XBP-1S and PCAF
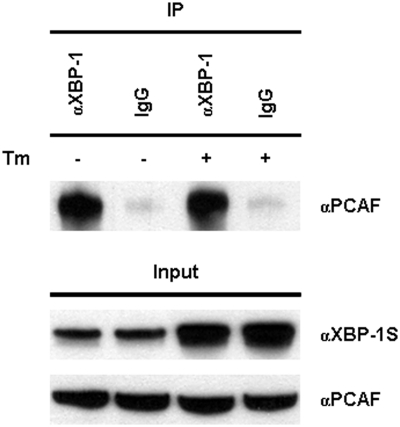
A recent study demonstrated that the association between XBP-1S and its binding protein could be UPR-dependent and such protein–protein interaction was disrupted after treating cells with UPR-inducing compound, Tm (32). We examined the influence of UPR on the PCAF-XBP-1S interaction by treating cells with Tm followed by IP analyses. No changes in the binding of PCAF to XBP-1S were detected under the treatment of Tm (Figure 9), suggesting the existence of the PCAF-XBP-1S protein complexes during UPR.
Figure 9.
Interaction between XBP-1S and PCAF under UPR. 293T cells were transiently transfected with a XBP-1S expression vector and incubated with 10 µg/ml Tm or 0.1% DMSO (i.e. the negative control) for 16 h. IP was performed using the cell lysates prepared from the transfected cells and the antibody against XBP-1. Normal IgG (IgG) was used as a negative control. The immunoprecipitated complexes and the protein inputs were analyzed by western blotting.
DISCUSSION
In this study, we investigate the molecular mechanism to elucidate the distinct functions between the inactive XBP-1U and active XBP-1S. Both isoforms have an identical N-terminus and an isoform-specific C-terminal region (Figure 2A). We identify PCAF as a novel XBP-1S binding protein and demonstrate the biological importance of PCAF in regulating the XBP-1S-mediated cellular and viral transcription. PCAF binds to XBP-1S through the interaction with the XBP-1S-specific C-terminal domain but fails to associate with the full-length XBP-1U or the XBP-1U-specific C-terminus (Figures 1 and 2), providing an explanation for the transactivating ability of XBP-1S on gene expression.
Basal transcription of HTLV-1 occurs after proviral integration into the host cell genome and induces the initial expression of HTLV-1 proteins, including the transactivator, Tax, followed by Tax transactivation to boost the synthesis of viral transcripts. Two HATs, PCAF and p300, have been shown to interact with Tax and play a role in Tax-activated viral transcription (26,33–35). Tax, which does not bind to DNA by itself, activates HTLV-1 transcription through three 21-bp repeats known as TRE, located within the promoter of HTLV-1. Each 21-bp TRE repeat contains a CREB/ATF binding site and is known to associate with CREB/ATF family proteins (36,37). Tax binds to TREs through the interaction with CREB/ATF family proteins (including XBP-1S, CREB1 and CREB2) and recruits PCAF/p300 to HTLV-1 promoter, resulting in Tax transactivation (26,33–35). We previously found that XBP-1S bound to Tax and induced stronger Tax transactivation than other CREB/ATF family proteins (26). Interestingly, XBP-1S also stimulated basal transcription of HTLV-1, while CREB1 and CREB2 did not show any activating effects, suggesting a crucial role for XBP-1S during the early phase of viral transcription as well (26). No interaction between PCAF and CREB1 was detected in Co-IP analyses (Figure 1A). This observation could explain why CREB1 and other CREB/ATF family proteins fail to up-regulate HTLV-1 transcription in the absence of Tax. In contrast, the requirement for PCAF in the XBP-1S-dependent HTLV-1 basal transcription was clearly demonstrated in the cell-based reporter assays (Figures 3A and 4A).
Functional significance of PCAF-XBP-1S interaction on the cellular target genes of XBP-1S, including BiP, CHOP and EDEM, was demonstrated in the PCAF overexpression and knockdown experiments (Figures 3–6). In addition, quantitative ChIP assays showed that XBP-1S recruited PCAF to the promoters of endogenous XBP-1S target genes in vivo, establishing direct functional connection between PCAF and XBP-1S (Figure 7). However, knockdown of PCAF by siRNA or shRNA did not completely inhibit the elevated transcription caused by XBP-1S (Figures 4 and 6). There are two possible explanations for these observations. First, both siRNA and shRNA against PCAF did not completely block the protein synthesis of PCAF (Figures 4A and 6A). Therefore, it is possible that the siRNA- and shRNA-transfected cells still have sufficient PCAF left to participate in the gene activation by XBP-1S. Secondly, PCAF may be one of the cellular co-factors responsible for XBP-1S-dependent transcription. Therefore, elimination of PCAF by RNA interference could only partially inhibit the transactivation of XBP-1S.
Discovery of the involvement of PCAF in the transcriptional regulation of BiP and EDEM genes is novel. PCAF has been identified as a co-factor of ATF4 (or CREB2) for the expression of CHOP (38). In response to amino acid starvation, ATF4 binds to the amino acid response element located in the CHOP promoter and recruits PCAF to the promoter, leading to the activation of CHOP transcription (38). Besides PCAF, ATF4 also interacts with other HATs, including p300 and CBP, through its N-terminal transactivation domain (39,40). As shown in Figure 1, XBP-1S shows more stringent protein binding than ATF4 and fails to associate with p300. Future study is required to investigate the interaction between XBP-1S and other HATs to further determine the binding specificity of the XBP-1S transactivation domain. Collectively, the findings by our and other groups point out that PCAF may play an important role in transcriptional activation of CHOP through the XBP-1S- as well as ATF4-dependent pathways.
It has been reported that p300 is recruited to the endogenous BiP promoter in the Tg-treated cells by ChIP assays (41). Co-overexpression of p300, YY1 and ATF6 showed synergistic activation of luciferase expression driven by the BiP promoter, suggesting that p300 might be required for YY1-/ATF6-mediated activation of BiP (41). Similar cell-based reporter assays (i.e. using the BiP-Luc reporter plasmid) were performed to determine the requirement of p300 for XBP-1S-mediated transactivation. Neither overexpression nor knockdown of p300 showed any significant effects on XBP-1S-dependent luciferase expression (Figures 3 and 4), suggesting that p300 might function in a XBP-1S-independent manner. These results were further supported by Co-IP data, in which no interaction between p300 and XBP-1S was detected (Figure 1C). Furthermore, we assessed the requirement of p300 for transcriptional activation of BiP and CHOP genes under UPR. In contrast to the report by Baumeister et al. (41), our results obtained from the quantitative ChIP analyses did not show any increased p300 binding to either BiP or CHOP promoters in the Tm- or Tg-stressed cells (data not shown), raising the questions regarding to the involvement of p300 in the regulation of XBP-1S target genes.
Under the ER stress-free condition, our data clearly indicated that PCAF was required for XBP-1S-mediated transcriptional regulation (Figures 3–7). However, results from QRT–PCR and quantitative ChIP showed that PCAF only exhibited limited involvement in the expression of XBP-1S target genes when UPR was induced (Figure 8). A recent study identified the regulatory subunit of phosphoinositide 3-kinase (PI3K) as a novel XBP-1S binding protein and demonstrated that the association between XBP-1S and the PI3K subunit could be UPR-dependent (32). We examined the XBP-1S-PCAF protein–protein interaction under the normal or Tm-stressed conditions. As shown in Figure 9, the interaction between XBP-1S and PCAF was not disrupted during UPR. On-going research focuses on the identification of the XBP-1S co-factor(s) required for the transactivation caused by XBP-1S once UPR is induced. GCN5, a HAT which shares 73% identity in amino acid sequence with PCAF (42), is a possible candidate for XBP-1S binding partner. The involvement of GCN5 in XBP-1S-dependent transcription and during UPR is currently under investigation.
The tumor microenvironment is hypoglycemic and hypoxic, resulting in induction of UPR and overexpression of XBP-1. Recent studies show the involvement of XBP-1 in tumorigenesis of various cancers and suggest XBP-1 as a potential target for anti-cancer therapeutics (10–12,43). Fujimoto et al. investigated the expression of XBP-1 in 11 primary breast cancers and five breast cancer cell lines, including MCF7. The increased expression of XBP-1 was detected in all breast cancers and cell lines examined, but not in the non-cancerous breast issue (44). In addition, clinical results showed that high levels of XBP-1S increased the survival of breast cancers (45). Our data presented here demonstrate the functional importance of PCAF in mediating the expression of XBP-1S target genes in MCF7 cells (Figures 6 and 7), suggesting a potential role of PCAF in XBP-1S-mediated tumerigenesis of breast cancers. Furthermore, PCAF may be an essential factor for other XBP-1S-mediated signaling pathways. For example, XBP-1S is one of the key components in the transcriptional program controlling plasma cell differentiation (1). It would be worthwhile to examine the importance of PCAF-XBP-1S during the development of plasma cells.
FUNDING
This work is supported by the Agency for Science, Technology and Research (A*STAR), Singapore. Funding for open access charge: A*STAR.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Kazutoshi Mori, Dr Hiderou Yoshida and Dr Arnold Rabson for providing the expression and reporter plasmids, Dr Niki Wong for critical review of the manuscript, and Ms Yi Ling Chia for expert technical assistance.
REFERENCES
- 1.Brewer JW, Hendershot LM. Building an antibody factory: a job for the unfolded protein response. Nat. Immunol. 2005;6:23–29. doi: 10.1038/ni1149. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 3.Uemura A, Oku M, Mori K, Yoshida H. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J. Cell Sci. 2009;122:2877–2886. doi: 10.1242/jcs.040584. [DOI] [PubMed] [Google Scholar]
- 4.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ku SC, Ng DT, Yap MG, Chao SH. Effects of overexpression of X-box binding protein 1 on recombinant protein production in Chinese hamster ovary and NS0 myeloma cells. Biotechnol. Bioeng. 2008;99:155–164. doi: 10.1002/bit.21562. [DOI] [PubMed] [Google Scholar]
- 7.Ku SC, Toh PC, Lee YY, Chusainow J, Yap MG, Chao SH. Regulation of XBP-1 signaling during transient and stable recombinant protein production in CHO cells. Biotechnol. Prog. 2010;26:517–526. doi: 10.1002/btpr.322. [DOI] [PubMed] [Google Scholar]
- 8.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 9.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 10.Shajahan AN, Riggins RB, Clarke R. The role of X-box binding protein-1 in tumorigenicity. Drug News Perspect. 2009;22:241–246. doi: 10.1358/dnp.2009.22.5.1378631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koong AC, Chauhan V, Romero-Ramirez L. Targeting XBP-1 as a novel anti-cancer strategy. Cancer Biol. Ther. 2006;5:756–759. doi: 10.4161/cbt.5.7.2973. [DOI] [PubMed] [Google Scholar]
- 12.Romero-Ramirez L, Cao H, Regalado MP, Kambham N, Siemann D, Kim JJ, Le QT, Koong AC. X box-binding protein 1 regulates angiogenesis in human pancreatic adenocarcinomas. Transl. Oncol. 2009;2:31–38. doi: 10.1593/tlo.08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenner RG, Maillard K, Cattini N, Weiss RA, Boshoff C, Wooster R, Kellam P. Kaposi's sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc. Natl Acad. Sci. USA. 2003;100:10399–10404. doi: 10.1073/pnas.1630810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medigeshi GR, Lancaster AM, Hirsch AJ, Briese T, Lipkin WI, Defilippis V, Fruh K, Mason PW, Nikolich-Zugich J, Nelson JA. West Nile virus infection activates the unfolded protein response leading to CHOP induction and apoptosis. J. Virol. 2007;81:10849–10860. doi: 10.1128/JVI.01151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su HL, Liao CL, Lin YL. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J. Virol. 2002;76:4162–4171. doi: 10.1128/JVI.76.9.4162-4171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tardif KD, Mori K, Siddiqui A. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 2002;76:7453–7459. doi: 10.1128/JVI.76.15.7453-7459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tardif KD, Mori K, Kaufman RJ, Siddiqui A. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J. Biol. Chem. 2004;279:17158–17164. doi: 10.1074/jbc.M312144200. [DOI] [PubMed] [Google Scholar]
- 18.Tirosh B, Iwakoshi NN, Lilley BN, Lee AH, Glimcher LH, Ploegh HL. Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J. Virol. 2005;79:2768–2779. doi: 10.1128/JVI.79.5.2768-2779.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isler JA, Skalet AH, Alwine JC. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 2005;79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu CY, Hsu YW, Liao CL, Lin YL. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J. Virol. 2006;80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan CP, Siu KL, Chin KT, Yuen KY, Zheng B, Jin DY. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2006;80:9279–9287. doi: 10.1128/JVI.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bechill J, Chen Z, Brewer JW, Baker SC. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J. Virol. 2008;82:4492–4501. doi: 10.1128/JVI.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao JR, Chang KC, Chen CW, Wu SY, Su IJ, Hsu MC, Jin YT, Tsai ST, Takada K, Chang Y. Endoplasmic reticulum stress triggers XBP-1-mediated up-regulation of an EBV oncoprotein in nasopharyngeal carcinoma. Cancer Res. 2009;69:4461–4467. doi: 10.1158/0008-5472.CAN-09-0277. [DOI] [PubMed] [Google Scholar]
- 24.Barry G, Fragkoudis R, Ferguson MC, Lulla A, Merits A, Kohl A, Fazakerley JK. Semliki Forest virus induced endoplasmic reticulum stress accelerates apoptotic death of mammalian cells. J. Virol. 2010;84:7369–7377. doi: 10.1128/JVI.02310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alefantis T, Mostoller K, Jain P, Harhaj E, Grant C, Wigdahl B. Secretion of the human T cell leukemia virus type I transactivator protein tax. J. Biol. Chem. 2005;280:17353–17362. doi: 10.1074/jbc.M409851200. [DOI] [PubMed] [Google Scholar]
- 26.Ku SC, Lee J, Lau J, Gurumurthy M, Ng R, Lwa SH, Lee J, Klase Z, Kashanchi F, Chao SH. XBP-1, a novel human T-lymphotropic virus type 1 (HTLV-1) tax binding protein, activates HTLV-1 basal and tax-activated transcription. J. Virol. 2008;82:4343–4353. doi: 10.1128/JVI.02054-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin HC, Hickey M, Hsu L, Medina D, Rabson AB. Activation of human T cell leukemia virus type 1 LTR promoter and cellular promoter elements by T cell receptor signaling and HTLV-1 Tax expression. Virology. 2005;339:1–11. doi: 10.1016/j.virol.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashanchi F, Brady JN. Transcriptional and post-transcriptional gene regulation of HTLV-1. Oncogene. 2005;24:5938–5951. doi: 10.1038/sj.onc.1208973. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 32.Winnay JN, Boucher J, Mori MA, Ueki K, Kahn CR. A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat. Med. 2010;16:438–445. doi: 10.1038/nm.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrod R, Kuo YL, Tang Y, Yao Y, Vassilev A, Nakatani Y, Giam CZ. p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J. Biol. Chem. 2000;275:11852–11857. doi: 10.1074/jbc.275.16.11852. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, Lu H, Schiltz RL, Pise-Masison CA, Ogryzko VV, Nakatani Y, Brady JN. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell Biol. 1999;19:8136–8145. doi: 10.1128/mcb.19.12.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwok RP, Laurance ME, Lundblad JR, Goldman PS, Shih H, Connor LM, Marriott SJ, Goodman RH. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 36.Brady J, Jeang KT, Duvall J, Khoury G. Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type I long terminal repeat. J. Virol. 1987;61:2175–2181. doi: 10.1128/jvi.61.7.2175-2181.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeang KT, Boros I, Brady J, Radonovich M, Khoury G. Characterization of cellular factors that interact with the human T-cell leukemia virus type I p40x-responsive 21-base-pair sequence. J. Virol. 1988;62:4499–4509. doi: 10.1128/jvi.62.12.4499-4509.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherasse Y, Maurin AC, Chaveroux C, Jousse C, Carraro V, Parry L, Deval C, Chambon C, Fafournoux P, Bruhat A. The p300/CBP-associated factor (PCAF) is a cofactor of ATF4 for amino acid-regulated transcription of CHOP. Nucleic Acids Res. 2007;35:5954–5965. doi: 10.1093/nar/gkm642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang G, Hai T. Characterization of human activating transcription factor 4, a transcriptional activator that interacts with multiple domains of cAMP-responsive element-binding protein (CREB)-binding protein. J. Biol. Chem. 1997;272:24088–24095. doi: 10.1074/jbc.272.38.24088. [DOI] [PubMed] [Google Scholar]
- 40.Lassot I, Estrabaud E, Emiliani S, Benkirane M, Benarous R, Margottin-Goguet F. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J. Biol. Chem. 2005;280:41537–41545. doi: 10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- 41.Baumeister P, Luo S, Skarnes WC, Sui G, Seto E, Shi Y, Lee AS. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol. Cell Biol. 2005;25:4529–4540. doi: 10.1128/MCB.25.11.4529-4540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 43.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, Zheng M, Mani M, Henderson J, Pinkus GS, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimoto T, Onda M, Nagai H, Nagahata T, Ogawa K, Emi M. Upregulation and overexpression of human X-box binding protein 1 (hXBP-1) gene in primary breast cancers. Breast Cancer. 2003;10:301–306. doi: 10.1007/BF02967649. [DOI] [PubMed] [Google Scholar]
- 45.Davies MP, Barraclough DL, Stewart C, Joyce KA, Eccles RM, Barraclough R, Rudland PS, Sibson DR. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. Int. J. Cancer. 2008;123:85–88. doi: 10.1002/ijc.23479. [DOI] [PubMed] [Google Scholar]