Abstract
RNA polymerases (RNAPs) require basal transcription factors to assist them during transcription initiation. One of these factors, TFIIB, combines promoter recognition, recruitment of RNAP, promoter melting, start site selection and various post-initiation functions. The ability of 381 site-directed mutants in the TFIIB ‘linker domain’ to stimulate abortive transcription was systematically quantitated using promoter-independent dinucleotide extension assays. The results revealed two distinct clusters (mjTFIIB E78-R80 and mjTFIIB R90-G94, respectively) that were particularly sensitive to substitutions. In contrast, a short sequence (mjTFIIB A81-K89) between these two clusters tolerated radical single amino acid substitutions; short deletions in that region even caused a marked increase in the ability of TFIIB to stimulate abortive transcription (‘superstimulation’). The superstimulating activity did, however, not correlate with increased recruitment of the TFIIB/RNAP complex because substitutions in a particular residue (mjTFIIB K87) increased recruitment by more than 5-fold without affecting the rate of abortive transcript stimulation. Our work demonstrates that highly localized changes within the TFIIB linker have profound, yet surprisingly disconnected, effects on RNAP recruitment, TFIIB/RNAP complex stability and the rate of transcription initiation. The identification of superstimulating TFIIB variants reveals the existence of a previously unknown rate-limiting step acting on the earliest stages of gene expression.
INTRODUCTION
Eukaryotic RNA polymerases (RNAPs) require a variety of basal transcription factors to guide them to their promoters and to assist with the initiation of transcription. In the RNAPII system, the basal transcription factor TFIIB plays an essential role during the formation of pre-initiation complexes. TFIIB stabilizes the binding of TATA-binding protein (TBP) to the TATA-box and provides additional sequence specificity through contacts with the B-recognition elements (BREs) flanking the TATA box on either site (1,2). Once stably bound, TFIIB recruits RNAPII (3–5). The domain structure of TFIIB reflects these functions; the highly conserved C-terminal domain contains all TBP-binding and promoter-recognition functions (6–8), and the Zn-ribbon and the linker domain are required for binding to RNAPII (Figure 1) (9–12). The Zn-ribbon domain associates with the ‘dock’ domain, whereas the TFIIB linker penetrates extensively into the catalytic site of RNAPII (Figure 1B) (13–15). The intimate association between the TFIIB linker with elements of the catalytic site, and possibly the nucleic acid and rNTP substrates, are the likely cause for the involvement of TFIIB in post-initiation transcription events, such as a potential clash between TFIIB and the nascent RNA transcript causing the release of abortive transcripts before they exceed a defined length (13). TFIIB also stabilizes and/or stimulates critical stages during the abortive transcription cycle (10).
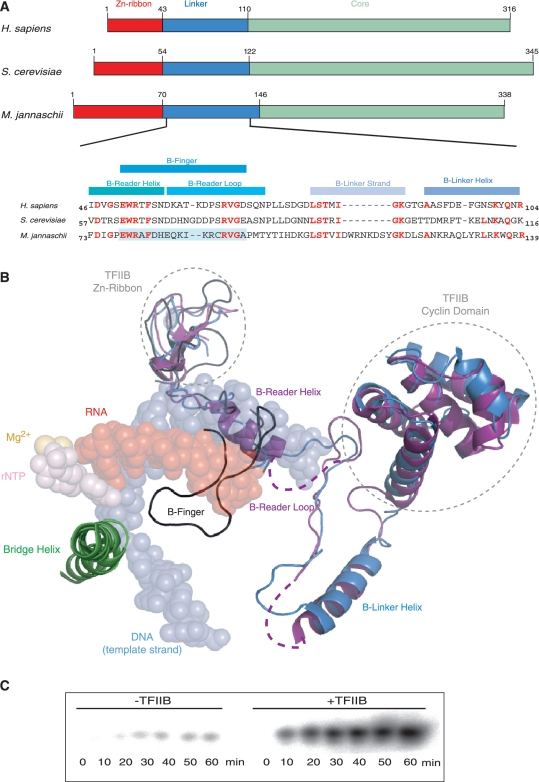
Figure 1.
Structural organization of TFIIB. (A) Schematic alignment of the human, yeast and M. jannaschii TFIIB orthologues [Zn-ribbon domain (red), C-terminal core domain (light green) and Linker region (blue)]. Part of the Linker domain primary amino acid sequence is shown with residues in the yeast and human orthologues identical to the residues found in M. jannaschii shown in red. The pale blue background identifies the sequence that was mutagenized in M. jannaschii TFIIB in the current study. (B) Alignment of the TFIIB structures of three yeast RNAP–TFIIB co-crystals [PDB codes 3K7A (blue) (14), 3K1F (purple) (15) and 1R5U (black) (13)] superimposed on the RNAP active centre domains and nucleic acids (PDB code: 2E2H). The structures differ in the position of the linker domain that either assumes the conformation of a B-Reader Helix/B-Reader Loop (15) or a hairpin-like B-finger structure (13). (C) Stimulatory effect of TFIIB on abortive transcription. The presence of TFIIB leads to a substantial stimulation (typically >10-fold) of abortive transcription. The assay shows a linear accumulation of abortive transcripts over a 60-min incubation period.
Three crystal structures showing the linker region in the context of a yeast RNAPII-TFIIB co-crystal are currently available at 4.5 (13), 4.3 (15) and 3.8 Å (14) resolution (Figure 1B). The relatively low resolutions of the structures have resulted in different interpretations of the electron density data, especially concerning the sub-domain that has been referred to as the ‘B-finger domain’ (13). An initial structural model of the TFIIB-linker RNAPII interactions suggested that the B-finger domain forms a loop projecting towards the catalytic centre (13). Surprisingly, the primary sequence of the B-finger tip region—which is assumed to be closest to the active site—is rather poorly conserved (Figure 1A). In vitro transcription assays also demonstrated that mutations in the B-finger tip had only minor consequences, whereas mutations in the lateral B-finger residues resulted in a severe reduction of transcription (16).
A more recently published structure proposes a mostly α-helical conformation for the B-finger domain [the ‘B-Reader Helix’; (15)]. In addition, more C-terminal parts of the linker region lie in proximity to RNAP domains (e.g. ‘Lid’, ‘Rudder’ and ‘Fork Loop 1’) that are unstructured in the absence of TFIIB, suggesting that the linker might stabilise them and additionally mediate clamp closure (14,15).
Despite the appearance of such new structural information, at least two problems remain. First, there is disagreement regarding the position and structure of the N-terminal portion of the linker domain relative to the active site [B-finger versus B-Reader Helix (13,15)]. Second, and probably more importantly, all the TFIIB/RNAPII crystals characterized up to now lack the substrates (template DNA–RNA hybrid and nucleotides), which are key components of active initiation complexes. The interactions between the TFIIB linker and the various substrates are anticipated to influence the structure of the linker region and its interactions with other domains within the catalytic site. We therefore do not regard the currently available structural information as sufficiently conclusive for deducing a reliable mechanism of the functional role of the TFIIB linker region during the transcription initiation process.
Over the last decade, we have developed an archaeal in vitro transcription system, based on the euryarchaeon Methanocaldococcus jannaschii, to serve as a model system for understanding the fundamental mechanisms of the eukaryotic RNAPII basal transcriptional machinery. Archaea contain an RNAPII-like polymerase, which resembles, in conjunction with TBP and TFIIB orthologues, the core components of the RNAPII basal transcriptional machinery (17–19). The absence of other basal factors, such as TFIIA, TFIIF, TFIIH, TBP-associated factors (TAFs) and mediator, allows the fundamental steps leading to transcription initiation to be studied in an exceptionally direct manner in archaea (10,20–22).
In the current study, we have taken advantage of the high degree of structural conservation of archaeal TFIIB orthologues to study the functional consequences of mutations in the linker region [although archaeal TFIIB orthologues have traditionally been named ‘TFB’ (23), we will refer to them as TFIIB to emphasize the high degree of structural similarities with eukaryotic TFIIB orthologues; Figure 1A]. The promoter-independent stimulation of abortive transcription reactions by the TFIIB orthologue of the archaeon M. jannaschii (‘mjTFIIB’) provides a sensitive assay capable of shedding more light on the roles of TFIIB during pre-initiation complex formation and transcription initiation (Figure 1C). In contrast to promoter-directed transcription activity assays (16), which rely on an unknown combination of recruitment and initiation stimulation, our experimental set-up allows these functionally highly intertwined molecular mechanisms to be dissected into separate processes that can be assayed independently of each other (10).
MATERIALS AND METHODS
Generation of mjTFIIB variants
The N-terminus of mjTFIIB in a pET21a or pET24a bacterial expression plasmid (10) was substituted by a synthetic codon-optimized construct (GeneScript). Mutants containing TFIIB linker mutations [eukaryotic or archaeal substitutions, deletion variants; sequential permutation libraries (GeneArt, Regensburg)] were generated by replacing the corresponding wild-type sequence (24).
Expression and automated purification of mjTFIIB variants
Bacterial cultures for the production of recombinant mjTFIIB (1.5 ml) were grown in a 24-well plate at 37°C shaking at 250 r.p.m. for ∼18 h in autoinduction medium (Overnight Express, Novagen) containing 100 µg/ml ampicillin (pET21a constructs) or 25 µg/ml kanamycin (pET24a constructs). All subsequent steps were carried out on a liquid handler (Theonyx, Aviso). Nine-hundred microlitres of cell culture was mixed with 100 µl of cell lysis solution [10 × FastBreak (Promega), 2 µl lysonase/sample (Novagen), 3 µl 0.5 M Mg-acetate] and 100 µl of a 5-fold diluted stock of magnetic nickel beads (‘MagneHis’, Promega). After a 30-min incubation, supernatants were removed from the plate on the magnet after each transfer step. The beads were washed three times with 500 µl of wash buffer (20 mM imidazole, 0.1% Triton X-100, 0.5 M NaCl, 20 mM Tris-acetate, pH 7.9, 10 mM Mg-acetate, 0.7 mM Zn-acetate, 10% glycerol). Finally, 100 µl of elution buffer (0.5 M imidazole, 0.1% Triton X-100, 0.5 M NaCl, 20 mM Tris-acetate, pH 7.9, 10 mM Mg-acetate, 0.7 mM Zn-acetate, 10% glycerol) was added to the beads and incubated for 30 min (1500 r.p.m.). The plate was then placed again on the magnet and the eluates transferred to a new 96-well plate.
Abortive initiation assays
Abortive initiation reactions were carried out in total volumes of 25 µl containing 1 × transcription buffer [250 mM Tris–Cl (pH 7.5), 375 mM KCl, 125 mM MgCl2] supplemented with 10 mM DTT, 600 ng ‘activated’ calf thymus DNA (Sigma, Type 15), 400 µM GpC dinucleotide, 10 µM unlabelled rUTP and 2.5 µCi α32P-rUTP (3000 Ci/mmol). Appropriate RNAP and TFIIB concentrations were experimentally determined (TFIIB: 30–70 ng/µl, RNAP: 10–20 ng/µl). Samples were incubated at 65°C for 30 min. Portions of the samples were boiled for 2 min in equal volumes of formamide loading buffer. Ten microlitres of each sample was loaded on a denaturing urea gel and run in 1 × TBE for 70–80 min at 225 V. The gels images were quantitated with a Fuji FLA5000-PhosphoImager and AIDA photoimaging software.
Electrophoretic mobility shift assays
A measure of 0.5 µl of 32P-end-labelled SSV T6 promoter probe (0.1 pmol) was mixed with 1 µl BSA (1 mg/ml in 100 mM DTT) in binding buffer (2.5 mM MgCl2, 5% glycerol, 0.1 mM EDTA, 40 mM HEPES pH 7.3, 250 mM NaCl, 5 mM β-mercaptoethanol); 130 ng/µl recombinant mjRNAP, 2 ng/µl mjTBP and 7–10 ng/µl of mjTFIIB or the respective mutants were added to the reaction mixtures. The reaction mixture was incubated at 65°C for 20 min, then 1 µl of heparin (0.5 mg/ml) was added to each reaction and the samples were incubated for another 20 min (at 65°C). For competition assays, unlabelled SSV T6 promoter was added in 400-fold excess after 20 min instead of heparin. Ten microlitres of each reaction was mixed with 6 µl native gel loading buffer and 10 µl was loaded on a 4–20% Tris-glycine gradient gel and run in 1× Tris-glycine buffer at 180 V for 1 h. Gels were quantitated on a Fuji FLA5000-PhosphoImager with AIDA photoimaging software.
RESULTS
The sequence between residues E78 and R92 is crucial for the stimulatory activity of mjTFIIB
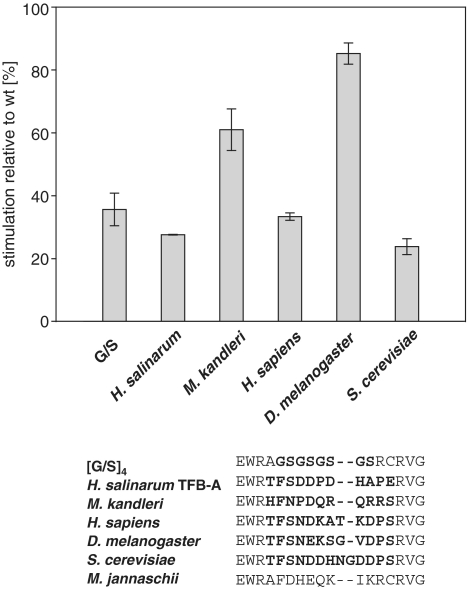
mjTFIIB massively stimulates abortive transcription by mjRNAP as measured in an in vitro dinucleotide extension assay (Figure 1C). Previous experiments demonstrated that certain recruitment functions and the stimulatory effect of TFIIB on the catalytic activity of RNAP are associated with the linker domain and, more precisely, with residues that are orthologous to the yeast TFIIB subdomain projecting into the RNAP II active centre cleft (10,13–15). A direct correlation between the stimulatory activity and the linker domain (10) allows us thus to dissect the stimulatory TFIIB function by site-directed mutagenesis. First, we replaced residues including W79 to C91 (the ‘B-finger’) by alternative sequences derived from various archaeal and eukaryotic species. The majority of TFIIB constructs containing such alternative B-fingers were severely impaired in their stimulatory efficiency, demonstrating that the identities of residues in this part of the TFIIB linker are essential for its stimulatory function (Figure 2). The results further suggest that the B-finger domains of different organisms display species-specific functional differences and/or adaptations to evolutionarily divergent structures in the active site (24).
Figure 2.
Replacement of TFIIB linker sequences. Stimulatory activities of a variety of artificial [glycine-serine (‘G-S’) repeats], archaeal (Halobacterium salinarum TFB-A; Methanopyrus kandleri) and eukaryotic (Saccharomyces cerevisiae; Drosophila melanogaster; Homo sapiens) TFIIB-linkers after replacement of the orthologous sequence of mjTFIIB. The primary sequences of the hybrids are shown below with the replaced TFIIB-linker sequence highlighted in bold. The activity of each mutant was calculated relative to the mjTFIIB wild-type stimulation rate valued as 100%. Each mutant was assessed in triplicate (error bars represent standard deviations).
The identities of mjTFIIB residues A81 to K89 are mostly irrelevant for transcriptional stimulation
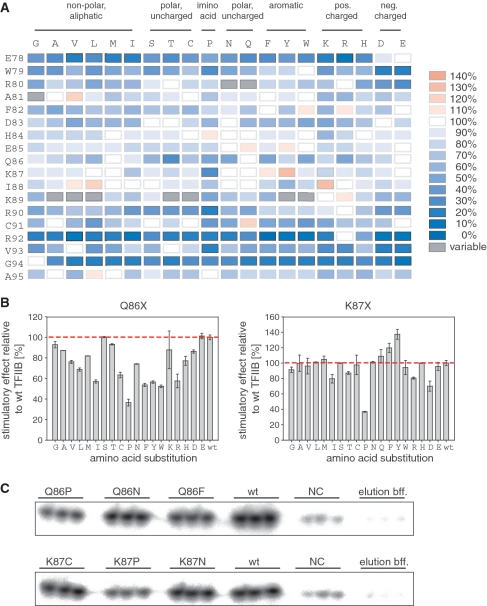
In order to learn more about the structural requirement of TFIIB linker residues we carried out high-density mutagenesis by substituting residues E78-A95 (i.e. all residues of the B-finger or most residues of the B-reader) by all 19 other amino acids. The results, based on averages calculated from abortive transcription assays carried out in triplicate, are summarized in form of a heat-map (Figure 3A and C). Substitutions of residues E78, W79, R80, Q86, R90, C91, R92, V93 and G94 resulted in particularly large numbers of mutants with a diminished ability to stimulate abortive transcription. Intriguingly, it was difficult to ascribe particular phenotypes to certain substitutions in R80, A81 and K89. Occasionally, R80-N, R80-Q, A81-G, K89-A, K89-V, K89-L, K89-T, K89-C, K89-Y and K89-W displayed stimulatory activities that exceeded wild-type TFIIB. However, even though triplicate assays within one experiment showed little variation, repeated testing of these mutants in separate experiments resulted in different phenotypes, including wild-type-like phenotypes or even significant defects in activity. Such marked discrepancies were not observed with the majority of other mutants assayed in independent experiments. Variability in the stimulation effect of some mutants is probably due to small fluctuations in experimental conditions that might cause a poorly structured domain to adopt alternative conformations and thus influence the stimulation properties of this class of mutants.
Figure 3.
High-throughput mutagenesis of the TFIIB linker. (A) mjTFIIB residues E78-A95 (listed on the left) were substituted by all 19 other amino acids (listed at the top and grouped according to their properties). Stimulatory effects of the mutants in abortive initiation assays are presented as a heat-map with shades of blue indicating defects in the stimulatory effects and shades of red indicating increased stimulation. The stimulatory effect of wild-type mjTFIIB is defined as 100% (white). Mutants with exceptionally high variability in their performance are depicted in grey. (B) Representative examples of the data obtained for full sets of substitutions of residues Q86 and K87. The activity of each mutant was calculated relative to the wild-type stimulation rate valued as 100% (red dotted line). Each mutant was assessed in triplicate (error bars represent standard deviations). (C) Representative examples of abortive initiation assay results for six mutants. Wild-type mjTFIIB was used as positive control and ‘elution buffer only’ samples were used to determine the background RNAP activity. Negative control samples (‘NC’; pET24a and medium only) confirmed the specificity of the signals obtained.
Consistent with the findings of an alanine scanning approach of human TFIIB (16), certain TFIIB-linker domain residues (A81 to K89) emerged as insensitive to most substitutions, with the sole exception of Q86. In particular, substitutions in residues A81, H84, E85 and I88 behaved similarly to wild-type TFIIB (in terms of their stimulatory effect on RNAP), no matter which amino acid substitution was made. For residue K87, only minor changes in the stimulatory activity were observed in the presence of most amino acid substitutions, with the exception of two mutations: the stimulatory effect of K87-P was only 30–40% of the wild-type level (thus making this one of the substitutions that impeded the transcription stimulatory effect to the greatest extent observed), whereas K87-Y consistently exceeded the wild-type activity level (∼135%) (Figure 3A and B).
In summary, our data identify two clusters of residues (E78-W79-R80 and R90-C91-R92-V93-G94) within the TFIIB linker that make key contributions towards the abortive transcription stimulation function. In contrast, the majority of residues in the sequence (A81 to K89) between these two clusters play only a minor functional role and even tolerate substantial structural disruptions introduced by various radical substitutions.
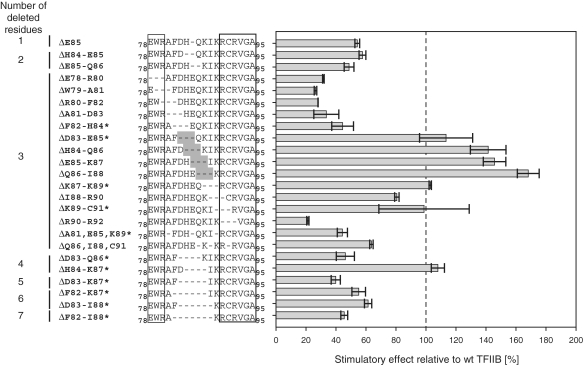
Precisely defined deletions in certain linker positions cause super-stimulation
To assess the influence of individual or groups of residues on the stimulation effect in more detail, a series of deletion mutants (ranging from 1 to 7 amino acids; Figure 4) were produced and tested in abortive initiation assays. The single deletion mutant ΔE85, as well as the double-deletion mutants ΔH84-E85 and ΔE85-Q86, significantly reduced the stimulating activity (Figure 4). A series of three-residue deletion mutants covering residues E78 to D83 generated phenotypes that were reproducibly impaired in their stimulatory effect on RNAP. Similarly, the stimulatory activity of ΔR90-R92 was significantly reduced (Figure 4). The activities of ΔF82-H84, ΔD83-E85, ΔK87-K89 and ΔK89-C91 were not reproducible i.e. fluctuating in a similar way as has been observed for some of the point mutants (see above). Strikingly, ΔH84-Q86, ΔE85-K87 and ΔQ86-I88 displayed stimulation ranging from 120% to ∼170% (Figure 4; see also below). We refer to this phenomenon as ‘superstimulation’ because the robust and reproducible nature of these phenotypes clearly distinguishes them from the normal level of stimulation achievable by wild-type mjTFIIB.
Figure 4.
Effect of deletions in the TFIIB linker domain. The stimulatory effect of mjTFIIB deletion mutants was calculated relative to wild-type TFIIB (dotted line). Three-residue deletions leading to super-stimulation are shaded in dark grey in the sequence alignments. Mutants with highly variable performance in independent experiments are indicated with an asterisk.
Some TFIIB-linker mutants disrupt both transcriptional stimulation- and recruitment-function of TFIIB
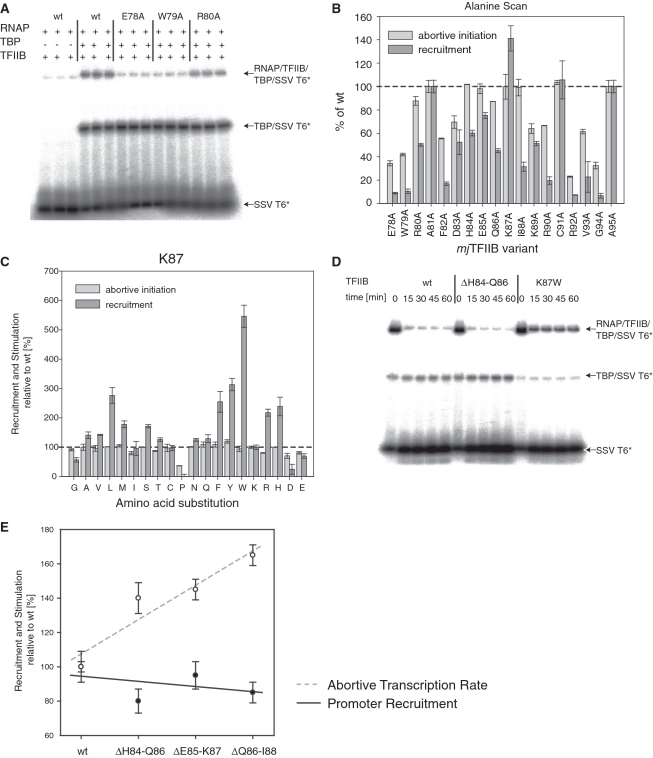
The TFIIB-linker region is not only involved in transcriptional stimulation, but also plays a key role in establishing specific protein–protein contacts with RNA polymerase (9, 10). Although the abortive transcription stimulation assays used for assessing the effect of the mutants in the linker region are promoter-independent, we were interested in establishing whether some of the loss- and gain-of-function phenotypes observed in our mutagenesis screen were due to changes in the affinity between the TFIIB-linker domain and RNAP. We therefore tested alanine substitution mutants of residues mjTFIIB E78 to A95 in promoter-directed recruitment assays [electrophoretic mobility shift assays (EMSAs); Figure 5A].
Figure 5.
Recruitment properties of TFIIB linker mutants. (A) Typical EMSA experiment to quantitate the RNAP recruitment activity of mjTFIIB mutants on radiolabelled SSV T6 core promoter (‘SSV T6*’). The arrows identify the various types of complexes formed. (B) Recruitment and stimulatory activities of the mjTFIIB alanine substitution series relative to wild-type mjTFIIB (dotted line). Mutants were tested in triplicate and error bars represent standard deviations. (C) Recruitment and stimulatory activities of the mjTFIIB K87 mutants relative to wild-type mjTFIIB (dotted line). Mutants were tested in triplicate and error bars represent standard deviations. (D) Wild-type TFIIB and TFIIB variants ΔH84-Q86 and K87W were incubated with RNAP, TBP and radiolabelled probe under the usual binding conditions as detailed in ‘Materials and Methods’ section. After 20 min, unlabelled SSV T6 core promoter was added at 400-fold excess as a competitor and samples taken after 0, 15, 30, 45 and 60 min. The addition of competitor DNA reduced the binding of RNAP/TFIIB/TBP to the radiolabelled promoter 5-fold less for mjTFIIB-K87W than for wild-type TFIIB or mjTFIIB ΔH84-Q86. (E) Recruitment (circles) and stimulatory activities (solid dots) of mjTFIIB deletion mutants were calculated relative to wild-type TFIIB (=100%). Mutants were tested in triplicate and error bars represent standard deviations. Multiple regression trendlines for both recruitment activity (black line) and stimulatory activity (dotted grey line) show that increased stimulatory functions of the deletion mutants are distinct from their recruitment activities that remain at or slightly below wild-type levels.
In a manner similar to that observed in the abortive initiation assays, the degree of residue conservation correlated roughly with their sensitivity to mutations. Mutations at most positions lead to more than 50% reduction in their recruitment activities with residues E78, W79, F82, R92, V93 and G94 being most affected (Figure 5B). Results from the abortive initiation and recruitment assays correlate approximately (Figure 5B), suggesting that in at least some instances the reduction of the capacity for stimulating abortive transcription is a consequence of structural alterations in the B-linker that also reduce its affinity for RNAP.
Enhanced recruitment phenotypes of K87
One of the alanine substitutions tested, K87-A, displayed a clearly enhanced recruitment activity (∼140% wild-type; Figure 5B). This observation was surprising, considering that substitutions of other amino acid residues tested resulted in defective or wild-type-like recruitment, and suggests a key role for K87 in the formation and/or stability of the TBP/TFIIB/RNAP promoter complex.
In order to investigate the role of this residue in the interaction with RNAP further, the full range of K87 substitutions was tested. The results show that the majority of substitutions displayed near-wild-type recruitment activity, although there are a few noteworthy exceptions (Figure 5B). Some radical substitutions, such as K87-P and K87-D, caused a substantial drop in RNAP recruitment, whereas substitutions of K87 with a range of large hydrophobic (L, F, Y and W) or basic residues (R and H) showed substantial increases.
The cause of this increased recruitment activity was investigated in more detail for the most remarkable mutant, K87-W. When tested in mobility shift assays, K87-W converted a proportion of TBP/promoter complexes into fully assembled pre-initiation complexes comparable to wild-type TFIIB (Figure 5D). However, once assembly was completed, the addition of excess promoter DNA showed that pre-initiation complexes containing mjTFIIB K87-W were around five times more resistant to competition than complexes containing wild-type TFIIB (Figure 5D). We therefore conclude that the K87-W substitution increases recruitment by substantially prolonging the half-life of the complex.
It is worthwhile to compare the remarkable stabilizing effects of some of the K87 mutations with their ability to stimulate abortive transcription. As can be seen in several instances in the alanine substitution series (Figure 5B), and especially with K87-P (Figure 5C), many substitutions that reduce the transcription-stimulating activity also appear to weaken the stability of pre-initiation complexes. The opposite effect (i.e. increased transcriptional stimulation coupled with increased stabilization of pre-initiation complexes) does, however, not apply; K87-W stimulates only with a rate comparable to wild-type TFIIB (Figure 3B). This suggests that even a substantial increase in pre-initiation complex formation/stability may not be sufficient to explain the superstimulation phenotypes observed with some of the linker deletion mutants described earlier. EMSAs carried out with three consistently superstimulating mutants (ΔH84-Q86, ΔE85-K87 and ΔQ86-I88) displayed indeed only wild-type-like, or even slightly reduced, rates of formation of pre-initiation complexes (ranging from ∼80% to 100%; Figure 5E).
In summary, the data presented above show that some types of mutants (e.g. K87-P) cause disturbances of the linker region that affect RNAP recruitment and/or post-initiation transcriptional stimulation. This class of mutants is relatively uninformative because they merely interfere with some fundamental processes in a rather non-specific manner. On the other hand, we observe distinct groups of mutants that either substantially increase the affinity of TFIIB to RNAP (e.g. K87-W), or increase the ability of the TFIIB linker region to stimulate the rate of abortive transcription (e.g. ΔQ86-I88). The functionally distinct nature of these two gain-of-function mutant classes allows these different TFIIB functionalities to be separated from each other with a degree of clarity that is unprecedented in any other transcriptional model system.
Superstimulating TFIIB mutants further enhance the transcription rate of superactive RNAPs
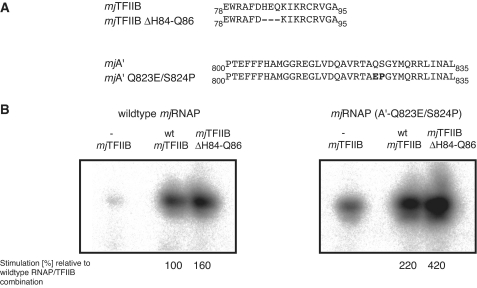
We have previously identified mutations in the Bridge Helix and Trigger Loop domains that cause a noticeable increase in the specific activity of RNAP (25). A subset of these ‘superactive’ mutants enhances the transcription rate by favouring particular conformational changes, such as kinking the C-terminal portion of the Bridge Helix (e.g. mjA′ Q823-E and S824-P), or by increasing the mobility of the base of the Trigger Loop (substitutions in mjA′′ G72 and I98). Combinations of superactive Bridge Helix and Trigger Loop mutations within the same enzyme do not increase the transcription rate beyond that achievable by either of these mutant classes on their own, suggesting that they affect the same mechanism and therefore do not display any additive or synergistic phenotypes (25).
We were curious to see to what extent such superactive RNAP mutants would react to stimulation by both wild-type TFIIB and superstimulating TFIIB variants. Wild-type and mutant RNAPs and TFIIBs (Figure 6A) were assayed in various combinations. Wild-type TFIIB, as well as the superstimulating mjTFIIB ΔH84-Q86 variant, enhance the activity of the superactive RNAP (mjA′Q823-E/S824-P) mutant in a clearly detectable manner. The combination of superactive RNAP—displaying an intrinsically ∼1.8-fold higher activity than wild-type RNAP—with superstimulating TFIIB resulted in a rate of abortive transcription that was around 4-fold higher than the rate achievable with wild-type-only proteins (Figure 6B). Such a remarkable increase in activity demonstrates that the superactive RNAP mutations and superstimulatory TFIIB variants target two distinct rate-limiting mechanisms. The simultaneous release of two independent catalytic constraints on the nucleotide addition cycle thus causes an additive combination of the functionally distinct superstimulation and superactivation activities.
Figure 6.
Additive effects of superactive RNAP and superstimulating TFIIB phenotypes. (A) Sequences of wild-type and mutant phenotypes used in the combinatorial experiments. The two substitutions in mjA′, Q823-E and S824-P are located in the C-terminal portion of the Bridge Helix domain. The mutated residues are highlighted in bold. (B) Stimulation of wild-type RNAP with superstimulating TFIIB ΔH84-Q86 results in an increased abortive transcription rate of ∼160% as compared to the stimulation with wild-type TFIIB (left panel). In contrast, stimulation with wild-type TFIIB increases the activity of superactive RNAP to ∼220% and the combination of superactive TFIIB with TFIIB ΔH84-Q86 increases this rate further to ∼420%.
DISCUSSION
The roles of TFIIB during transcription initiation in the RNAPII basal transcriptional machinery are exemplified by several distinct activities: sequence-specific recognition of core promoter elements (BRE) (2), stabilization of TBP binding through protein–protein contacts (26), specific recruitment of RNAPII (9–12,27) and start site selection (13,15,28–30). The archaeal orthologue of TFIIB carries out the same spectrum of functions, along with the additional function of strong stimulation of abortive transcription (10), which has thus far not been demonstrated directly in an eukaryotic system. It is nevertheless very likely that TFIIB also has a stimulatory role in controlling the initiation rate in eukaryotes, but additional basal factors (e.g. TFIIH) that carry out activities that are potentially rate limiting (e.g. promoter melting) may obscure this important contribution of eukaryotic TFIIBs. The archaeal system therefore offers a more direct route towards studying the functional interplay between structural elements of TFIIB with the catalytic site of RNAP.
We have previously identified the linker region of archaeal TFIIB as a key domain involved in the stimulation of abortive transcription (10). The extraordinarily high degree of structural identity between archaeal and eukaryotic TFIIBs allows an essentially one-to-one correlation of these proteins on the primary amino acid sequence level (Figure 1A). Although there is some controversy concerning the detailed path of the TFIIB linker domain through the active site (Figure 1B), it is clear that the residues mutagenized in our study are close to the catalytic site and have the potential to interact directly with the DNA–RNA hybrid, functional domains of the catalytic site (such as the Bridge Helix and Trigger Loop) and/or with other domains regulating promoter melting and clamp position (13–15).
Two clusters of residues are important for the TFIIB stimulatory function
Upon mutation, two clusters of residues (E78-W79-R80 and R90-C91-R92-V93-G94) were most significantly impaired in stimulatory functions and in several cases (E78, W79, R92 and G94) only the wild-type residue supported the highest level of activity (Figure 3A). These results match data obtained in the yeast or human systems: in both of these systems, orthologous residues have been identified as crucial for transcription initiation (16) or transcription start-site selection (28,30). Direct interactions between residues of the linker domain and template DNA at and near the transcription start site have been suggested (13) and are consistent with data obtained here. In sharp contrast, the region (A81–K89) between these two clusters is mostly highly resistant to substitutions, often of the most radical nature. The ability to stimulate abortive transcription is typically not affected by mutations that dramatically change the size, hydrophobicity or charge of the amino acid side chain present in these residues. This region plays nevertheless an important alternative role because some substitutions of K87 reveal astonishingly high increases in RNAP recruitment levels (exceeding 500% of wild-type activity for K87-W; Figure 5C). We therefore conclude that two major TFIIB functions, RNAP recruitment and abortive transcription stimulation, are controlled in a functionally distinct manner by the B-linker domain (31,32).
A new class of TFIIB superstimulators
Some of the most unexpected observations emerged from the deletion studies within the TFIIB linker region. A systematic deletion scan of three adjacent amino acid residues across the E78 to R82 sequence reflects in part the results obtained in the high-throughput point mutant screen, again identifying two clusters (E78-W79-R80 and R90-C91-R92-V93-G94) as essential. Intriguingly, deletions within the region separating these two clusters cause a marked increase in the ability of these mutants to stimulate abortive transcription that increases the levels to ∼170% relative to the wild-type level. These ‘superstimulatory’ phenotypes appear at first glance to be structurally relatively non-specific because the corresponding deletions are distributed over a relatively large target area (D83 to I88; Figure 4), suggesting that removal of any amino acids in that particular region will cause a similar effect. We demonstrated, however, that superstimulation is restricted to mutants lacking precisely three adjacent amino acids. Such a tightly defined size-constraint, combined with the spread of a shared phenotype over a six amino acid target area, strongly suggests an underlying structural impact on RNAP.
The currently most up-to-date structures of TFIIB provide two alternative models for the interpretation of our data. If the TFIIB linker takes up the B-finger conformation (13), the deletions cause a shortening of the loop protruding towards the catalytic centre. Although such an event would be expected to diminish potentially stabilizing contacts between the B-finger tip and the incoming rNTP, an overall stimulatory effect could be due to other domain movements becoming less restrained. Point mutants in the Bridge Helix and Trigger Loop, which are predicted to increase the flexibility of certain molecular hinge regions, result in a corresponding increase in the rate of the nucleotide addition cycle [‘superactivity’, (25)]. It is therefore conceivable that the additional space resulting from a shortening of the B-finger projecting towards the Bridge Helix (Figure 1C) increases the abortive transcription rate by allowing, for example, a higher rate of Bridge Helix kinking. Results obtained from combining superactive RNAP with the superstimulatory TFIIB mutants are, in principle, compatible by showing the additive effect predicted by such a model (Figure 6B).
An alternative structural model suggests that the D83 to I88 region overlaps the junction between the B-Reader Helix and the B-Reader Loop (15). Superficially, the location of the identified region at the junction between two different motifs appears to be difficult to reconcile with the structural data, but the observed spatial restrictions of the deletion window to three (ΔD83-E85, ΔH84-Q86, ΔE85-K87 and ΔQ86-I88) or four (ΔH84-K87) amino acids suggest a possible explanation. The region immediately C-terminal of the B-Reader Helix appears either unstructured or lacking in a distinct secondary structure (13–15). It is therefore plausible that the strict spatial restrictions identified in our deletion screen are indicative of a metastable extension of the B-Reader Helix. Deletion of three or four residues, corresponding approximately to one complete α-(or 3–10) helical turn, would result in a shortening of such an extended B-Reader Helix, but minimize any variation in the overall topology of its C-terminal connection to the B-Reader Loop. The corollary of such an interpretation is that transitions between unstructured and α-helical conformations of the C-terminal extension of the B-Reader Helix play an important role in determining the rate of abortive transcription.
CONCLUDING REMARKS
The intimate structural association between the TFIIB linker region with domains surrounding the catalytic site of RNAP results in a fascinating functional interplay between these proteins during transcriptional initiation. The extensive collection of mutant phenotypes described here proves that key parameters controlling recruitment (or half-life) of the TFIIB–RNAP complex are strongly influenced by a single amino acid residue. Similarly, the abortive transcription rate can be regulated by the length of a polypeptide stretch within the B-linker region. Up to now it has always been puzzling why the TFIIB linker region displays a higher rate of variation in length and primary amino acid sequence than some of the flanking TFIIB domains. Our data suggest that many of the natural variations observed reflect functional differences that alter the kinetic parameters of transcription initiation machinery and are therefore subject to natural selection affecting the function of the basal transcriptional machinery on promoters.
FUNDING
A Wellcome project grant (078043/Z/05/Z to R.O.J.W.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Peter Blattmann and Nicole Bidmon for preparing some of the mutants and Martin Buck, Finn Werner and Patricia Burrows for their helpful comments on the manuscript.
REFERENCES
- 1.Deng W, Roberts SG. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawadogo M, Roeder RG. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 4.Reinberg D, Roeder RG. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of initiation factors IIB and IIE. J. Biol. Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 5.Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 6.Barberis A, Muller CW, Harrison SC, Ptashne M. Delineation of two functional regions of transcription factor TFIIB. Proc. Natl Acad. Sci. USA. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malik S, Lee DK, Roeder RG. Potential RNA polymerase II-induced interactions of transcription factor TFIIB. Mol. Cell. Biol. 1993;13:6253–6259. doi: 10.1128/mcb.13.10.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratowski S, Zhou H. Functional domains of transcription factor TFIIB. Proc. Natl Acad. Sci. USA. 1993;90:5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 10.Werner F, Weinzierl RO. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol. Cell. Biol. 2005;25:8344–8355. doi: 10.1128/MCB.25.18.8344-8355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HT, Hahn S. Binding of TFIIB to RNA polymerase II: mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell. 2003;12:437–447. doi: 10.1016/s1097-2765(03)00306-x. [DOI] [PubMed] [Google Scholar]
- 12.Elsby LM, O'Donnell AJ, Green LM, Sharrocks AD, Roberts SG. Assembly of transcription factor IIB at a promoter in vivo requires contact with RNA polymerase II. EMBO Rep. 2006;7:898–903. doi: 10.1038/sj.embor.7400767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2009;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 16.Thompson NE, Glaser BT, Foley KM, Burton ZF, Burgess RR. Minimal promoter systems reveal the importance of conserved residues in the B-finger of human transcription factor IIB (TFIIB) J. Biol. Chem. 2009;284:24754–24766. doi: 10.1074/jbc.M109.030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werner F, Weinzierl RO. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell. 2002;10:635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 18.Bell SD, Jackson SP. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 1998;6:222–228. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 19.Zillig W, Stetter KO, Janekovic D. DNA-dependent RNA polymerase from the archaebacterium Sulfolobus acidocaldarius. Eur. J. Biochem. 1979;96:597–604. doi: 10.1111/j.1432-1033.1979.tb13074.x. [DOI] [PubMed] [Google Scholar]
- 20.Werner F. Structure and function of archaeal RNA polymerases. Mol. Microbiol. 2007;65:1395–1404. doi: 10.1111/j.1365-2958.2007.05876.x. [DOI] [PubMed] [Google Scholar]
- 21.Qureshi SA, Bell SD, Jackson SP. Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell SD, Kosa PL, Sigler PB, Jackson SP. Orientation of the transcription preinitiation complex in archaea. Proc. Natl Acad. Sci. USA. 1999;96:13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gohl HP, Grondahl B, Thomm M. Promoter recognition in archaea is mediated by transcription factors: identification of transcription factor aTFB from Methanococcus thermolithotrophicus as archaeal TATA-binding protein. Nucleic Acids Res. 1995;23:3837–3841. doi: 10.1093/nar/23.19.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werner F, Wiesler S, Nottebaum S, Weinzierl RO. Modulation of RNA polymerase core functions by basal transcription factor TFB/TFIIB. Biochem. Soc. Symp. 2006:49–58. doi: 10.1042/bss0730049. [DOI] [PubMed] [Google Scholar]
- 25.Tan L, Wiesler S, Trzaska D, Carney HC, Weinzierl RO. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J. Biol. 2008;7:40. doi: 10.1186/jbiol98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai FT, Sigler PB. Structural basis of preinitiation complex assembly on human pol II promoters. EMBO J. 2000;19:25–36. doi: 10.1093/emboj/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magill CP, Jackson SP, Bell SD. Identification of a conserved archaeal RNA polymerase subunit contacted by the basal transcription factor TFB. J. Biol. Chem. 2001;276:46693–46696. doi: 10.1074/jbc.C100567200. [DOI] [PubMed] [Google Scholar]
- 28.Pinto I, Wu WH, Na JG, Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- 29.Bangur CS, Pardee TS, Ponticelli AS. Mutational analysis of the D1/E1 core helices and the conserved N-terminal region of yeast transcription factor IIB (TFIIB): identification of an N-terminal mutant that stabilizes TATA-binding protein-TFIIB-DNA complexes. Mol. Cell. Biol. 1997;17:6784–6793. doi: 10.1128/mcb.17.12.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faitar SL, Brodie SA, Ponticelli AS. Promoter-specific shifts in transcription initiation conferred by yeast TFIIB mutations are determined by the sequence in the immediate vicinity of the start sites. Mol. Cell. Biol. 2001;21:4427–4440. doi: 10.1128/MCB.21.14.4427-4440.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho EJ, Buratowski S. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J. Biol. Chem. 1999;274:25807–25813. doi: 10.1074/jbc.274.36.25807. [DOI] [PubMed] [Google Scholar]
- 32.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]